Heavy Metal Tolerance of Microorganisms Isolated from Coastal Marine Sediments and Their Lead Removal Potential
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area and Sample Collection
2.2. Microorganism Isolation and Culture
2.3. Cadmium (Cd2+), Lead (Pb2+), and Zinc (Zn2+) Tolerance Screening
2.4. Molecular Identification of Promising Bacteria
2.5. Viability Test of Promising Bacteria
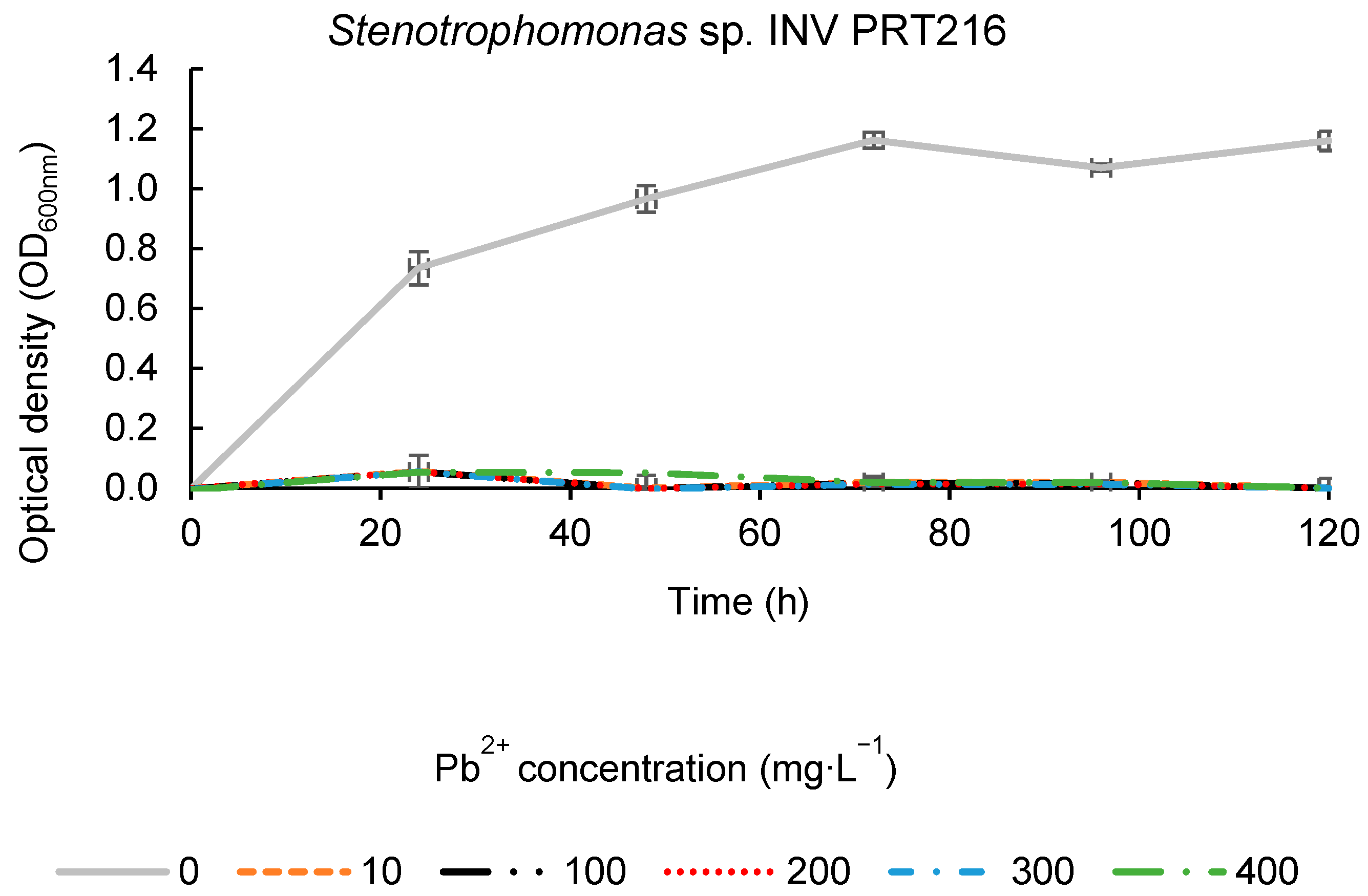
2.6. Bacterial Growth Curves of Promising Isolates at Different Pb2+ Concentrations
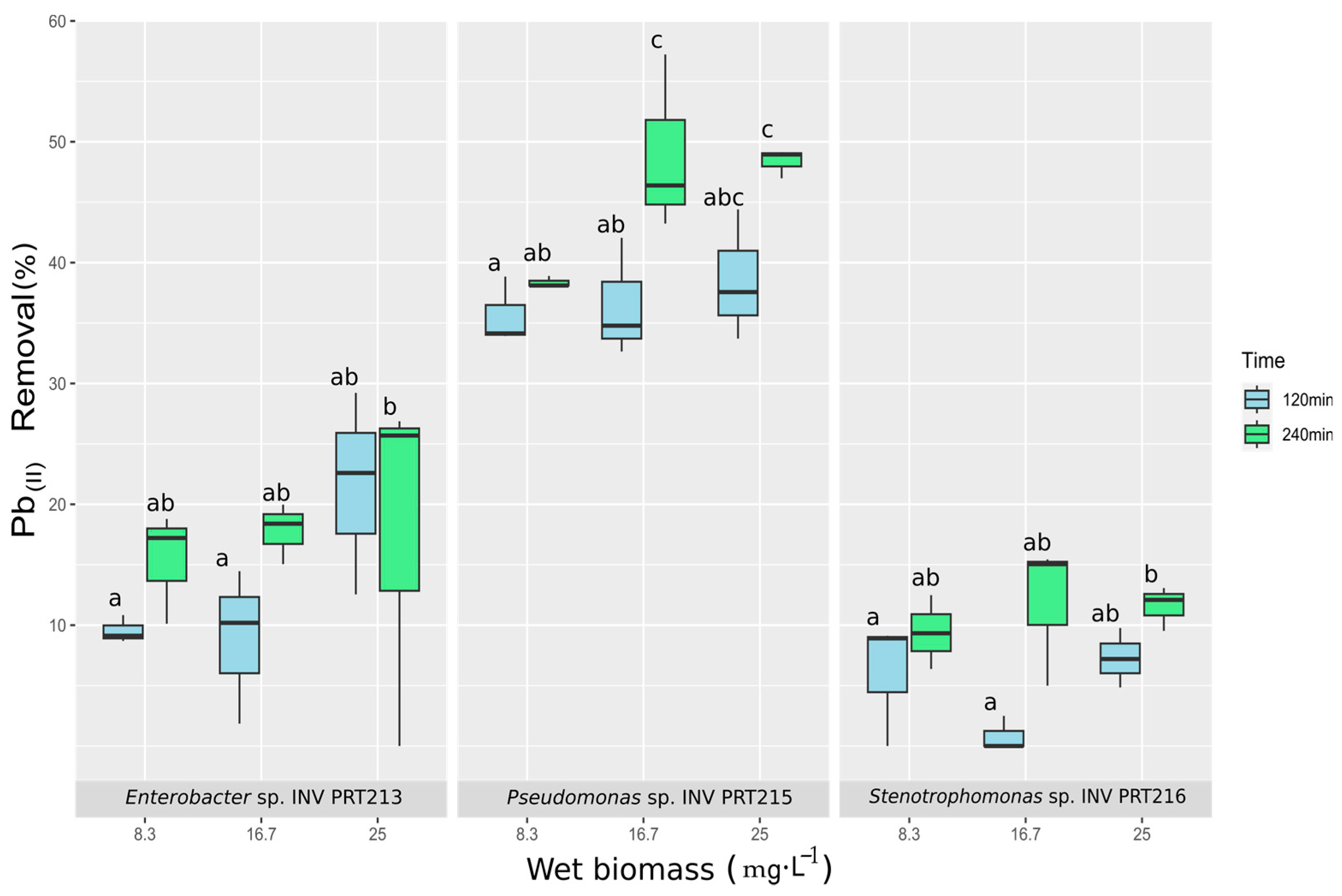
2.7. Pb2+ Removal Experiments
