Tolerance Evaluation of Celery Commercial Cultivars and Genetic Variability of Fusarium oxysporum f. sp. apii
Abstract
:1. Introduction
2. Materials and Methods
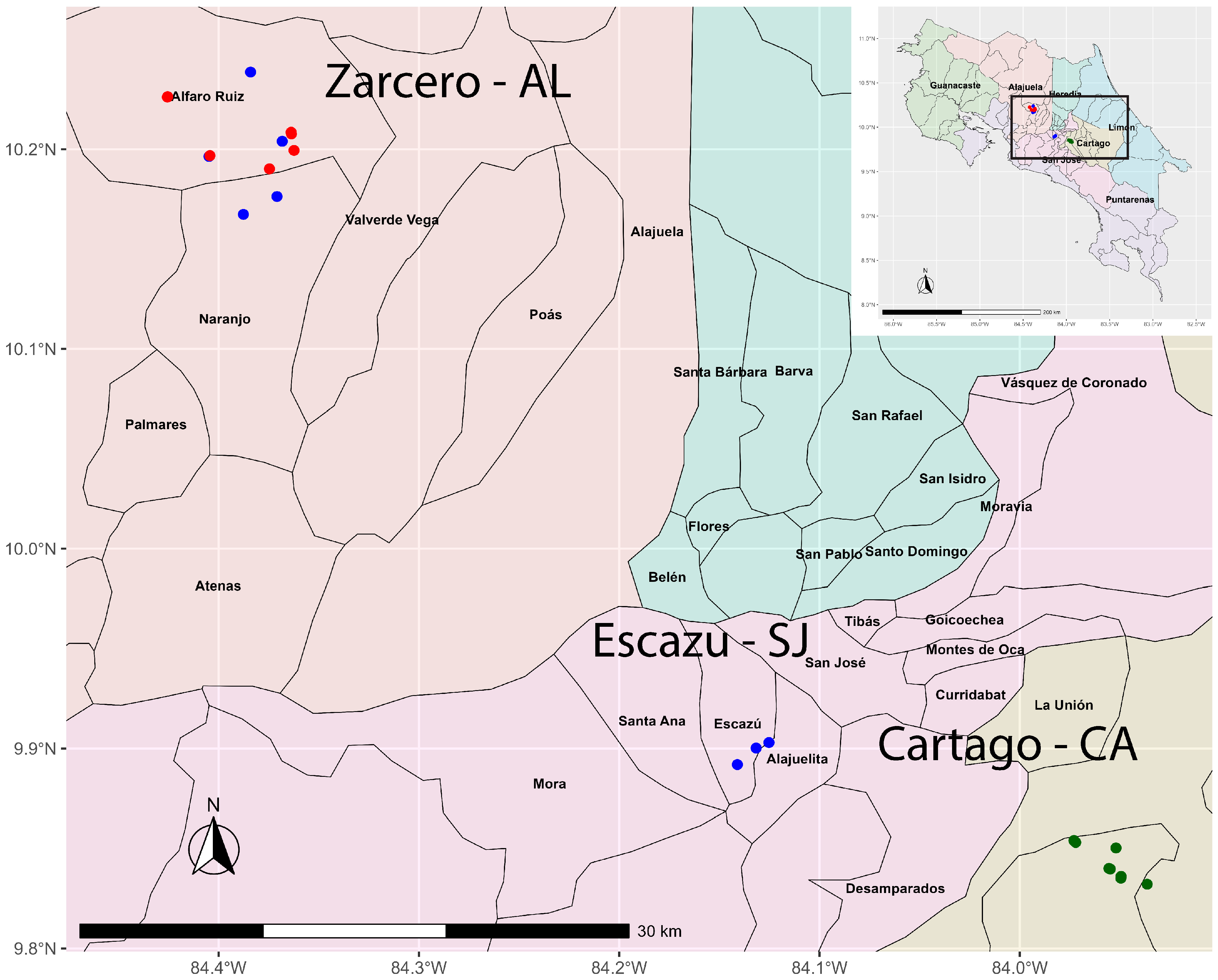
2.1. Collection and Isolation of Fusarium spp.
2.2. DNA Extraction and Procedures
2.3. Phylogenetic Analysis
2.4. Disease Tolerance Evaluation
2.4.1. Celery Cultivars
2.4.2. Pathogen Isolates and Inoculum Production
2.4.3. Greenhouse Conditions
2.4.4. Experimental Design and Data Analysis
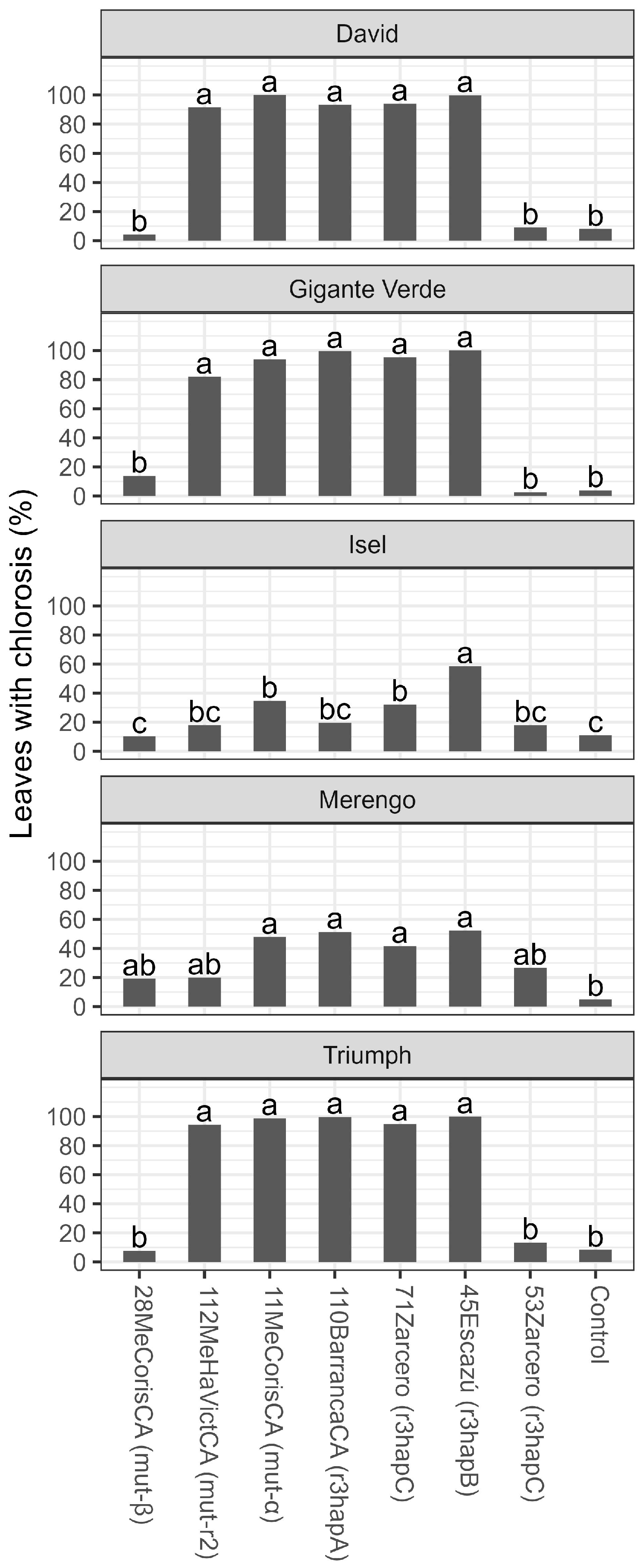
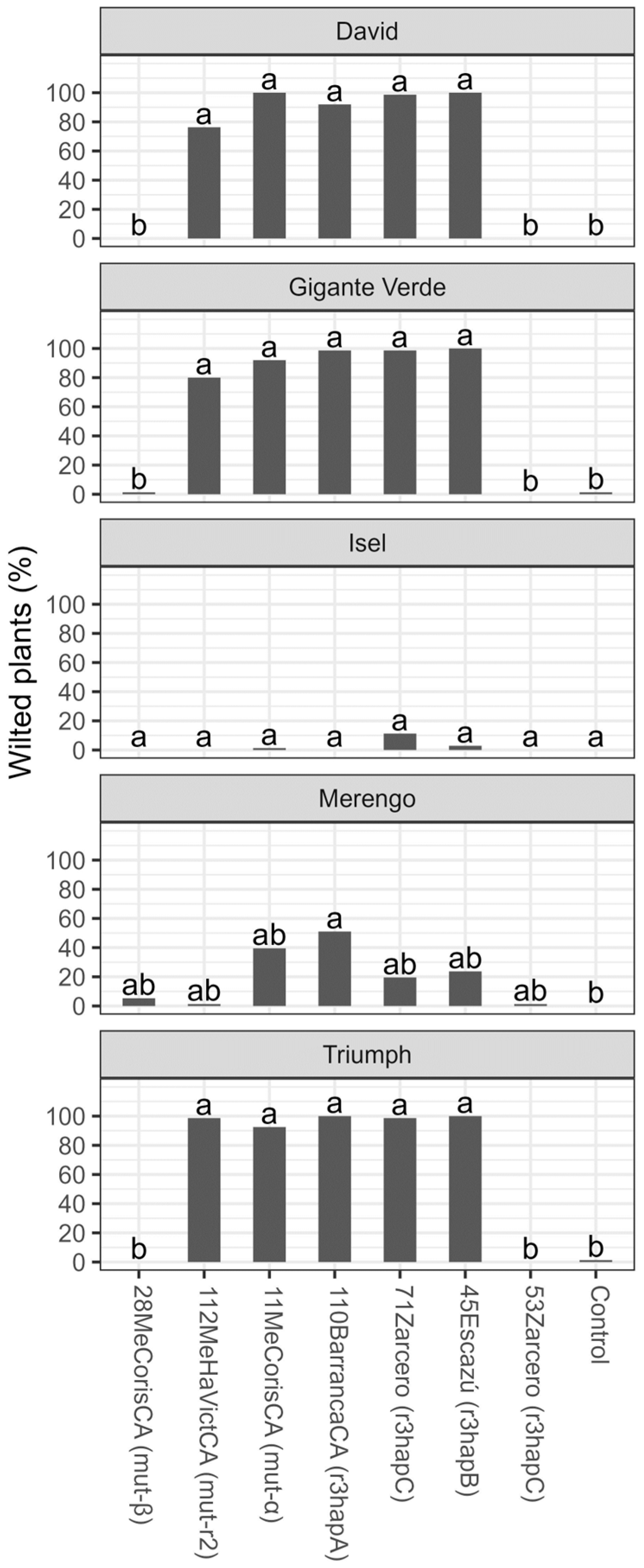
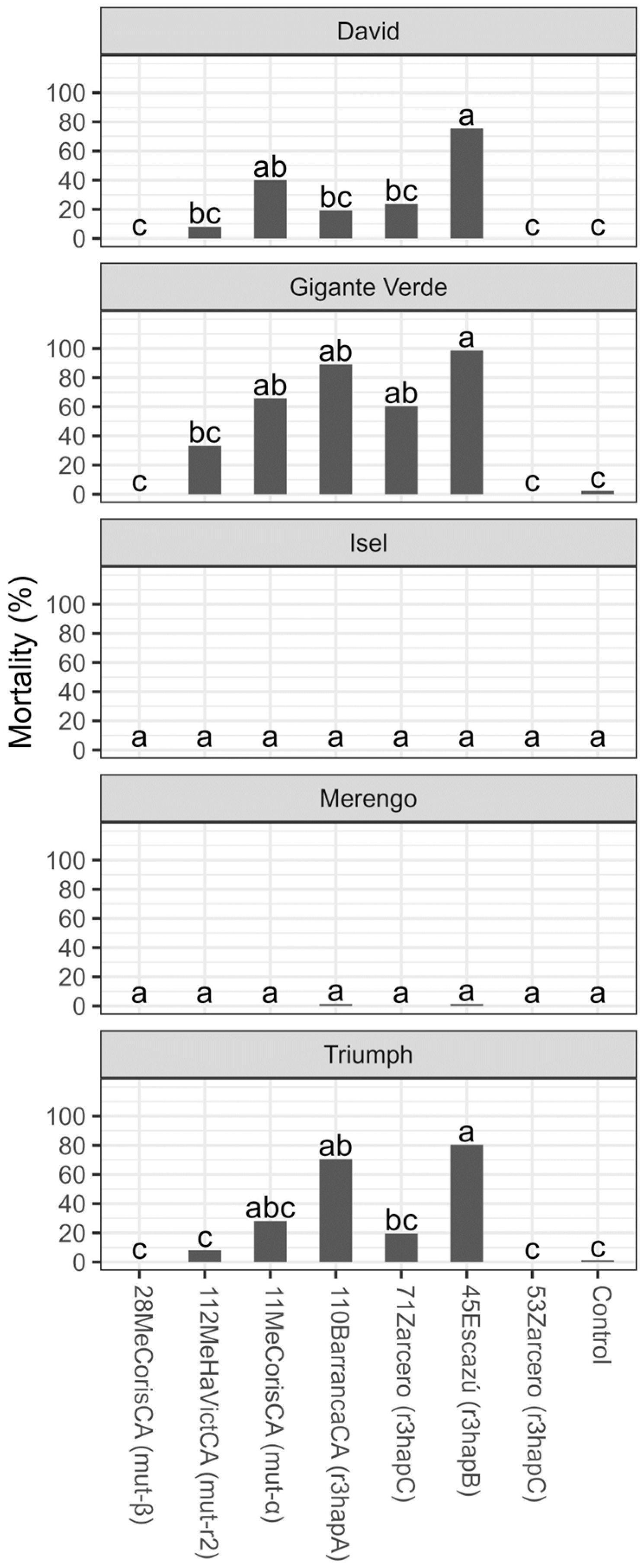
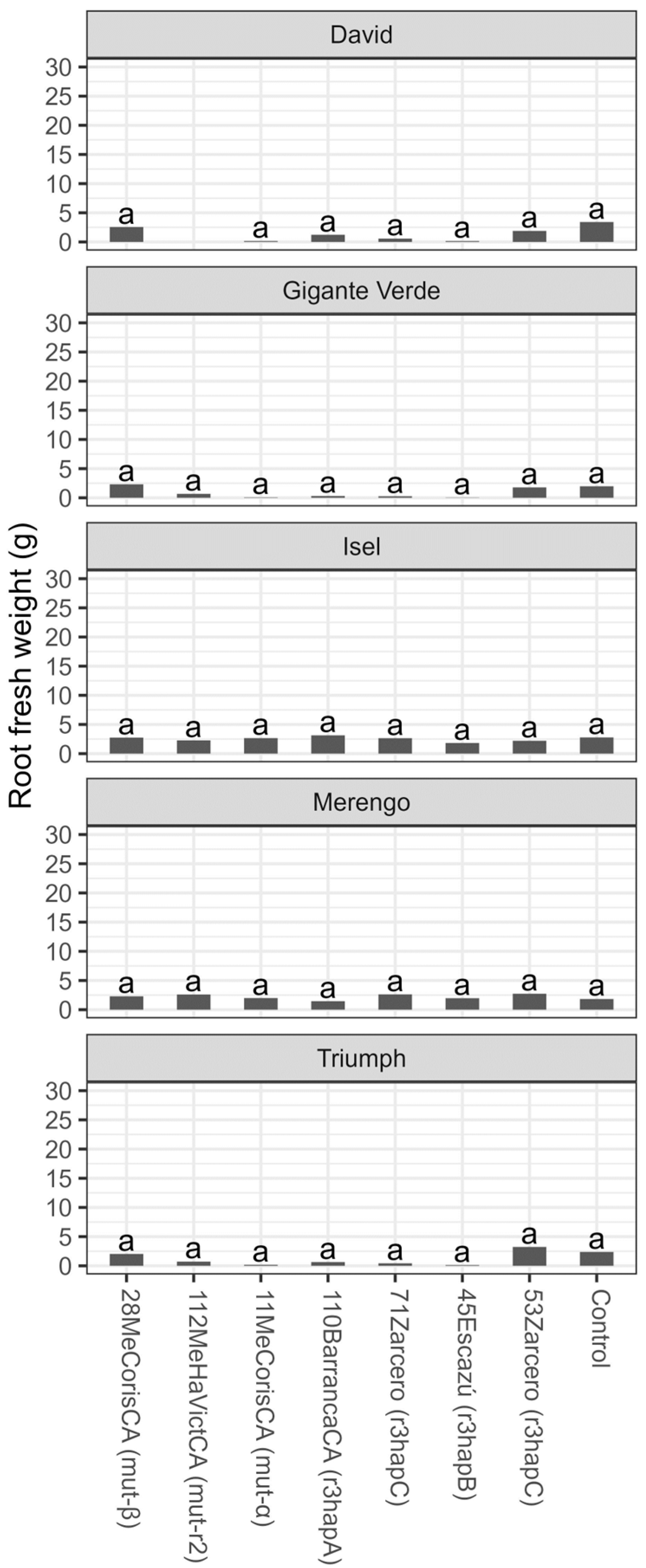
3. Results
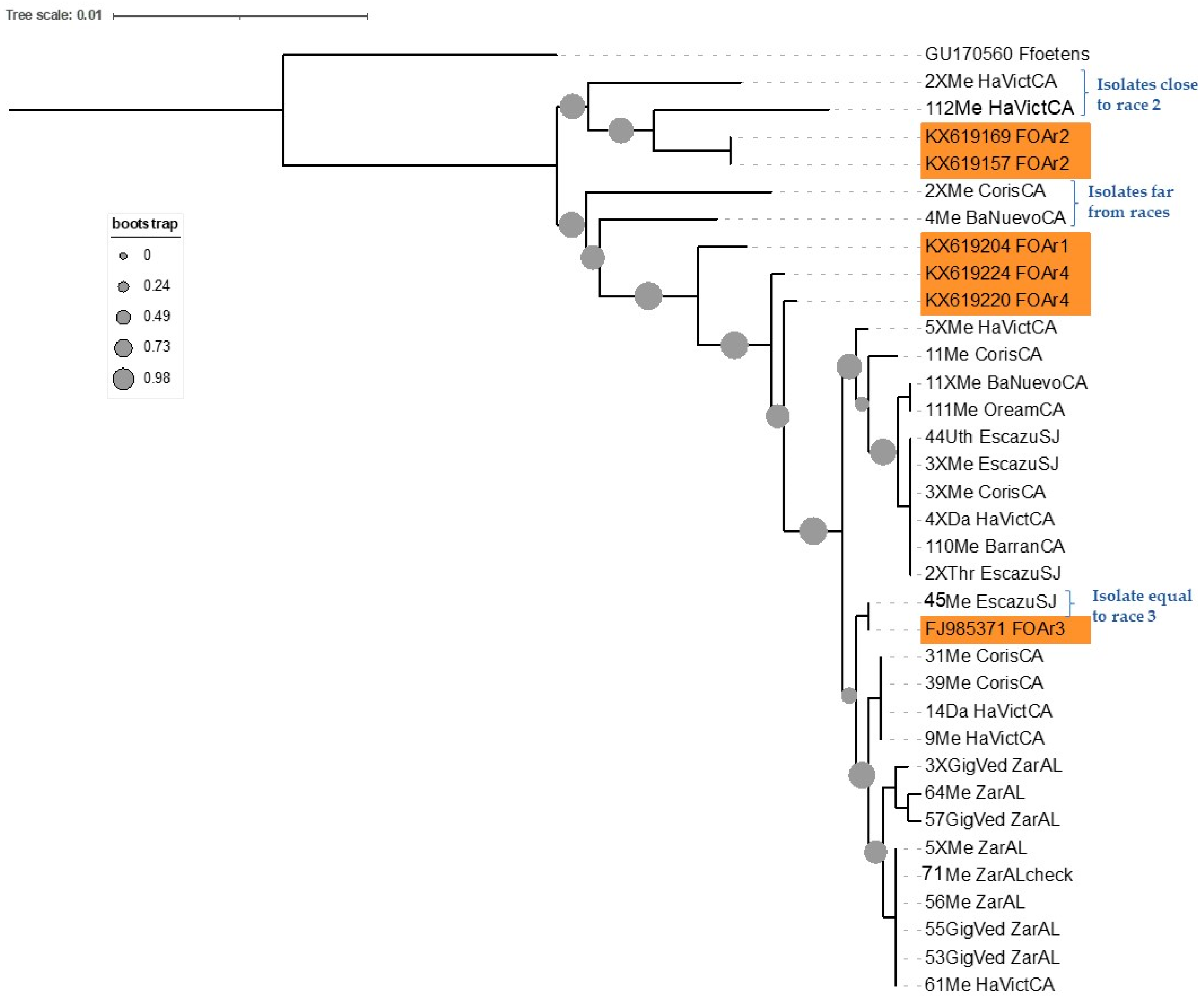
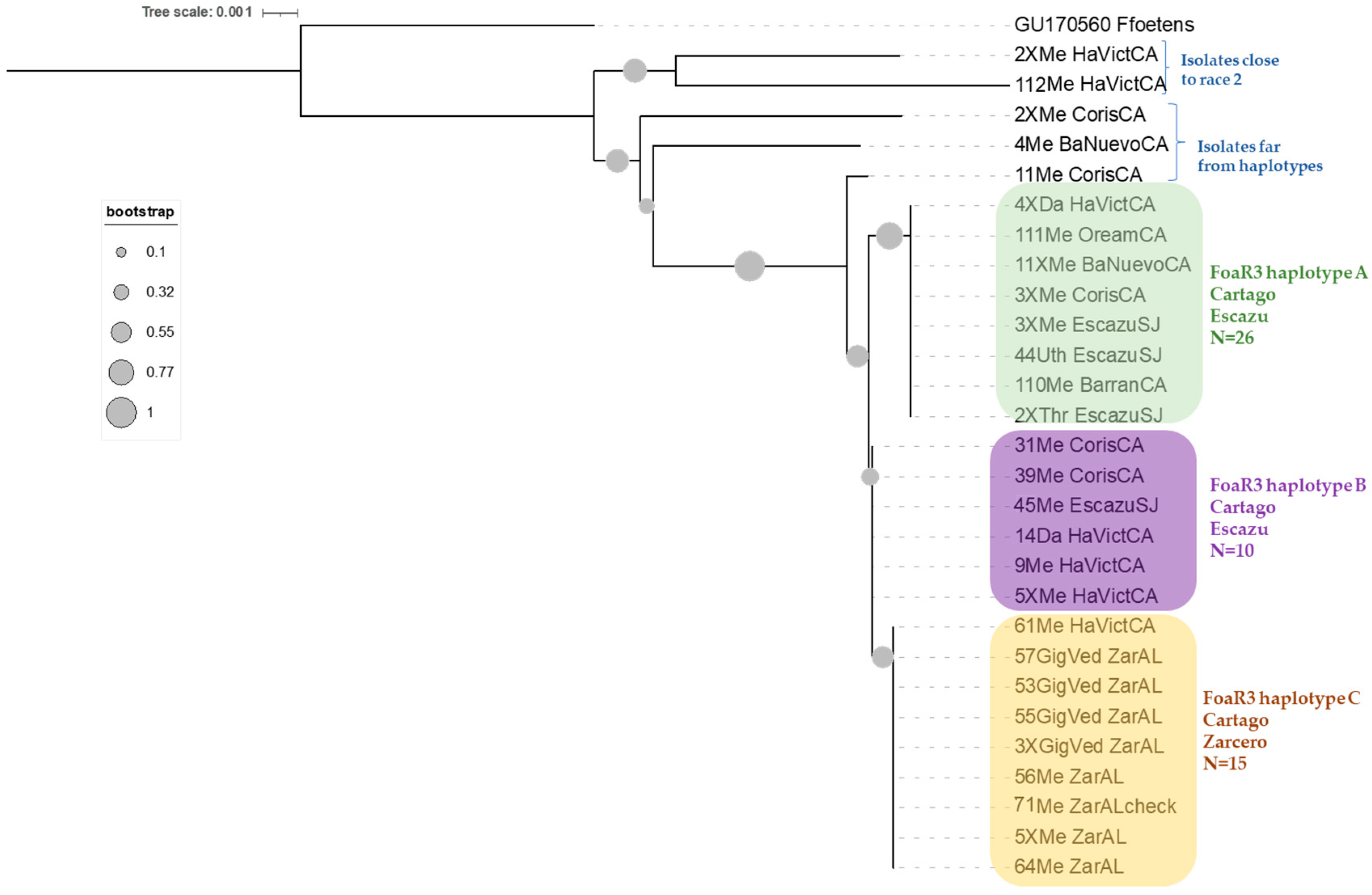
3.1. Phylogenetic Analysis
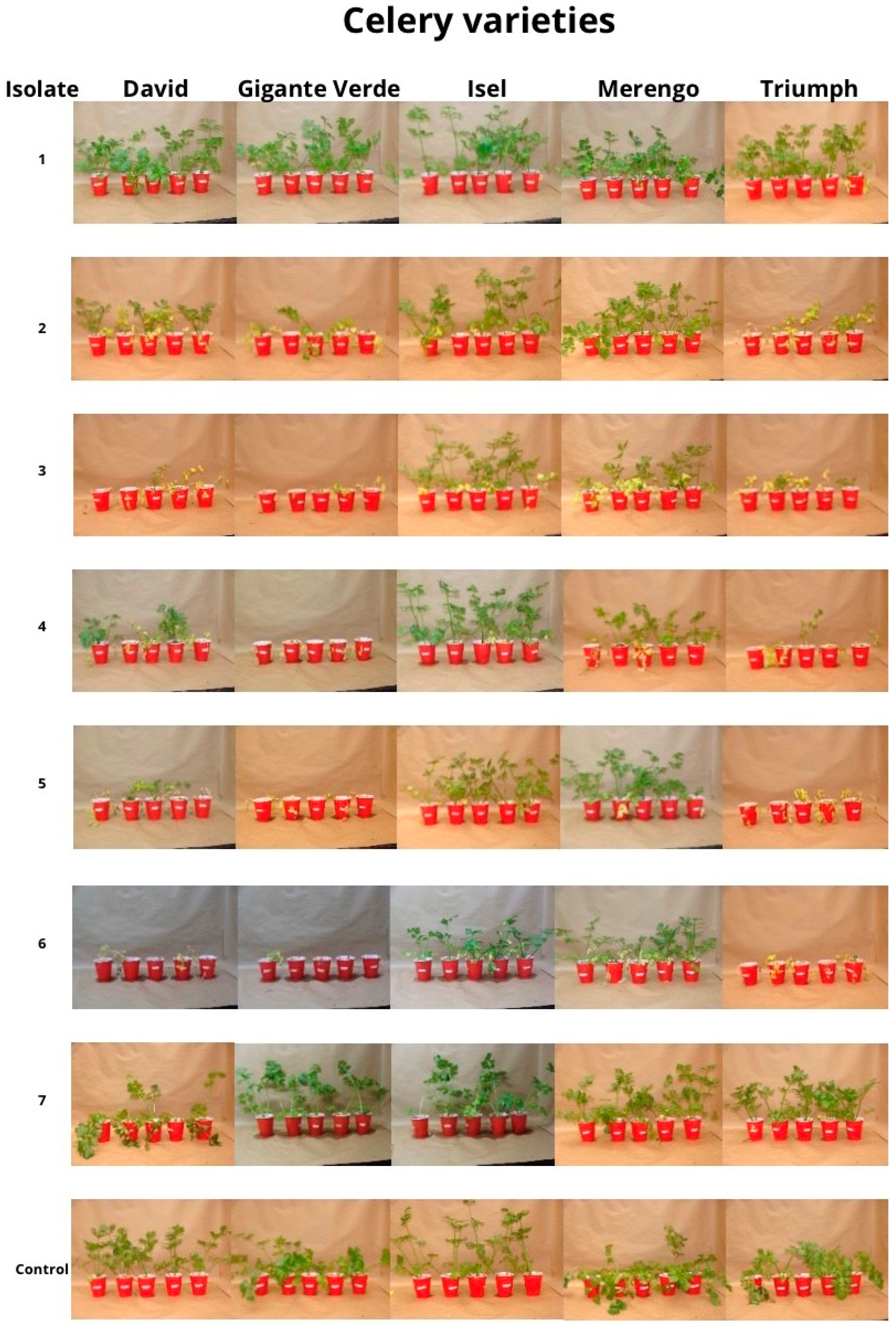
3.2. Tolerance Evaluation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Khalil, A.; Nawaz, H.; Ghania, J.B.; Rehman, R.; Nadeem, F. Value Added Products, Chemical Constituents and Medicinal Uses of Celery (Apium Graveolens L.)—A Review. Int. J. Chem. Biochem. Sci. 2015, 10, 40–48. [Google Scholar]
