Holistic View and Novel Perspective on Ruminal and Extra-Gastrointestinal Methanogens in Cattle
Abstract
:1. Introduction
2. Brief Overview of Methanogens
3. Main Methanogenic Species Present in the Rumen
4. Pro-and Anti-Methanogenic Ruminal Microorganisms
5. Methanogens in the Reproductive Tract: Vagina, Uterus, and Semen
6. Methanogens in the Respiratory Tract
7. Methanogens in the Udder
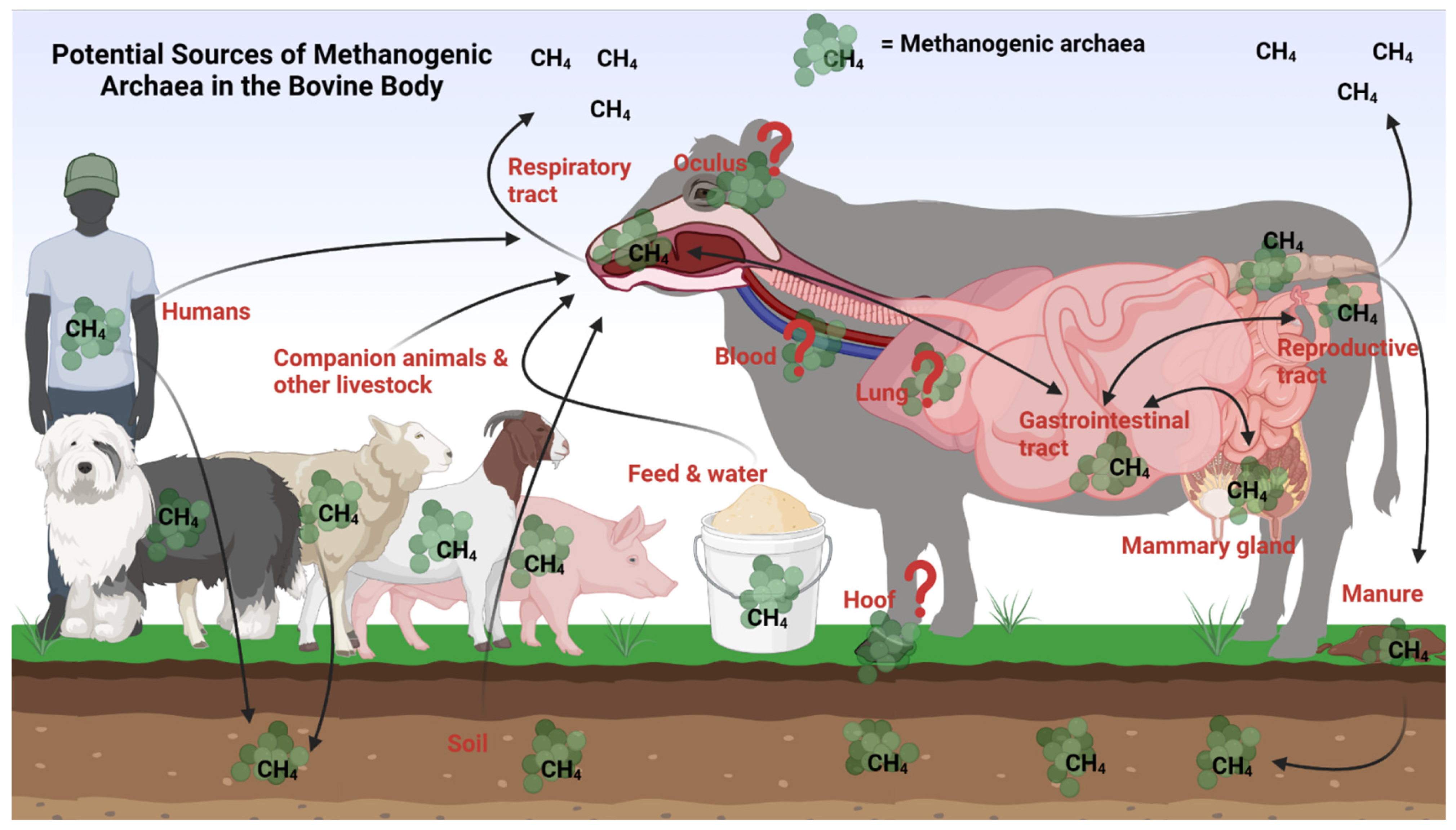
8. Potential Seeding Sources of the Ruminal Methanogens
8.1. Within the Bovine Body
8.2. Other Sources
9. Challenges Associated with Studying the Ruminal Methanogens and Future Directions
10. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- World Bank Climate Explainer: Food Security and Climate Change. Available online: https://www.worldbank.org/en/news/feature/2022/10/17/what-you-need-to-know-about-food-security-and-climate-change (accessed on 17 May 2023).
- IPCC. IPCC—Intergovernmental Panel on Climate Change 2022. Available online: https://www.clientearth.org/latest/latest-updates/news/the-latest-ipcc-report-a-wake-up-call-for-climate-action/?gclid=Cj0KCQiAo7KqBhDhARIsAKhZ4ugobwC7Py9J-ef1tTHY_MN1TaHp3-ctlWLgrOuIX-eQI15bu9U8vxQaAvR2EALw_wcB (accessed on 2 July 2023).
- Ripple, W.J.; Smith, P.; Haberl, H.; Montzka, S.A.; McAlpine, C.; Boucher, D.H. Ruminants, Climate Change and Climate Policy. Nat. Clim. Chang. 2014, 4, 2–5. [Google Scholar] [CrossRef]
- FAO Sustainable Agriculture|Sustainable Development Goals|Food and Agriculture Organization of the United Nations. Available online: https://www.fao.org/sustainable-development-goals/overview/fao-and-the-2030-agenda-for-sustainable-development/sustainable-agriculture/en/ (accessed on 21 February 2023).
- Clark, M.; Tilman, D. Comparative Analysis of Environmental Impacts of Agricultural Production Systems, Agricultural Input Efficiency, and Food Choice. Environ. Res. Lett. 2017, 12, 064016. [Google Scholar] [CrossRef]
- US EPA. Overview of Greenhouse Gases. Available online: https://www.epa.gov/ghgemissions/overview-greenhouse-gases (accessed on 20 June 2023).