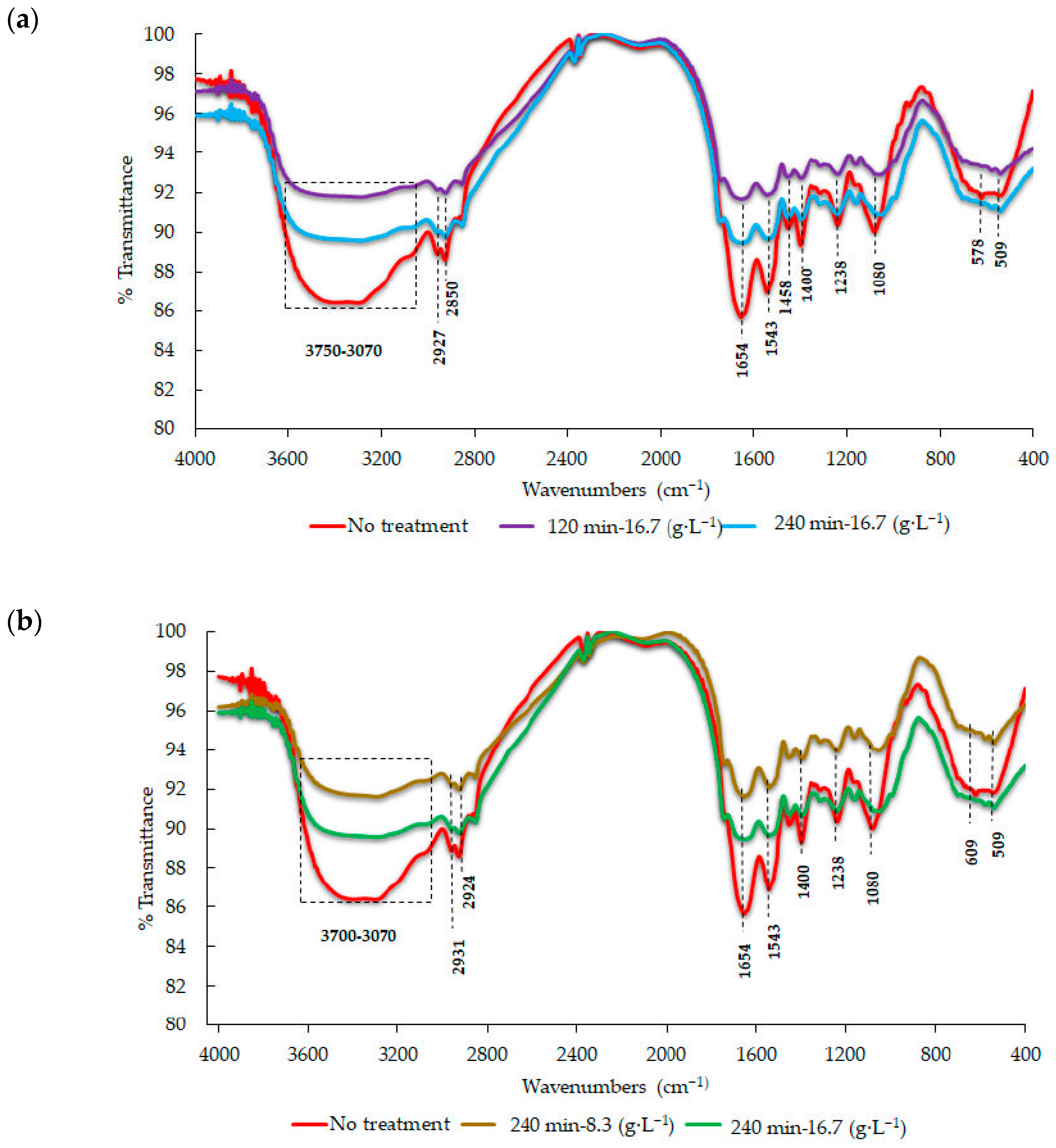
2.8. Fourier Transform Infrared (FT-IR) Spectroscopy Analysis of Bacterial Biomass
2.9. Statistical Analysis
3. Results
3.1. Study Area and Sample Collection
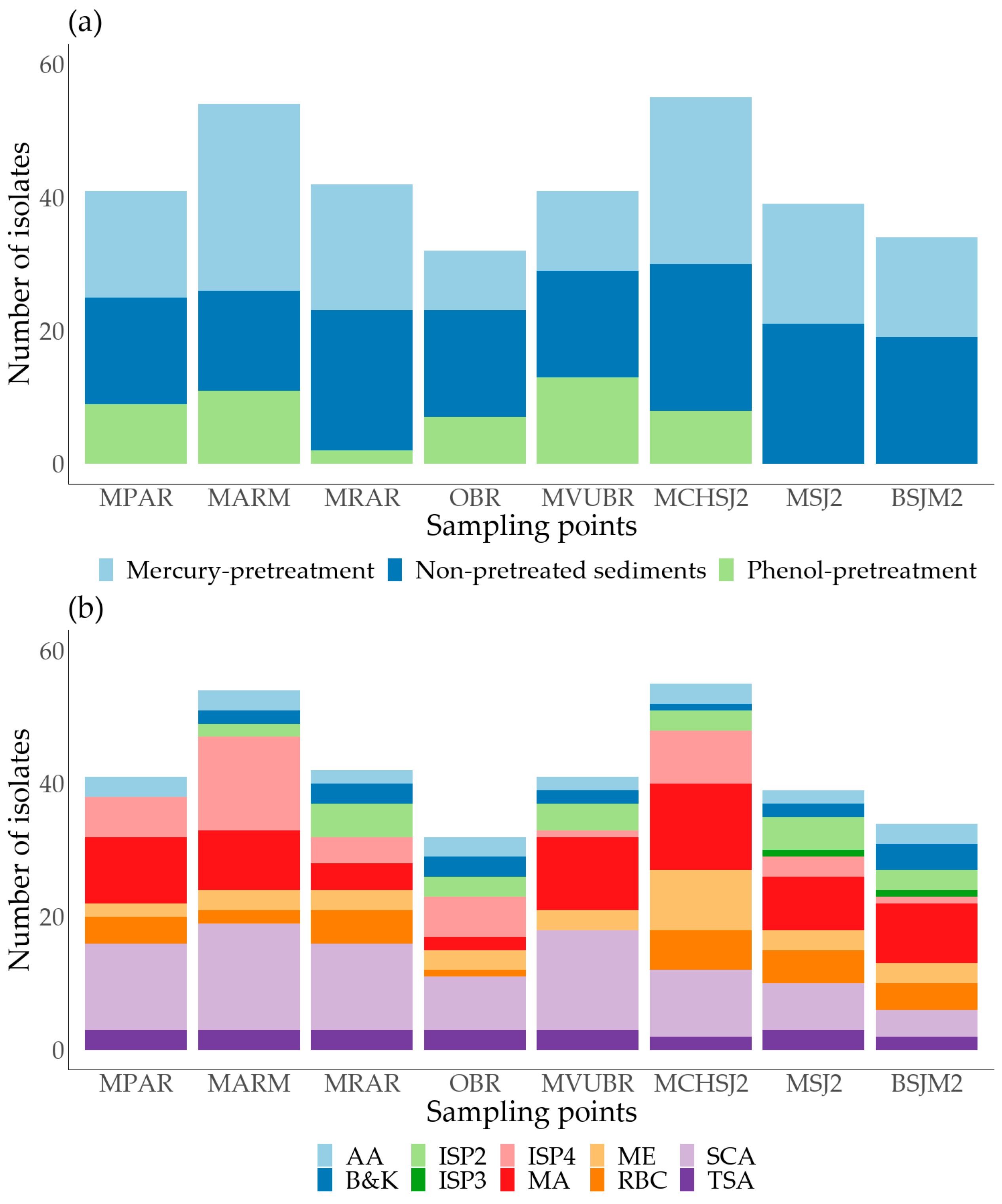
3.2. Microorganism Isolation and Culture
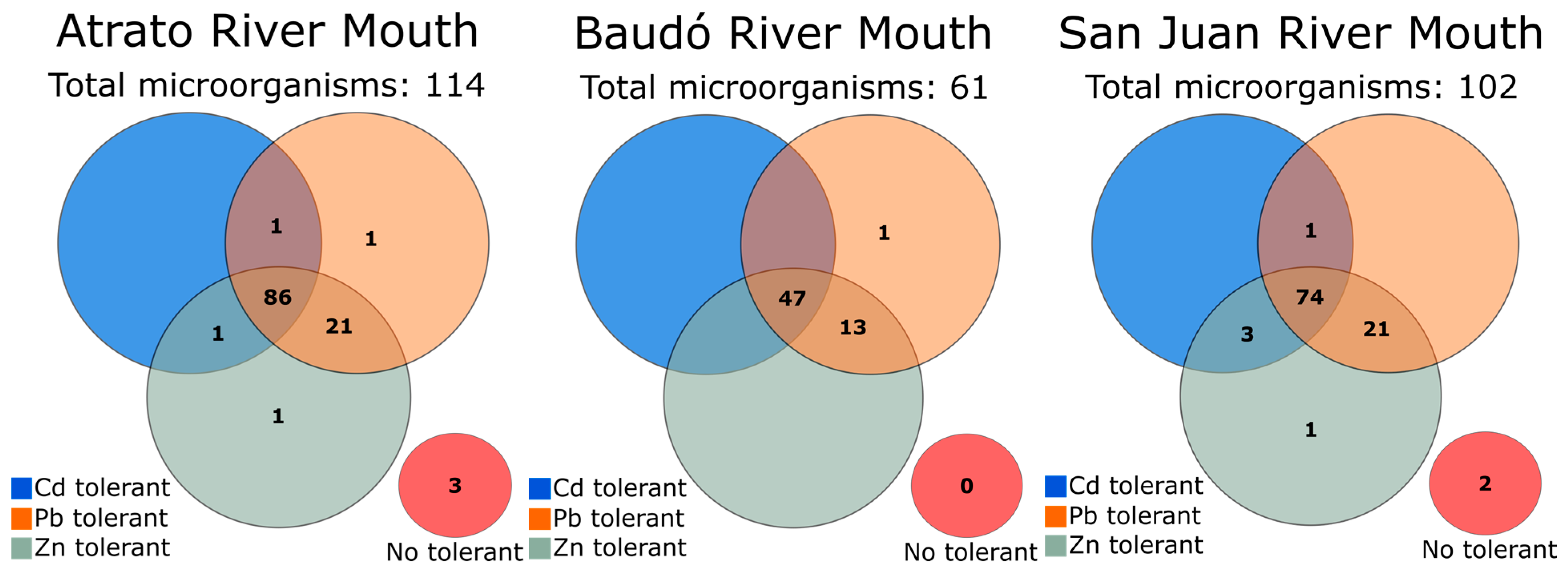
3.3. Cd, Pb, and Zn Tolerance Screening
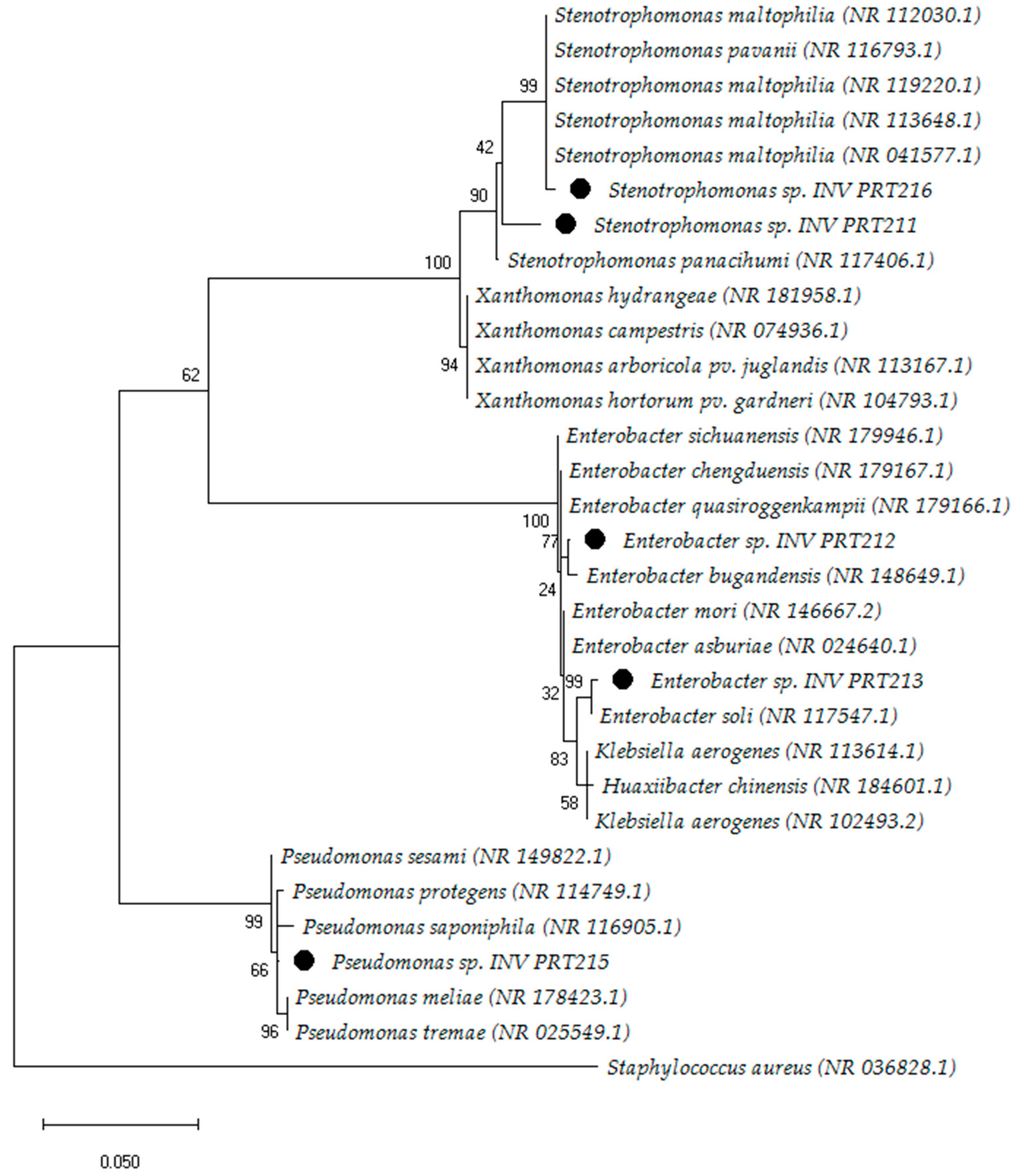
3.4. Molecular Identification and Phylogenetic Analysis
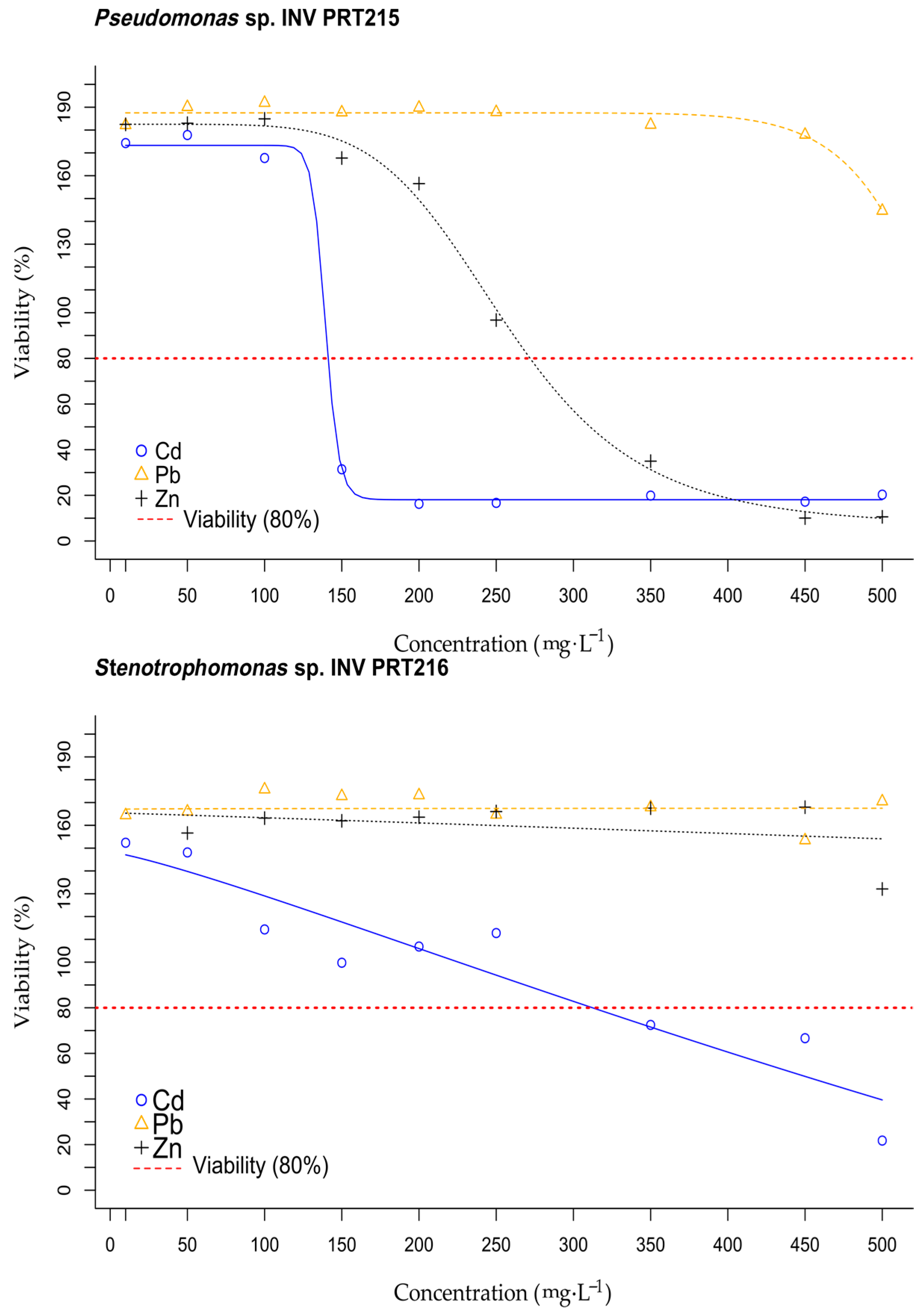
3.5. Viability Test of Enterobacter sp. INV PRT213, Pseudomonas sp. INV PRT215, and Stenotrophomonas sp. INV PRT216