- Frese, L.; Bönisch, M.; Nachtigall, M.; Schirmak, U. Patterns of Genetic Diversity and Implications for In Situ Conservation of Wild Celery (Apium graveolens L. ssp. graveolens). Agriculture 2018, 8, 129. [Google Scholar] [CrossRef]
- Li, M.-Y.; Hou, X.-L.; Wang, F.; Tan, G.-F.; Xu, Z.-S.; Xiong, A.-S. Advances in the Research of Celery, an Important Apiaceae Vegetable Crop. Crit. Rev. Biotechnol. 2018, 38, 172–183. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Nara, U.; Kaur, K.; Rani, N.; Jaswal, C. Genetic, Genomic and Biochemical Insights of Celery (Apium graveolens L.) in the Era of Molecular Breeding. J. Appl. Res. Med. Aromat. Plants 2022, 31, 100420. [Google Scholar] [CrossRef]
- Kaur, S.; Barakat, R.; Kaur, J.; Epstein, L. The Effect of Temperature on Disease Severity and Growth of Fusarium oxysporum f. sp. apii Races 2 and 4 in Celery. Phytopathology® 2022, 112, 364–372. [Google Scholar] [CrossRef]
- Elmer, W.H. Seeds as Vehicles for Pathogen Importation. Biological Invasions 2001, 3, 263–271. [Google Scholar] [CrossRef]
- Shi, Y.; Jin, Z.; Meng, X.; Wang, L.; Xie, X.; Chai, A.; Li, B. Development and Evaluation of a Loop-Mediated Isothermal Amplification Assay for the Rapid Detection and Identification of Pectobacterium carotovorum on Celery in the Field. Hortic. Plant J. 2020, 6, 313–320. [Google Scholar] [CrossRef]
- Koike, S.T.; Daugovish, O.; Downer, J.A. Sclerotinia Petiole and Crown Rot of Celery Caused by Sclerotinia minor in California. Plant Dis. 2006, 90, 829. [Google Scholar] [CrossRef]
- Groenewald, M.; Groenewald, J.Z.; Braun, U.; Crous, P.W. Host Range of Cercospora apii and C. beticola and Description of C. apiicola, a Novel Species from Celery. Mycologia 2006, 98, 275–285. [Google Scholar] [CrossRef]
- Lori, G.A.; Malbran, I.; Mourelos, C.A.; Wolcan, S.M. First Report of Fusarium oxysporum f. sp. apii Race 2 Causing Fusarium Yellows on Celery in Argentina. Plant Dis. 2016, 100, 1020. [Google Scholar] [CrossRef]