- Buddle, B.M.; Denis, M.; Attwood, G.T.; Altermann, E.; Janssen, P.H.; Ronimus, R.S.; Pinares-Patiño, C.S.; Muetzel, S.; Neil Wedlock, D. Strategies to Reduce Methane Emissions from Farmed Ruminants Grazing on Pasture. Vet. J. 2011, 188, 11–17. [Google Scholar] [CrossRef]
- Kumar, S.; Choudhury, P.K.; Carro, M.D.; Griffith, G.W.; Dagar, S.S.; Puniya, M.; Calabro, S.; Ravella, S.R.; Dhewa, T.; Upadhyay, R.C.; et al. New Aspects and Strategies for Methane Mitigation from Ruminants. Appl. Microbiol. Biotechnol. 2014, 98, 31–44. [Google Scholar] [CrossRef]
- Lan, W.; Yang, C. Ruminal Methane Production: Associated Microorganisms and the Potential of Applying Hydrogen-Utilizing Bacteria for Mitigation. Sci. Total Environ. 2019, 654, 1270–1283. [Google Scholar] [CrossRef]
- Johnson, K.A.; Johnson, D.E. Methane Emissions from Cattle. J. Anim. Sci. 1995, 73, 2483–2492. [Google Scholar] [CrossRef]
- Beauchemin, K.A.; Ungerfeld, E.M.; Eckard, R.J.; Wang, M. Review: Fifty Years of Research on Rumen Methanogenesis: Lessons Learned and Future Challenges for Mitigation. Animal 2020, 14, s2–s16. [Google Scholar] [CrossRef]
- Jeyanathan, J.; Martin, C.; Morgavi, D.P. Screening of Bacterial Direct-Fed Microbials for Their Antimethanogenic Potential in Vitro and Assessment of Their Effect on Ruminal Fermentation and Microbial Profiles in Sheep. J. Anim. Sci. 2016, 94, 739–750. [Google Scholar] [CrossRef]
- Tedeschi, L.O.; Fox, D.G.; Tylutki, T.P. Potential Environmental Benefits of Ionophores in Ruminant Diets. J. Environ. Qual. 2003, 32, 1591–1602. [Google Scholar] [CrossRef]
- Huws, S.A.; Creevey, C.J.; Oyama, L.B.; Mizrahi, I.; Denman, S.E.; Popova, M.; Muñoz-Tamayo, R.; Forano, E.; Waters, S.M.; Hess, M.; et al. Addressing Global Ruminant Agricultural Challenges Through Understanding the Rumen Microbiome: Past, Present, and Future. Front. Microbiol. 2018, 9, 2161. [Google Scholar] [CrossRef]
- Króliczewska, B.; Pecka-Kiełb, E.; Bujok, J. Strategies Used to Reduce Methane Emissions from Ruminants: Controversies and Issues. Agriculture 2023, 13, 602. [Google Scholar] [CrossRef]
- Malik, P.K.; Bhatta, R.; Gagen, E.J.; Sejian, V.; Soren, N.M.; Prasad, C.S. Alternate H2 Sinks for Reducing Rumen Methanogenesis. In Climate Change Impact on Livestock: Adaptation and Mitigation; Sejian, V., Gaughan, J., Baumgard, L., Prasad, C., Eds.; Springer: New Delhi, India, 2015; pp. 303–320. ISBN 978-81-322-2265-1. [Google Scholar]
- de Haas, Y.; Veerkamp, R.F.; de Jong, G.; Aldridge, M.N. Selective Breeding as a Mitigation Tool for Methane Emissions from Dairy Cattle. Animal 2021, 15, 100294. [Google Scholar] [CrossRef] [PubMed]
- Jalil Sarghale, A.; Moradi Shahrebabak, M.; Moradi Shahrebabak, H.; Nejati Javaremi, A.; Saatchi, M.; Khansefid, M.; Miar, Y. Genome-Wide Association Studies for Methane Emission and Ruminal Volatile Fatty Acids Using Holstein Cattle Sequence Data. BMC Genet. 2020, 21, 129. [Google Scholar] [CrossRef]
- Zetouni, L.; Kargo, M.; Norberg, E.; Lassen, J. Genetic Correlations between Methane Production and Fertility, Health, and Body Type Traits in Danish Holstein Cows. J. Dairy Sci. 2018, 101, 2273–2280. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Zhao, G. Decreasing Ruminal Methane Production through Enhancing the Sulfate Reduction Pathway. Anim. Nutr. 2022, 9, 320–326. [Google Scholar] [CrossRef]
- Amat, S.; Holman, D.B.; Schmidt, K.; Menezes, A.C.B.; Baumgaertner, F.; Winders, T.; Kirsch, J.D.; Liu, T.; Schwinghamer, T.D.; Sedivec, K.K.; et al. The Nasopharyngeal, Ruminal, and Vaginal Microbiota and the Core Taxa Shared across These Microbiomes in Virgin Yearling Heifers Exposed to Divergent In Utero Nutrition during Their First Trimester of Gestation and in Pregnant Beef Heifers in Response to Mineral Supplementation. Microorganisms 2021, 9, 2011. [Google Scholar] [CrossRef]
- Kroeger, M.E.; Meredith, L.K.; Meyer, K.M.; Webster, K.D.; de Camargo, P.B.; de Souza, L.F.; Tsai, S.M.; van Haren, J.; Saleska, S.; Bohannan, B.J.M.; et al. Rainforest-to-Pasture Conversion Stimulates Soil Methanogenesis across the Brazilian Amazon. ISME J. 2021, 15, 658–672. [Google Scholar] [CrossRef] [PubMed]
- Guindo, C.O.; Drancourt, M.; Grine, G. Dairy Products as Sources of Methanogens for Humans 2021. bioRxiv 2021. [Google Scholar] [CrossRef]
- de Souza, J.; Leskinen, H.; Lock, A.L.; Shingfield, K.J.; Huhtanen, P. Between-Cow Variation in Milk Fatty Acids Associated with Methane Production. PLoS ONE 2020, 15, e0235357. [Google Scholar] [CrossRef] [PubMed]
- Winders, T.M.; Holman, D.B.; Schmidt, K.N.; Luecke, S.M.; Smith, D.J.; Neville, B.W.; Dahlen, C.R.; Swanson, K.C.; Amat, S. Feeding Hempseed Cake Alters the Bovine Gut, Respiratory and Reproductive Microbiota. Sci. Rep. 2023, 13, 8121. [Google Scholar] [CrossRef]
- Welch, C.B.; Ryman, V.E.; Pringle, T.D.; Lourenco, J.M. Utilizing the Gastrointestinal Microbiota to Modulate Cattle Health through the Microbiome-Gut-Organ Axes. Microorganisms 2022, 10, 1391. [Google Scholar] [CrossRef] [PubMed]
- Guzman, C.E.; Wood, J.L.; Egidi, E.; White-Monsant, A.C.; Semenec, L.; Grommen, S.V.H.; Hill-Yardin, E.L.; De Groef, B.; Franks, A.E. A Pioneer Calf Foetus Microbiome. Sci. Rep. 2020, 10, 17712. [Google Scholar] [CrossRef] [PubMed]
- Buan, N.R. Methanogens: Pushing the Boundaries of Biology. Emerg. Top. Life Sci. 2018, 2, 629–646. [Google Scholar] [CrossRef] [PubMed]
- Lyu, Z.; Rotaru, A.-E.; Pimentel, M.; Zhang, C.-J.; Rittmann, S.K.-M.R. Editorial: The Methane Moment-Cross-Boundary Significance of Methanogens: Preface. Front. Microbiol. 2022, 13, 1055494. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.M.; de Lurdes Nunes Enes Dapkevicius, M.; Borba, A.E.S. Alternative Pathways for Hydrogen Sink Originated from the Ruminal Fermentation of Carbohydrates: Which Microorganisms Are Involved in Lowering Methane Emission? Anim. Microbiome 2022, 4, 5. [Google Scholar] [CrossRef]
- Liu, Y. Taxonomy of Methanogens. In Handbook of Hydrocarbon and Lipid Microbiology; Timmis, K.N., Ed.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 547–558. ISBN 978-3-540-77587-4. [Google Scholar]
- Steinfeld, H.; Gerber, P.; Wassenaar, T.; Castel, V.; Rosales, M.; de Haan, C. Livestock’s Long Shadow. 2006. Available online: https://www.fao.org/3/a0701e/a0701e.pdf (accessed on 23 June 2023).
- Gribaldo, S.; Brochier-Armanet, C. The Origin and Evolution of Archaea: A State of the Art. Philos. Trans. R. Soc. B Biol. Sci. 2006, 361, 1007–1022. [Google Scholar] [CrossRef]