3.6. Bacterial Growth Curves of Promising Isolates at Different Pb2+ Concentrations
3.7. Evaluation of Pb2+ Removal Capacity of Promising Isolates Enterobacter sp. INV PRT213, Pseudomonas sp. INV PRT215, and Stenotrophomonas sp. INV PRT216
3.8. Analysis of Pseudomonas sp. INV PRT215 Biomass via Infrared Spectroscopy (FT-IR)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Prasad Ahirvar, B.; Das, P.; Srivastava, V.; Kumar, M. Perspectives of heavy metal pollution indices for soil, sediment, and water pollution evaluation: An insight. Total Environ. Res. Themes 2023, 6, 100039. [Google Scholar] [CrossRef]
- Keshavarzi, A.; Kumar, V.; Ertunç, G.; Brevik, E.C. Ecological risk assessment and source apportionment of heavy metals contamination: An appraisal based on the Tellus soil survey. Environ. Geochem. Health 2021, 43, 2121–2142. [Google Scholar] [CrossRef]
- Dehghani-Sanij, A.R.; Tharumalingam, E.; Dusseault, M.B.; Fraser, R. Study of energy storage systems and environmental challenges of batteries. Renew. Sustain. Energy Rev. 2019, 104, 192–208. [Google Scholar] [CrossRef]
- Turner, A.; Filella, M. Hazardous metal additives in plastics and their environmental impacts. Environ. Int. 2021, 156, 106622. [Google Scholar] [CrossRef]