- Retana, K.; Ramirez, A.; Castro, O.; Blanco-Meneses, M. Morphological and Molecular Characterization of Fusarium oxysporum f. sp. apii in Costa Rica. Agron. Costarric. 2018, 42, 115–126. [Google Scholar] [CrossRef]
- Edel-Hermann, V.; Lecomte, C. Current Status of Fusarium oxysporum Formae Speciales and Races. Phytopathology® 2019, 109, 512–530. [Google Scholar] [CrossRef]
- Henry, P.; Kaur, S.; Pham, Q.A.T.; Barakat, R.; Brinker, S.; Haensel, H.; Daugovish, O.; Epstein, L. Genomic Differences between the New Fusarium oxysporum f. sp. apii (Foa) Race 4 on Celery, the Less Virulent Foa Races 2 and 3, and the Avirulent on Celery f. sp. coriandrii. BMC Genom. 2020, 21, 730. [Google Scholar] [CrossRef]
- Elmer, W.H.; Lacy, M.L. Effects of crop residues and colonization of plant tissues on propagule survival and soil populations of Fusarium oxysporum f. sp. apii race 2. Phytopathology 1987, 77, 381–387. [Google Scholar] [CrossRef]
- Puhalla, J.E. A Visual Indicator of Heterokaryosis in Fusarium oxysporum from Celery. Can. J. Bot. 1984, 62, 540–545. [Google Scholar] [CrossRef]