- Lyu, Z.; Liu, Y. Diversity and Taxonomy of Methanogens. In Biogenesis of Hydrocarbons; Stams, A.J.M., Sousa, D.Z., Eds.; Handbook of Hydrocarbon and Lipid Microbiology; Springer International Publishing: Cham, Switzerland, 2019; pp. 19–77. ISBN 978-3-319-78108-2. [Google Scholar]
- Berghuis, B.A.; Yu, F.B.; Schulz, F.; Blainey, P.C.; Woyke, T.; Quake, S.R. Hydrogenotrophic Methanogenesis in Archaeal Phylum Verstraetearchaeota Reveals the Shared Ancestry of All Methanogens. Proc. Natl. Acad. Sci. USA 2019, 116, 5037–5044. [Google Scholar] [CrossRef]
- Vanwonterghem, I.; Evans, P.N.; Parks, D.H.; Jensen, P.D.; Woodcroft, B.J.; Hugenholtz, P.; Tyson, G.W. Methylotrophic Methanogenesis Discovered in the Archaeal Phylum Verstraetearchaeota. Nat. Microbiol. 2016, 1, 16170. [Google Scholar] [CrossRef] [PubMed]
- Gaci, N.; Borrel, G.; Tottey, W.; O’Toole, P.W.; Brugère, J.-F. Archaea and the Human Gut: New Beginning of an Old Story. World J. Gastroenterol. WJG 2014, 20, 16062–16078. [Google Scholar] [CrossRef]
- Cheng, L.; Wu, K.; Zhou, L.; Tahon, G.; Liu, L.; Li, J.; Zhang, J.; Zheng, F.; Deng, C.; Han, W.; et al. Isolation of a Methyl-Reducing Methanogen Outside the Euryarchaeota. Res. Sequare 2023. [Google Scholar] [CrossRef]
- Ferry, J.G.; Kastead, K.A. Methanogenesis. In Archaea; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2007; pp. 288–314. ISBN 978-1-68367-168-8. [Google Scholar]
- Kurade, M.B.; Saha, S.; Salama, E.-S.; Patil, S.M.; Govindwar, S.P.; Jeon, B.-H. Acetoclastic Methanogenesis Led by Methanosarcina in Anaerobic Co-Digestion of Fats, Oil and Grease for Enhanced Production of Methane. Bioresour. Technol. 2019, 272, 351–359. [Google Scholar] [CrossRef]
- Mei, R.; Kaneko, M.; Imachi, H.; Nobu, M.K. The Origin and Evolution of Methanogenesis and Archaea Are Intertwined. PNAS Nexus 2023, 2, pgad023. [Google Scholar] [CrossRef]
- Noel, S.J.; Højberg, O.; Urich, T.; Poulsen, M. Draft Genome Sequence of “Candidatus Methanomethylophilus” Sp. 1R26, Enriched from Bovine Rumen, a Methanogenic Archaeon Belonging to the Methanomassiliicoccales Order. Genome Announc. 2016, 4, e01734-15. [Google Scholar] [CrossRef]
- Knief, C. Diversity of Methane Cycling Microorganisms in Soils and Their Relation to Oxygen. Curr. Issues Mol. Biol. 2019, 33, 23–56. [Google Scholar] [CrossRef]
- Narrowe, A.B.; Borton, M.A.; Hoyt, D.W.; Smith, G.J.; Daly, R.A.; Angle, J.C.; Eder, E.K.; Wong, A.R.; Wolfe, R.A.; Pappas, A.; et al. Uncovering the Diversity and Activity of Methylotrophic Methanogens in Freshwater Wetland Soils. mSystems 2019, 4, e00320-19. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.J.; Angle, J.C.; Solden, L.M.; Borton, M.A.; Morin, T.H.; Daly, R.A.; Johnston, M.D.; Stefanik, K.C.; Wolfe, R.; Gil, B.; et al. Members of the Genus Methylobacter Are Inferred To Account for the Majority of Aerobic Methane Oxidation in Oxic Soils from a Freshwater Wetland. mBio 2018, 9, e00815-18. [Google Scholar] [CrossRef]
- Greening, C.; Geier, R.; Wang, C.; Woods, L.C.; Morales, S.E.; McDonald, M.J.; Rushton-Green, R.; Morgan, X.C.; Koike, S.; Leahy, S.C.; et al. Diverse Hydrogen Production and Consumption Pathways Influence Methane Production in Ruminants. ISME J. 2019, 13, 2617–2632. [Google Scholar] [CrossRef]
- Evans, P.N.; Boyd, J.A.; Leu, A.O.; Woodcroft, B.J.; Parks, D.H.; Hugenholtz, P.; Tyson, G.W. An Evolving View of Methane Metabolism in the Archaea. Nat. Rev. Microbiol. 2019, 17, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Enzmann, F.; Mayer, F.; Rother, M.; Holtmann, D. Methanogens: Biochemical Background and Biotechnological Applications. AMB Express 2018, 8, 1. [Google Scholar] [CrossRef] [PubMed]
- Tapio, I.; Snelling, T.J.; Strozzi, F.; Wallace, R.J. The Ruminal Microbiome Associated with Methane Emissions from Ruminant Livestock. J. Anim. Sci. Biotechnol. 2017, 8, 7. [Google Scholar] [CrossRef]
- Hoedt, E.C.; Parks, D.H.; Volmer, J.G.; Rosewarne, C.P.; Denman, S.E.; McSweeney, C.S.; Muir, J.G.; Gibson, P.R.; Cuív, P.Ó.; Hugenholtz, P.; et al. Culture- and Metagenomics-Enabled Analyses of the Methanosphaera Genus Reveals Their Monophyletic Origin and Differentiation According to Genome Size. ISME J. 2018, 12, 2942–2953. [Google Scholar] [CrossRef]
- Poehlein, A.; Schneider, D.; Soh, M.; Daniel, R.; Seedorf, H. Comparative Genomic Analysis of Members of the Genera Methanosphaera and Methanobrevibacter Reveals Distinct Clades with Specific Potential Metabolic Functions. Archaea 2018, 2018, 7609847. [Google Scholar] [CrossRef] [PubMed]
- Guzman, C.E.; Bereza-Malcolm, L.T.; Groef, B.D.; Franks, A.E. Presence of Selected Methanogens, Fibrolytic Bacteria, and Proteobacteria in the Gastrointestinal Tract of Neonatal Dairy Calves from Birth to 72 Hours. PLoS ONE 2015, 10, e0133048. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Bi, S.S.; Wang, W.W.; Zhou, M.; Neves, A.L.A.; Degen, A.A.; Guan, L.L.; Long, R.J. Maternal Rumen and Milk Microbiota Shape the Establishment of Early-Life Rumen Microbiota in Grazing Yak Calves. J. Dairy Sci. 2023, 106, 2054–2070. [Google Scholar] [CrossRef]
- Janssen, P.H. Influence of Hydrogen on Rumen Methane Formation and Fermentation Balances through Microbial Growth Kinetics and Fermentation Thermodynamics. Anim. Feed Sci. Technol. 2010, 160, 1–22. [Google Scholar] [CrossRef]
- Gunsalus, R.P.; Cook, L.E.; Crable, B.; Rohlin, L.; McDonald, E.; Mouttaki, H.; Sieber, J.R.; Poweleit, N.; Zhou, H.; Lapidus, A.L.; et al. Complete Genome Sequence of Methanospirillum Hungatei Type Strain JF1. Stand. Genom. Sci. 2016, 11, 2. [Google Scholar] [CrossRef]
- Lee, J.-H.; Kumar, S.; Lee, G.-H.; Chang, D.-H.; Rhee, M.-S.; Yoon, M.-H.; Kim, B.-C. Methanobrevibacter boviskoreani sp. nov., Isolated from the Rumen of Korean Native Cattle. Int. J. Syst. Evol. Microbiol. 2013, 63, 4196–4201. [Google Scholar] [CrossRef]
- Snelling, T.J.; Genç, B.; McKain, N.; Watson, M.; Waters, S.M.; Creevey, C.J.; Wallace, R.J. Diversity and Community Composition of Methanogenic Archaea in the Rumen of Scottish Upland Sheep Assessed by Different Methods. PLoS ONE 2014, 9, e106491. [Google Scholar] [CrossRef]
- Danielsson, R.; Dicksved, J.; Sun, L.; Gonda, H.; Müller, B.; Schnürer, A.; Bertilsson, J. Methane Production in Dairy Cows Correlates with Rumen Methanogenic and Bacterial Community Structure. Front. Microbiol. 2017, 8, 226. [Google Scholar] [CrossRef]
- Koziol, J.H.; Sheets, T.; Wickware, C.L.; Johnson, T.A. Composition and Diversity of the Seminal Microbiota in Bulls and Its Association with Semen Parameters. Theriogenology 2022, 182, 17–25. [Google Scholar] [CrossRef]