- Lucero Rincón, C.H.; Peña Salamanca, E.J.; Cantera Kintz, J.R.; Lizcano, O.V.; Cruz-Quintana, Y.; Neira, R. Assessment of mercury and lead contamination using the bivalve Anadara tuberculosa (Arcidae) in an estuary of the Colombian Pacific. Mar. Pollut. Bull. 2023, 187, 114519. [Google Scholar] [CrossRef]
- Briffa, J.; Sinagra, E.; Blundell, R. Heavy metal pollution in the environment and their toxicological effects on humans. Heliyon 2020, 6, e04691. [Google Scholar] [CrossRef]
- Ren, H.; Sang, S.; Wei, X.; Peng, C.; Zhu, K. Activity coefficients, phase diagram and separation process design for NaNO3 and Pb(NO3)2 of waste liquor in the production of lead–chromium pigments. J. Chem. Thermodyn. 2023, 187, 107146. [Google Scholar] [CrossRef]
- Gaur, V.K.; Sharma, P.; Gaur, P.; Varjani, S.; Ngo, H.H.; Guo, W.; Chaturvedi, P.; Singhania, R.R. Sustainable mitigation of heavy metals from effluents: Toxicity and fate with recent technological advancements. Bioengineered 2021, 12, 7297–7313. [Google Scholar] [CrossRef]
- Azhar, U.; Ahmad, H.; Shafqat, H.; Babar, M.; Shahzad Munir, H.M.; Sagir, M.; Arif, M.; Hassan, A.; Rachmadona, N.; Rajendran, S.; et al. Remediation techniques