- Epstein, L.; Kaur, S.; Chang, P.L.; Carrasquilla-Garcia, N.; Lyu, G.; Cook, D.R.; Subbarao, K.V.; O’Donnell, K. Races of the Celery Pathogen Fusarium oxysporum f. sp. apii Are Polyphyletic. Phytopathology® 2017, 107, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Epstein, L.; Kaur, S.; Henry, P.M. The Emergence of Fusarium oxysporum f. sp. apii Race 4 and Fusarium oxysporum f. sp. coriandrii Highlights Major Obstacles Facing Agricultural Production in Coastal California in a Warming Climate: A Case Study. Front. Plant Sci. 2022, 13, 921516. [Google Scholar] [CrossRef]
- Epstein, L.; Kaur, S. Apium graveolens PI 181714 is a Source of Resistance to Fusarium oxysporum f. sp. apii Race 4 in Celery (A. Graveolens var. dulce). Plant Breed. 2023, 142, 109–117. [Google Scholar] [CrossRef]
- Murray, M.G.; Thompson, W.F. Rapid isolation of high molecular weight plant DNA. Nucleic Acid Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef]
- O’Donnell, K.; Kistler, H.C.; Cigelnik, E.; Ploetz, R.C. Multiple Evolutionary Origins of the Fungus Causing Panama Disease of Banana: Concordant Evidence from Nuclear and Mitochondrial Gene Genealogies. Proc. Natl. Acad. Sci. USA 1998, 95, 2044–2049. [Google Scholar] [CrossRef]
- O’Donnell, K.; Gueidan, C.; Sink, S.; Johnston, P.R.; Crous, P.W.; Glenn, A.; Riley, R.; Zitomer, N.C.; Colyer, P.; Waalwijk, C.; et al. A Two-Locus DNA Sequence Database for Typing Plant and Human Pathogens within the Fusarium oxysporum Species Complex. Fungal Genet. Biol. 2009, 46, 936–948. [Google Scholar] [CrossRef] [PubMed]
- Penn, O.; Privman, E.; Ashkenazy, H.; Landan, G.; Graur, D.; Pupko, T. GUIDANCE: A Web Server for Assessing Alignment Confidence Scores. Nucleic Acids Res. 2010, 38, W23–W28. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. JModelTest 2: More Models, New Heuristics and Parallel Computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
- Koike, S.T.; Gordon, T.R. First Report of Fusarium Wilt of Cilantro Caused by Fusarium oxysporum in California. Plant Dis. 2005, 89, 1130. [Google Scholar] [CrossRef]