- Guindo, C.O.; Davoust, B.; Drancourt, M.; Grine, G. Diversity of Methanogens in Animals’ Gut. Microorganisms 2020, 9, 13. [Google Scholar] [CrossRef] [PubMed]
- Lyons, T.; Bielak, A.; Doyle, E.; Kuhla, B. Variations in Methane Yield and Microbial Community Profiles in the Rumen of Dairy Cows as They Pass through Stages of First Lactation. J. Dairy Sci. 2018, 101, 5102–5114. [Google Scholar] [CrossRef]
- Jarvis, G.N.; Strömpl, C.; Burgess, D.M.; Skillman, L.C.; Moore, E.R.B.; Joblin, K.N. Isolation and Identification of Ruminal Methanogens from Grazing Cattle. Curr. Microbiol. 2000, 40, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Belanche, A.; Kingston-Smith, A.H.; Griffith, G.W.; Newbold, C.J. A Multi-Kingdom Study Reveals the Plasticity of the Rumen Microbiota in Response to a Shift From Non-Grazing to Grazing Diets in Sheep. Front. Microbiol. 2019, 10, 122. [Google Scholar] [CrossRef] [PubMed]
- Lambie, S.C.; Kelly, W.J.; Leahy, S.C.; Li, D.; Reilly, K.; McAllister, T.A.; Valle, E.R.; Attwood, G.T.; Altermann, E. The Complete Genome Sequence of the Rumen Methanogen Methanosarcina Barkeri CM1. Stand. Genom. Sci. 2015, 10, 57. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.; Kang, K.; Wang, H.; Wang, Z.; Xue, B.; Wang, L.; Xu, F.; Peng, Q. Effects of Dietary Supplementation of Active Dried Yeast on Fecal Methanogenic Archaea Diversity in Dairy Cows. Anaerobe 2017, 44, 78–86. [Google Scholar] [CrossRef]
- Ngetich, D.K.; Bett, R.C.; Gachuiri, C.K.; Kibegwa, F.M. Diversity of Gut Methanogens and Functional Enzymes Associated with Methane Metabolism in Smallholder Dairy Cattle. Arch. Microbiol. 2022, 204, 608. [Google Scholar] [CrossRef]
- Webb, E.M.; Holman, D.B.; Schmidt, K.N.; Crouse, M.S.; Dahlen, C.R.; Cushman, R.A.; Snider, A.P.; McCarthy, K.L.; Amat, S. A Longitudinal Characterization of the Seminal Microbiota and Antibiotic Resistance in Yearling Beef Bulls Subjected to Different Rates of Gain. Microbiol. Spectr. 2023, 11, e0518022. [Google Scholar] [CrossRef]
- Triantafyllou, K.; Chang, C.; Pimentel, M. Methanogens, Methane and Gastrointestinal Motility. J. Neurogastroenterol. Motil. 2014, 20, 31–40. [Google Scholar] [CrossRef]
- Schink, B. Energetics of Syntrophic Cooperation in Methanogenic Degradation. Microbiol. Mol. Biol. Rev. 1997, 61, 262–280. [Google Scholar]
- Hao, L.; Michaelsen, T.Y.; Singleton, C.M.; Dottorini, G.; Kirkegaard, R.H.; Albertsen, M.; Nielsen, P.H.; Dueholm, M.S. Novel Syntrophic Bacteria in Full-Scale Anaerobic Digesters Revealed by Genome-Centric Metatranscriptomics. ISME J. 2020, 14, 906–918. [Google Scholar] [CrossRef]
- Ruiz, B.; Chávez, A.; Forero, A.; García-Huante, Y.; Romero, A.; Sánchez, M.; Rocha, D.; Sánchez, B.; Rodríguez-Sanoja, R.; Sánchez, S.; et al. Production of Microbial Secondary Metabolites: Regulation by the Carbon Source. Crit. Rev. Microbiol. 2010, 36, 146–167. [Google Scholar] [CrossRef] [PubMed]
- Rotaru, A.-E.; Calabrese, F.; Stryhanyuk, H.; Musat, F.; Shrestha, P.M.; Weber, H.S.; Snoeyenbos-West, O.L.O.; Hall, P.O.J.; Richnow, H.H.; Musat, N.; et al. Conductive Particles Enable Syntrophic Acetate Oxidation between Geobacter and Methanosarcina from Coastal Sediments. mBio 2018, 9, e00226-18. [Google Scholar] [CrossRef] [PubMed]
- Hinsley, A.P.; Berks, B.C. Specificity of Respiratory Pathways Involved in the Reduction of Sulfur Compounds by Salmonella Enterica. Microbiol. Read. Engl. 2002, 148, 3631–3638. [Google Scholar] [CrossRef]
- van Zijderveld, S.M.; Gerrits, W.J.J.; Apajalahti, J.A.; Newbold, J.R.; Dijkstra, J.; Leng, R.A.; Perdok, H.B. Nitrate and Sulfate: Effective Alternative Hydrogen Sinks for Mitigation of Ruminal Methane Production in Sheep. J. Dairy Sci. 2010, 93, 5856–5866. [Google Scholar] [CrossRef] [PubMed]
- Granja-Salcedo, Y.T.; Fernandes, R.M.; de Araujo, R.C.; Kishi, L.T.; Berchielli, T.T.; de Resende, F.D.; Berndt, A.; Siqueira, G.R. Long-Term Encapsulated Nitrate Supplementation Modulates Rumen Microbial Diversity and Rumen Fermentation to Reduce Methane Emission in Grazing Steers. Front. Microbiol. 2019, 10, 614. [Google Scholar] [CrossRef]
- Doyle, N.; Mbandlwa, P.; Kelly, W.J.; Attwood, G.; Li, Y.; Ross, R.P.; Stanton, C.; Leahy, S. Use of Lactic Acid Bacteria to Reduce Methane Production in Ruminants, a Critical Review. Front. Microbiol. 2019, 10, 2207. [Google Scholar] [CrossRef]
- Ramayo-Caldas, Y.; Zingaretti, L.; Popova, M.; Estellé, J.; Bernard, A.; Pons, N.; Bellot, P.; Mach, N.; Rau, A.; Roume, H.; et al. Identification of Rumen Microbial Biomarkers Linked to Methane Emission in Holstein Dairy Cows. J. Anim. Breed. Genet. 2019, 137, 49–59. [Google Scholar] [CrossRef]
- Pope, P.B.; Smith, W.; Denman, S.E.; Tringe, S.G.; Barry, K.; Hugenholtz, P.; McSweeney, C.S.; McHardy, A.C.; Morrison, M. Isolation of Succinivibrionaceae Implicated in Low Methane Emissions from Tammar Wallabies. Science 2011, 333, 646–648. [Google Scholar] [CrossRef]
- Choudhury, P.K.; Jena, R.; Tomar, S.K.; Puniya, A.K. Reducing Enteric Methanogenesis through Alternate Hydrogen Sinks in the Rumen. Methane 2022, 1, 320–341. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Huang, X.; Ma, R.; Li, J.; Wang, F.; Jiao, N.; Zhang, R. Potential Metabolic and Genetic Interaction among Viruses, Methanogen and Methanotrophic Archaea, and Their Syntrophic Partners. ISME Commun. 2022, 2, 50. [Google Scholar] [CrossRef] [PubMed]
- Namonyo, S.; Wagacha, M.; Maina, S.; Wambua, L.; Agaba, M. A Metagenomic Study of the Rumen Virome in Domestic Caprids. Arch. Virol. 2018, 163, 3415–3419. [Google Scholar] [CrossRef] [PubMed]
- Pfister, P.; Wasserfallen, A.; Stettler, R.; Leisinger, T. Molecular Analysis of Methanobacterium Phage ΨM2. Mol. Microbiol. 1998, 30, 233–244. [Google Scholar] [CrossRef]
- Hanson, R.S.; Hanson, T.E. Methanotrophic Bacteria. Microbiol. Rev. 1996, 60, 439–471. [Google Scholar] [CrossRef] [PubMed]
- Auffret, M.; Stewart, R.; Dewhurst, R.; Duthie, C.-A.; Rooke, J.; Wallace, R.; Freeman, T.; Snelling, T.; Watson, M.; Roehe, R. Identification, Comparison and Validation of Robust Rumen Microbial Biomarkers for Methane Emissions Using Diverse Bos Taurus Breeds and Basal Diets. Front. Microbiol. 2018, 8, 2642. [Google Scholar] [CrossRef]
- Li, Y.; Meng, Z.; Xu, Y.; Shi, Q.; Ma, Y.; Aung, M.; Cheng, Y.; Zhu, W. Interactions between Anaerobic Fungi and Methanogens in the Rumen and Their Biotechnological Potential in Biogas Production from Lignocellulosic Materials. Microorganisms 2021, 9, 190. [Google Scholar] [CrossRef]