- Song, X.; Sun, P.; Yuan, J.; Gong, K.; Li, N.; Meng, F.; Zhang, Z.; Li, X.; Hu, J.; Wang, J.; et al. The Celery Genome Sequence Reveals Sequential Paleo-Polyploidizations, Karyotype Evolution and Resistance Gene Reduction in Apiales. Plant Biotechnol. J. 2021, 19, 731–744. [Google Scholar] [CrossRef]
- Lori, G.A.; Wolcan, S.M.; Larran, S. Fusarium Yellows of Celery Caused by Fusarium oxysporum f. sp. apii in Argentina. J. Plant Pathol. 2008, 90, 173–178. [Google Scholar]

| p Value | |||||
|---|---|---|---|---|---|
| Factor | Chlorosis 1 | Wilting 1 | Mortality 1 | Plant Fresh Weight | Root Fresh Weight |
| Variety | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.4423 |
| Treatment | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.4484 |
| Variety*Treatment | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.4933 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blanco-Meneses, M.; Serrano-Porras, M.; Calderón-Abarca, A.; Sebiani-Calvo, A.; Vargas, G.; Castro-Zúñiga, O. Tolerance Evaluation of Celery Commercial Cultivars and Genetic Variability of Fusarium oxysporum f. sp. apii. Microorganisms 2023, 11, 2732. https://doi.org/10.3390/microorganisms11112732
Blanco-Meneses M, Serrano-Porras M, Calderón-Abarca A, Sebiani-Calvo A, Vargas G, Castro-Zúñiga O. Tolerance Evaluation of Celery Commercial Cultivars and Genetic Variability of Fusarium oxysporum f. sp. apii. Microorganisms. 2023; 11(11):2732. https://doi.org/10.3390/microorganisms11112732
Chicago/Turabian StyleBlanco-Meneses, Mónica, Mauricio Serrano-Porras, Anny Calderón-Abarca, Alejandro Sebiani-Calvo, Gabriel Vargas, and Oscar Castro-Zúñiga. 2023. "Tolerance Evaluation of Celery Commercial Cultivars and Genetic Variability of Fusarium oxysporum f. sp. apii" Microorganisms 11, no. 11: 2732. https://doi.org/10.3390/microorganisms11112732
APA StyleBlanco-Meneses, M., Serrano-Porras, M., Calderón-Abarca, A., Sebiani-Calvo, A., Vargas, G., & Castro-Zúñiga, O. (2023). Tolerance Evaluation of Celery Commercial Cultivars and Genetic Variability of Fusarium oxysporum f. sp. apii. Microorganisms, 11(11), 2732. https://doi.org/10.3390/microorganisms11112732