- Matthews, C.; Crispie, F.; Lewis, E.; Reid, M.; O’Toole, P.W.; Cotter, P.D. The Rumen Microbiome: A Crucial Consideration When Optimising Milk and Meat Production and Nitrogen Utilisation Efficiency. Gut Microbes 2019, 10, 115–132. [Google Scholar] [CrossRef]
- Luecke, S.M.; Webb, E.M.; Dahlen, C.R.; Reynolds, L.P.; Amat, S. Seminal and Vagino-Uterine Microbiome and Their Individual and Interactive Effects on Cattle Fertility. Front. Microbiol. 2022, 13, 1029128. [Google Scholar] [CrossRef]
- Adnane, M.; Chapwanya, A. A Review of the Diversity of the Genital Tract Microbiome and Implications for Fertility of Cattle. Anim. Open Access J. 2022, 12, 460. [Google Scholar] [CrossRef]
- Laguardia-Nascimento, M.; Branco, K.M.G.R.; Gasparini, M.R.; Giannattasio-Ferraz, S.; Leite, L.R.; Araujo, F.M.G.; de Matos Salim, A.C.; Nicoli, J.R.; de Oliveira, G.C.; Barbosa-Stancioli, E.F. Vaginal Microbiome Characterization of Nellore Cattle Using Metagenomic Analysis. PLoS ONE 2015, 10, e0143294. [Google Scholar] [CrossRef]
- Cojkic, A.; Niazi, A.; Guo, Y.; Hallap, T.; Padrik, P.; Morrell, J.M. Identification of Bull Semen Microbiome by 16S Sequencing and Possible Relationships with Fertility. Microorganisms 2021, 9, 2431. [Google Scholar] [CrossRef]
- Ekman, L.; Bagge, E.; Nyman, A.; Waller, K.P.; Pringle, M.; Segerman, B. A Shotgun Metagenomic Investigation of the Microbiota of Udder Cleft Dermatitis in Comparison to Healthy Skin in Dairy Cows. PLoS ONE 2020, 15, e0242880. [Google Scholar] [CrossRef] [PubMed]
- Amat, S.; Holman, D.B.; Timsit, E.; Schwinghamer, T.; Alexander, T.W. Evaluation of the Nasopharyngeal Microbiota in Beef Cattle Transported to a Feedlot, With a Focus on Lactic Acid-Producing Bacteria. Front. Microbiol. 2019, 10, 1988. [Google Scholar] [CrossRef] [PubMed]
- Belay, N.; Mukhopadhyay, B.; Conway de Macario, E.; Galask, R.; Daniels, L. Methanogenic Bacteria in Human Vaginal Samples. J. Clin. Microbiol. 1990, 28, 1666–1668. [Google Scholar] [CrossRef] [PubMed]
- Grine, G.; Drouet, H.; Fenollar, F.; Bretelle, F.; Raoult, D.; Drancourt, M. Detection of Methanobrevibacter Smithii in Vaginal Samples Collected from Women Diagnosed with Bacterial Vaginosis. Eur. J. Clin. Microbiol. Infect. Dis. 2019, 38, 1643–1649. [Google Scholar] [CrossRef]
- Khelaifia, S.; Raoult, D.; Drancourt, M. A Versatile Medium for Cultivating Methanogenic Archaea. PLoS ONE 2013, 8, e61563. [Google Scholar] [CrossRef]
- Swartz, J.D.; Lachman, M.; Westveer, K.; O’Neill, T.; Geary, T.; Kott, R.W.; Berardinelli, J.G.; Hatfield, P.G.; Thomson, J.M.; Roberts, A.; et al. Characterization of the Vaginal Microbiota of Ewes and Cows Reveals a Unique Microbiota with Low Levels of Lactobacilli and Near-Neutral pH. Front. Vet. Sci. 2014, 1, 19. [Google Scholar] [CrossRef]
- Zang, X.; Wang, W.; Gu, S.; Gu, T.; Yang, H.; Zheng, E.; Xu, Z.; Huang, S.; Li, Z.; Cai, G.; et al. Interaction between Microbes and Host in Sow Vaginas in Early Pregnancy. mSystems 2023, 8, e01192-22. [Google Scholar] [CrossRef]
- Abou Chacra, L.; Fenollar, F. Exploring the Global Vaginal Microbiome and Its Impact on Human Health. Microb. Pathog. 2021, 160, 105172. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, J.; Li, H.; Fu, K.; Pang, B.; Yang, Y.; Liu, Y.; Tian, W.; Cao, R. Characterization of the Cervical Bacterial Community in Dairy Cows with Metritis and during Different Physiological Phases. Theriogenology 2018, 108, 306–313. [Google Scholar] [CrossRef]
- Galvão, K.N.; Bicalho, R.C.; Jeon, S.J. Symposium Review: The Uterine Microbiome Associated with the Development of Uterine Disease in Dairy Cows. J. Dairy Sci. 2019, 102, 11786–11797. [Google Scholar] [CrossRef] [PubMed]
- Ault-Seay, T.B.; Moorey, S.E.; Mathew, D.J.; Schrick, F.N.; Pohler, K.G.; McLean, K.J.; Myer, P.R. Importance of the Female Reproductive Tract Microbiome and Its Relationship with the Uterine Environment for Health and Productivity in Cattle: A Review. Front. Anim. Sci. 2023, 4, 1111636. [Google Scholar] [CrossRef]
- Pascottini, O.B.; Van Schyndel, S.J.; Spricigo, J.F.W.; Rousseau, J.; Weese, J.S.; LeBlanc, S.J. Dynamics of Uterine Microbiota in Postpartum Dairy Cows with Clinical or Subclinical Endometritis. Sci. Rep. 2020, 10, 12353. [Google Scholar] [CrossRef]
- Webb, E.M.; Holman, D.B.; Schmidt, K.N.; Pun, B.; Sedivec, K.K.; Hurlbert, J.L.; Bochantin, K.A.; Ward, A.K.; Dahlen, C.R.; Amat, S. Sequencing and Culture-Based Characterization of the Vaginal and Uterine Microbiota in Beef Cattle That Became Pregnant or Non-Pregnant via Artificial Insemination. Microbiol. Spectr. 2023, e0273223. [Google Scholar] [CrossRef]
- Patra, A.; Park, T.; Kim, M.; Yu, Z. Rumen Methanogens and Mitigation of Methane Emission by Anti-Methanogenic Compounds and Substances. J. Anim. Sci. Biotechnol. 2017, 8, 13. [Google Scholar] [CrossRef]
- Luecke, S.M.; Holman, D.B.; Schmidt, K.N.; Gzyl, K.E.; Hurlbert, J.L.; Menezes, A.C.B.; Bochantin, K.A.; Kirsch, J.D.; Baumgaertner, F.; Sedivec, K.K.; et al. Whole-Body Microbiota of Newborn Calves and Their Response to Prenatal Vitamin and Mineral Supplementation. Front. Microbiol. 2023, 14, 1207601. [Google Scholar] [CrossRef]
- Dahlen, C.R.; Stoltenow, C.L. The PregCard Study: Assessing the Impact of Routine Management Strategies on Reproductive Performance of Beef Herds in the Upper Great Plains. Bov. Pract. 2015, 49, 152–155. [Google Scholar] [CrossRef]
- Alexander, T.W.; Timsit, E.; Amat, S. The Role of the Bovine Respiratory Bacterial Microbiota in Health and Disease. Anim. Health Res. Rev. 2020, 21, 168–171. [Google Scholar] [CrossRef] [PubMed]
- McMullen, C.; Alexander, T.W.; Orsel, K.; Timsit, E. Progression of Nasopharyngeal and Tracheal Bacterial Microbiotas of Feedlot Cattle during Development of Bovine Respiratory Disease. Vet. Microbiol. 2020, 248, 108826. [Google Scholar] [CrossRef]
- Timsit, E.; McMullen, C.; Amat, S.; Alexander, T.W. Respiratory Bacterial Microbiota in Cattle: From Development to Modulation to Enhance Respiratory Health. Vet. Clin. N. Am. Food Anim. Pract. 2020, 36, 297–320. [Google Scholar] [CrossRef]
- Zeineldin, M.; Lowe, J.; Aldridge, B. Contribution of the Mucosal Microbiota to Bovine Respiratory Health. Trends Microbiol. 2019, 27, 753–770. [Google Scholar] [CrossRef] [PubMed]
- Timsit, E.; Hallewell, J.; Booker, C.; Tison, N.; Amat, S.; Alexander, T.W. Prevalence and Antimicrobial Susceptibility of Mannheimia Haemolytica, Pasteurella Multocida, and Histophilus Somni Isolated from the Lower Respiratory Tract of Healthy Feedlot Cattle and Those Diagnosed with Bovine Respiratory Disease. Vet. Microbiol. 2017, 208, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Hassani, Y.; Brégeon, F.; Aboudharam, G.; Drancourt, M.; Grine, G. Detection of Methanobrevobacter Smithii and Methanobrevibacter Oralis in Lower Respiratory Tract Microbiota. Microorganisms 2020, 8, 1866. [Google Scholar] [CrossRef] [PubMed]
- McMullen, C.; Orsel, K.; Alexander, T.W.; van der Meer, F.; Plastow, G.; Timsit, E. Evolution of the Nasopharyngeal Bacterial Microbiota of Beef Calves from Spring Processing to 40 Days after Feedlot Arrival. Vet. Microbiol. 2018, 225, 139–148. [Google Scholar] [CrossRef]
- Kedzia, A.; Kwapisz, E.; Wierzbowska, M. Incidence of anaerobic bacteria in respiratory tract infections. Pneumonol. Alergol. Pol. 2003, 71, 68–73. [Google Scholar]
- Alsayed, A.; Al-Doori, A.; Al-Dulaimi, A.; Alnaseri, A.; Abuhashish, J.; Aliasin, K.; Alfayoumi, I. Influences of Bovine Colostrum on Nasal Swab Microbiome and Viral Upper Respiratory Tract Infections—A Case Report. Respir. Med. Case Rep. 2020, 31, 101189. [Google Scholar] [CrossRef]
- Centeno-Martinez, R.E.; Glidden, N.; Mohan, S.; Davidson, J.L.; Fernández-Juricic, E.; Boerman, J.P.; Schoonmaker, J.; Pillai, D.; Koziol, J.; Ault, A.; et al. Identification of Bovine Respiratory Disease through the Nasal Microbiome. Anim. Microbiome 2022, 4, 15. [Google Scholar] [CrossRef]
- Davey, M.E.; O’toole, G.A. Microbial Biofilms: From Ecology to Molecular Genetics. Microbiol. Mol. Biol. Rev. 2000, 64, 847–867. [Google Scholar] [CrossRef]
- Muhammad, M.H.; Idris, A.L.; Fan, X.; Guo, Y.; Yu, Y.; Jin, X.; Qiu, J.; Guan, X.; Huang, T. Beyond Risk: Bacterial Biofilms and Their Regulating Approaches. Front. Microbiol. 2020, 11, 928. [Google Scholar] [CrossRef]
- Spoering, A.L.; Lewis, K. Biofilms and Planktonic Cells of Pseudomonas Aeruginosa Have Similar Resistance to Killing by Antimicrobials. J. Bacteriol. 2001, 183, 6746–6751. [Google Scholar] [CrossRef]
- Derakhshani, H.; Fehr, K.B.; Sepehri, S.; Francoz, D.; De Buck, J.; Barkema, H.W.; Plaizier, J.C.; Khafipour, E. Invited Review: Microbiota of the Bovine Udder: Contributing Factors and Potential Implications for Udder Health and Mastitis Susceptibility. J. Dairy Sci. 2018, 101, 10605–10625. [Google Scholar] [CrossRef] [PubMed]
- Togo, A.H.; Grine, G.; Khelaifia, S.; des Robert, C.; Brevaut, V.; Caputo, A.; Baptiste, E.; Bonnet, M.; Levasseur, A.; Drancourt, M.; et al. Culture of Methanogenic Archaea from Human Colostrum and Milk. Sci. Rep. 2019, 9, 18653. [Google Scholar] [CrossRef]
- Drancourt, M.; Djemai, K.; Gouriet, F.; Grine, G.; Loukil, A.; Bedotto, M.; Levasseur, A.; Lepidi, H.; Bou-Khalil, J.; Khelaifia, S.; et al. Methanobrevibacter Smithii Archaemia in Febrile Patients With Bacteremia, Including Those With Endocarditis. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2021, 73, e2571–e2579. [Google Scholar] [CrossRef] [PubMed]
- Bartenslager, A.C.; Althuge, N.D.; Loy, J.D.; Hille, M.M.; Spangler, M.L.; Fernando, S.C. Longitudinal Assessment of the Bovine Ocular Bacterial Community Dynamics in Calves. Anim. Microbiome 2021, 3, 16. [Google Scholar] [CrossRef] [PubMed]
- Caddey, B.; Orsel, K.; Naushad, S.; Derakhshani, H.; De Buck, J. Identification and Quantification of Bovine Digital Dermatitis-Associated Microbiota across Lesion Stages in Feedlot Beef Cattle. mSystems 2021, 6, e00708-21. [Google Scholar] [CrossRef]
- Guerrero-Cruz, S.; Vaksmaa, A.; Horn, M.A.; Niemann, H.; Pijuan, M.; Ho, A. Methanotrophs: Discoveries, Environmental Relevance, and a Perspective on Current and Future Applications. Front. Microbiol. 2021, 12, 678057. [Google Scholar] [CrossRef]
- Wu, D.; Zhao, C.; Bai, H.; Feng, F.; Sui, X.; Sun, G. Characteristics and Metabolic Patterns of Soil Methanogenic Archaea Communities in the High-latitude Natural Forested Wetlands of China. Ecol. Evol. 2021, 11, 10396–10408. [Google Scholar] [CrossRef]
- Ayotte, J.B.; Parker, K.L.; Gillingham, M.P. Use of Natural Licks by Four Species of Ungulates in Northern British Columbia. J. Mammal. 2008, 89, 1041–1050. [Google Scholar] [CrossRef]
- Mahaney, W.C.; Krishnamani, R. Understanding Geophagy in Animals: Standard Procedures for Sampling Soils. J. Chem. Ecol. 2003, 29, 1503–1523. [Google Scholar] [CrossRef]
- Tong, C.; Cadillo-Quiroz, H.; Zeng, Z.H.; She, C.X.; Yang, P.; Huang, J.F. Changes of Community Structure and Abundance of Methanogens in Soils along a Freshwater–Brackish Water Gradient in Subtropical Estuarine Marshes. Geoderma 2017, 299, 101–110. [Google Scholar] [CrossRef]
- Mhuireach, G.Á.; Dietz, L.; Gillett, T. One or Many? Multi-Species Livestock Grazing Influences Soil Microbiome Community Structure and Antibiotic Resistance Potential. Front. Sustain. Food Syst. 2022, 6, 926824. [Google Scholar] [CrossRef]
- Angel, R.; Claus, P.; Conrad, R. Methanogenic Archaea Are Globally Ubiquitous in Aerated Soils and Become Active under Wet Anoxic Conditions. ISME J. 2012, 6, 847–862. [Google Scholar] [CrossRef]
- Radl, V.; Gattinger, A.; Chroňáková, A.; Němcová, A.; Čuhel, J.; Šimek, M.; Munch, J.C.; Schloter, M.; Elhottová, D. Effects of Cattle Husbandry on Abundance and Activity of Methanogenic Archaea in Upland Soils. ISME J. 2007, 1, 443–452. [Google Scholar] [CrossRef]
- Gattinger, A.; Höfle, M.G.; Schloter, M.; Embacher, A.; Böhme, F.; Munch, J.C.; Labrenz, M. Traditional Cattle Manure Application Determines Abundance, Diversity and Activity of Methanogenic Archaea in Arable European Soil. Environ. Microbiol. 2007, 9, 612–624. [Google Scholar] [CrossRef] [PubMed]
- Healy, W.B. Ingestion of Soil by Dairy Cows. N. Z. J. Agric. Res. 1968, 11, 487–499. [Google Scholar] [CrossRef]
- Jurjanz, S.; Feidt, C.; Pérez-Prieto, L.A.; Ribeiro Filho, H.M.N.; Rychen, G.; Delagarde, R. Soil Intake of Lactating Dairy Cows in Intensive Strip Grazing Systems. Anim. Int. J. Anim. Biosci. 2012, 6, 1350–1359. [Google Scholar] [CrossRef] [PubMed]
- Bønløkke, J.H.; Duchaine, C.; Schlünssen, V.; Sigsgaard, T.; Veillette, M.; Basinas, I. Archaea and Bacteria Exposure in Danish Livestock Farmers. Ann. Work Expo. Health 2019, 63, 965–974. [Google Scholar] [CrossRef]
- Lagier, J.-C.; Khelaifia, S.; Alou, M.T.; Ndongo, S.; Dione, N.; Hugon, P.; Caputo, A.; Cadoret, F.; Traore, S.I.; Seck, E.H.; et al. Culture of Previously Uncultured Members of the Human Gut Microbiota by Culturomics. Nat. Microbiol. 2016, 1, 16203. [Google Scholar] [CrossRef]
- Eckburg, P.B.; Bik, E.M.; Bernstein, C.N.; Purdom, E.; Dethlefsen, L.; Sargent, M.; Gill, S.R.; Nelson, K.E.; Relman, D.A. Diversity of the Human Intestinal Microbial Flora. Science 2005, 308, 1635–1638. [Google Scholar] [CrossRef]
- Miller, T.L.; Wolin, M.J. Stability of Methanobrevibacter Smithii Populations in the Microbial Flora Excreted from the Human Large Bowel. Appl. Environ. Microbiol. 1983, 45, 317–318. [Google Scholar] [CrossRef]
- Battumur, U.; Lee, M.; Bae, G.S.; Kim, C.-H. Isolation and Characterization of a New Methanoculleus Bourgensis Strain KOR-2 from the Rumen of Holstein Steers. Asian-Australas. J. Anim. Sci. 2018, 32, 241–248. [Google Scholar] [CrossRef]
- Kumar, S.; Dagar, S.S.; Puniya, A.K. Isolation and Characterization of Methanogens from Rumen of Murrah Buffalo. Ann. Microbiol. 2012, 62, 345–350. [Google Scholar] [CrossRef]
- McCartney, C.A.; Bull, I.D.; Dewhurst, R.J. Chemical Markers for Rumen Methanogens and Methanogenesis. Animal 2013, 7, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Knights, D.; Kuczynski, J.; Charlson, E.S.; Zaneveld, J.; Mozer, M.C.; Collman, R.G.; Bushman, F.D.; Knight, R.; Kelley, S.T. Bayesian Community-Wide Culture-Independent Microbial Source Tracking. Nat. Methods 2011, 8, 761–763. [Google Scholar] [CrossRef]
- McGhee, J.J.; Rawson, N.; Bailey, B.A.; Fernandez-Guerra, A.; Sisk-Hackworth, L.; Kelley, S.T. Meta-SourceTracker: Application of Bayesian Source Tracking to Shotgun Metagenomics. PeerJ 2020, 8, e8783. [Google Scholar] [CrossRef] [PubMed]
- André, A.C.; Debande, L.; Marteyn, B.S. The Selective Advantage of Facultative Anaerobes Relies on Their Unique Ability to Cope with Changing Oxygen Levels during Infection. Cell. Microbiol. 2021, 23, e13338. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Imlay, J.A. When Anaerobes Encounter Oxygen: Mechanisms of Oxygen Toxicity, Tolerance and Defence. Nat. Rev. Microbiol. 2021, 19, 774–785. [Google Scholar] [CrossRef] [PubMed]
- Zakem, E.J.; Mahadevan, A.; Lauderdale, J.M.; Follows, M.J. Stable Aerobic and Anaerobic Coexistence in Anoxic Marine Zones. ISME J. 2020, 14, 288–301. [Google Scholar] [CrossRef]
- Rackaityte, E.; Halkias, J.; Fukui, E.M.; Mendoza, V.F.; Hayzelden, C.; Crawford, E.D.; Fujimura, K.E.; Burt, T.D.; Lynch, S.V. Viable Bacterial Colonization Is Highly Limited in the Human Intestine in Utero. Nat. Med. 2020, 26, 599–607. [Google Scholar] [CrossRef]
- Al Alam, D.; Danopoulos, S.; Grubbs, B.; Ali, N.A.B.M.; MacAogain, M.; Chotirmall, S.H.; Warburton, D.; Gaggar, A.; Ambalavanan, N.; Lal, C.V. Human Fetal Lungs Harbor a Microbiome Signature. Am. J. Respir. Crit. Care Med. 2020, 201, 1002–1006. [Google Scholar] [CrossRef]
- Kimura, I.; Miyamoto, J.; Ohue-Kitano, R.; Watanabe, K.; Yamada, T.; Onuki, M.; Aoki, R.; Isobe, Y.; Kashihara, D.; Inoue, D.; et al. Maternal Gut Microbiota in Pregnancy Influences Offspring Metabolic Phenotype in Mice. Science 2020, 367, eaaw8429. [Google Scholar] [CrossRef] [PubMed]
- Vuong, H.E.; Pronovost, G.N.; Williams, D.W.; Coley, E.J.L.; Siegler, E.L.; Qiu, A.; Kazantsev, M.; Wilson, C.J.; Rendon, T.; Hsiao, E.Y. The Maternal Microbiome Modulates Fetal Neurodevelopment in Mice. Nature 2020, 586, 281–286. [Google Scholar] [CrossRef] [PubMed]
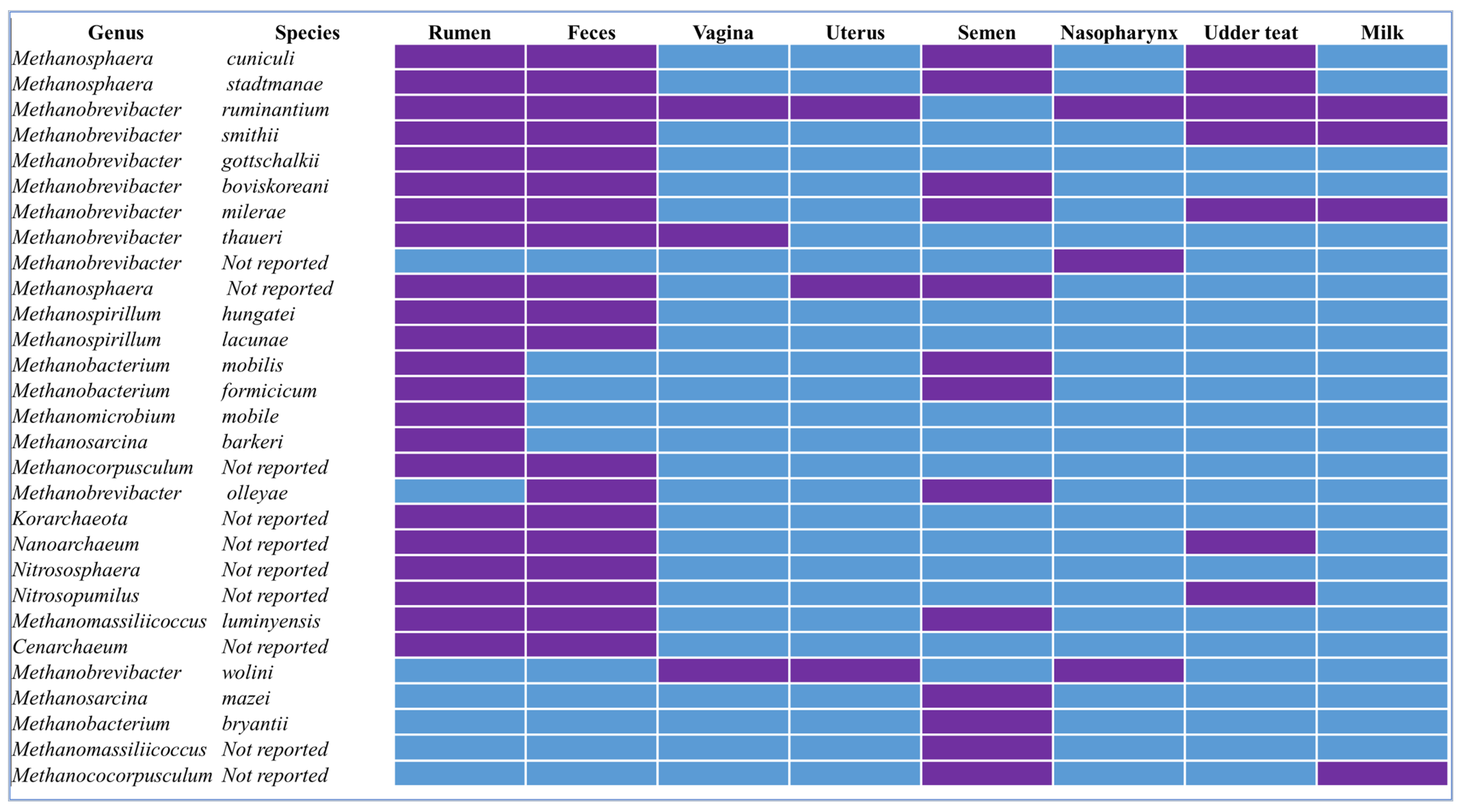
| Phylum | Family | Genus | Species | Sequencing Method | Host | Reference |
|---|---|---|---|---|---|---|
| Rumen | ||||||
| Euryarchaeota | Methanobacteriaceae | Methanobrevibacter | ruminantium, smithii, gottschalkii, boviskoreani, milerae | 16S rRNA Sequencing V4 | Korean native cattle | [56] |
| Euryarchaeota | Methanobacteriaceae | Methanobrevibacter | ruminantium, smithii, gottschalkii, boviskoreani, milerae | 16S rRNA Sequencing V6–V8 | Sheep | [57] |
| Euryarchaeota | Methanobacteriaceae | Methanobrevibacter | ruminantium, smithii, gottschalkii, boviskoreani, milerae, thaueri | 16S rRNA Sequencing V4 | Dairy cow | [58] |
| Euryarchaeota | Methanobacteriaceae | Methanobrevibacter | Ruminantium | 16S rRNA Sequencing V4 | Beef heifers | [21] |
| Euryarchaeota | Methanobacteriaceae | Methanosphaera | Not reported | 16S rRNA | Ruminants | [50] |
| Euryarchaeota | Methanobacteriaceae | Methanosphaera | stadtmanae, cuniculi | 16S rRNA Sequencing V4 | Breeding bulls | [59] |
| Thermoplasmatota | Methanomassiliicoccaceae | Methanomassiliicoccus | Luminyensis | qPCR | Sheep, cow | [60] |
| Euryarchaeota | Methanospirillaceae | Methanospirillum | hungatei and lacunae | 16S rRNA Sequencing V4 | Dairy cow | [61] |
| Euryarchaeota | Methanobacteriales | Methanobacterium | mobilis, formicicum, barkaeri | 16S &18S | Grazing cattle | [62] |
| Euryarchaeota | Methanobacteriaceae | Methanomicrobium | Mobile | 16S rRNA Sequencing V1–V2, V2–V3, culturing | Sheep | [63] |
| Euryarchaeota | Methanosarcinaceae | Methanosarcina | Barkeri | 16S rRNA Sequencing, culturing | Dairy cow | [64] |
| Euryarchaeota | Methanocorpusculaceae | Methanocorpusculum | Not reported | 16S rRNA Sequencing V3–V4 | Dairy cow | [65] |
| Nitrososphaerota | Nitrososphaeraceae | Nitrososphaera | Not reported | 16S rRNA Sequencing V3–V4 | Dairy cow | [65] |
| Thermoproteota | Cenarchaeaceae | Cenarchaeum | Not reported | Shotgun metagenomics | Dairy cattle | [66] |
| Nitrososphaerota | Nitrosopumilaceae | Nitrosopumilus | Not reported | Shotgun metagenomics | Dairy cattle | [66] |
| Korarchaeota | Korarchaeales | Korarchaeota | Not reported | Shotgun metagenomics | Dairy cattle | [66] |
| Nanoarchaeota | Nanoarchaeaceae | Nanoarchaeum | Not reported | Shotgun metagenomics | Dairy cattle | [66] |
| Feces | ||||||
| Euryarchaeota | Methanobacteriaceae | Methanosphaera | stadtmanae, cuniculi | 16S rRNA Sequencing V4 | Breeding bulls | [59] |
| Euryarchaeota | Methanospirillaceae | Methanospirillum | hungatei and lacunae | 16S rRNA Sequencing V4 | Dairy cow | [60] |
| Euryarchaeota | Methanocorpusculaceae | Methanocorpusculum | Not reported | 16S rRNA Sequencing V3–V4 | Sika deer, Dairy cow | [61] |
| Euryarchaeota | Methanobacteriaceae | Methanobrevibacter | smithii, millaerae, labreanum, aggregans | PCR | Sheep | [57] |
| Euryarchaeota | Methanobacteriaceae | Methanobrevibacter | smithii, millaerae, labreanum, aggregans, thaueri | PCR | Dairy cow | [65] |
| Euryarchaeota | Methanobacteriaceae | Methanobrevibacter | boviskoreoni, millerae, olleyae, ruminantium, wolini | 16S rRNA Sequencing V3–V4 | Dairy cow | [66] |
| Euryarchaeota | Methanobacteriaceae | Methanobrevibacter | Not reported | 16S rRNA Sequencing V3–V4 | Dairy cow | [66] |
| Euryarchaeota | Methanobacteriaceae | Methanosphaera | cuniculi | 16S rRNA Sequencing V3–V4 | Dairy cow | [66] |
| Euryarchaeota | Methanobacteriaceae | Methanosphaera | Not reported | 16S rRNA Sequencing V3–V4 | Dairy cow | [66] |
| Euryarchaeota | Methanobacteriaceae | Not reported | Not reported | 16S rRNA Sequencing V3–V4 | Dairy cow | [66] |
| Euryarchaeota | Methanosarcinaceae | Methanosarcina | Mazei | 16S rRNA Sequencing V3–V4 | Dairy cow | [60] |
| Halobacterota | Methanomicrobia | Methanococorpusculum | Not reported | 16S rRNA Sequencing V3–V4 | Dairy cow | [60] |
| Korarchaeota | Korarchaeales | Korarchaeota | Not reported | Shotgun metagenomics | Dairy cow | [66] |
| Nanoarchaeota | Nanoarchaeaceae | Nanoarchaeum | Not reported | Shotgun metagenomics | Dairy cow | [66] |
| Nitrososphaerota | Nitrososphaeraceae | Nitrososphaera | Not reported | 16S rRNA Sequencing V4 | Dairy cow | [65] |
| Nitrososphaerota | Nitrososphaeraceae | Nitrososphaera | Not reported | 16S rRNA Sequencing V3–V4 | Dairy cow | [65] |
| Nitrososphaerota | Nitrosopumilaceae | Nitrosopumilus | Not reported | Shotgun metagenomics | Dairy cow | [65] |
| Thermoplasmatota | Methanomassiliicoccaceae | Methanomassiliicoccus | luminyensis | qPCR | Sheep, cow | [60] |
| Thermoplasmatota | Methanomethylophilaceae | Methanomassiliicoccus | Not reported | 16S rRNA Sequencing V3–V4 | Beef bull | [67] |
| Thermoproteota | Cenarchaeaceae | Cenarchaeum | Not reported | Shotgun metagenomics | Dairy cattle | [66] |
| Phylum | Family | Genus | Species | Sequencing Platform | Host | Reference |
|---|---|---|---|---|---|---|
| Vagina | ||||||
| Euryarchaeota | Methanobacteriaceae | Methanobrevibacter | ruminantium | 16S rRNA Sequencing V3–V4 | Beef heifers | [21] |
| Euryarchaeota | Methanobacteriaceae | Methanobrevibacter | ruminantium | 16S rRNA Sequencing V3–V4 | Beef heifers | [25] |
| Euryarchaeota | Methanobacteriaceae | Methanobrevibacter | wolini | 16S rRNA Sequencing V3–V4 | Beef heifers | [25] |
| Euryarchaeota | Methanobacteriaceae | Methanobrevibacter | not reported | 16S rRNA Sequencing V3 | Beef heifers | [89] |
| Uterus | ||||||
| Euryarchaeota | Methanobacteriaceae | Methanobrevibacter | ruminantium | 16S rRNA Sequencing V3–V4 | Beef heifers | [25] |
| Euryarchaeota | Methanobacteriaceae | Methanobrevibacter | wolini | 16S rRNA Sequencing V3–V4 | Beef heifers | [25] |
| Euryarchaeota | Methanobacteriaceae | Methanosphaera | not reported | 16S rRNA Sequencing V3–V4 | Beef heifers | [25] |
| Semen | ||||||
| Euryarchaeota | Methanobacteriaceae | Methanosphaera | stadtmanae, cuniculi | 16S rRNA Sequencing V4 | Beef bulls | [59] |
| Thermoplasmatota | Methanomassiliicoccaceae | Methanomassiliicoccus | luminyensis | 16S rRNA Sequencing V4 | Beef bulls | [59] |
| Crenarchaeota | Nitrososphaeraceae | Not reported | Not reported | 16S rRNA Sequencing V3–V4 | Beef bulls | [67] |
| Euryarchaeota | Methanobacteriaceae | Methanobacterium | mobilis, formicicum, bryantii, | 16S rRNA Sequencing V4 | Beef bulls | [59] |
| Euryarchaeota | Methanobacteriaceae | Methanobrevibacter | boviskoreoni, millerae, olleyae, ruminatium, wolini | 16S rRNA Sequencing V3–V4 | Beef bulls | [67] |
| Euryarchaeota | Methanobacteriaceae | Methanobrevibacter | Not reportd | 16S rRNA Sequencing V3–V4 | Beef bulls | [67] |
| Euryarchaeota | Methanobacteriaceae | Methanosphaera | cuniculi | 16S rRNA Sequencing V3–V4 | Beef bulls | [67] |
| Euryarchaeota | Methanobacteriaceae | Methanosphaera | Not reported | 16S rRNA Sequencing V3–V4 | Beef bulls | [67] |
| Euryarchaeota | Methanobacteriaceae | Not reported | Not reported | 16S rRNA Sequencing V3–V4 | Beef bulls | [67] |
| Euryarchaeota | Methanosarcinaceae | Methanosarcina | mazei | 16S rRNA Sequencing V3–V4 | Beef bulls | [67] |
| Thermoplasmatota | Methanomethylophilaceae | Methanomassiliicoccus | Not reported | 16S rRNA Sequencing V3–V4 | Beef bulls | [67] |
| Halobacterota | Methanomicrobia | Methanococorpusculum | Not reported | 16S rRNA Sequencing V3–V4 | Beef bulls | [67] |
| Halobacterota | Methanomicrobia | Methanococorpusculum | Not reported | 16S rRNA Sequencing V3–V4 | Beef bulls | [90] |
| Milk | ||||||
| Euryarchaeota | Methanobacteriaceae | Methanobrevibacter | ruminantium, smithii, milerae | Shotgun metagenomics | Dairy cow | [60,91] |
| Euryarchaeota | Methanocorpusculaceae | Methanocorpusculum | Not reported | Shotgun metagenomics | Dairy cow | [91] |
| Nasopharynx | ||||||
| Euryarchaeota | Methanobacteriaceae | Methanobrevibacter | ruminantium | 16S rRNA Sequencing V3–V4 | Beef heifers | [25] |
| Euryarchaeota | Methanobacteriaceae | Methanobrevibacter | wolinii | 16S rRNA Sequencing V3–V5 | Beef heifers | [25] |
| Euryarchaeota | Methanobacteriaceae | Methanobrevibacter | Not reported | 16S rRNA Sequencing V4 | Beef steers | [92] |
| Udder teat | ||||||
| Euryarchaeota | Methanobacteriaceae | Methanobrevibacter | ruminantium, smithii, milerae | Shotgun metagenomics | Dairy cow | [60,91] |
| Euryarchaeota | Methanobacteriaceae | Methanosphaera | stadtmanae, cuniculi | Shotgun metagenomics | Dairy cow | [91] |
| Nitrososphaerota | Nitrosopumilaceae | Nitrosopumilus | Not reported | Shotgun metagenomics | Yak calves | [53] |
| Nanoarchaeota | Nanoarchaeaceae | Nanoarchaeum | Not reported | Shotgun metagenomics | Yak calves | [53] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aryee, G.; Luecke, S.M.; Dahlen, C.R.; Swanson, K.C.; Amat, S. Holistic View and Novel Perspective on Ruminal and Extra-Gastrointestinal Methanogens in Cattle. Microorganisms 2023, 11, 2746. https://doi.org/10.3390/microorganisms11112746
Aryee G, Luecke SM, Dahlen CR, Swanson KC, Amat S. Holistic View and Novel Perspective on Ruminal and Extra-Gastrointestinal Methanogens in Cattle. Microorganisms. 2023; 11(11):2746. https://doi.org/10.3390/microorganisms11112746
Chicago/Turabian StyleAryee, Godson, Sarah M. Luecke, Carl R. Dahlen, Kendall C. Swanson, and Samat Amat. 2023. "Holistic View and Novel Perspective on Ruminal and Extra-Gastrointestinal Methanogens in Cattle" Microorganisms 11, no. 11: 2746. https://doi.org/10.3390/microorganisms11112746
APA StyleAryee, G., Luecke, S. M., Dahlen, C. R., Swanson, K. C., & Amat, S. (2023). Holistic View and Novel Perspective on Ruminal and Extra-Gastrointestinal Methanogens in Cattle. Microorganisms, 11(11), 2746. https://doi.org/10.3390/microorganisms11112746