for elimination of heavy metal pollutants from soil: A review. Environ. Res. 2022, 214, 113918. [Google Scholar] [CrossRef]
- Tufail, M.A.; Iltaf, J.; Zaheer, T.; Tariq, L.; Amir, M.B.; Fatima, R.; Asbat, A.; Kabeer, T.; Fahad, M.; Naeem, H.; et al. Recent advances in bioremediation of heavy metals and persistent organic pollutants: A review. Sci. Total Environ. 2022, 850, 157961. [Google Scholar] [CrossRef]
- Guzmán, C.; Hurtado, A.; Carreño, C.; Casos, I. Production of polyhydroxyalkanoates by native halophilic bacteria using Solanum tuberosum L. shell starch. Sci. Agropecu. 2017, 8, 109–118. [Google Scholar] [CrossRef]
- Priyadarshanee, M.; Das, S. Biosorption and removal of toxic heavy metals by metal tolerating bacteria for bioremediation of metal contamination: A comprehensive review. J. Environ. Chem. Eng. 2021, 9, 104686. [Google Scholar] [CrossRef]
- Hassan, A.; Hamid, F.S.; Pariatamby, A.; Suhaimi, N.S.M.; Razali Noor Maiza binti, M.; Ling, K.N.H.; Mohan, P. Bioaugmentation-assisted bioremediation and biodegradation mechanisms for PCB in contaminated environments: A review on sustainable clean-up technologies. J. Environ. Chem. Eng. 2023, 11, 110055. [Google Scholar] [CrossRef]
- Zhang, H.; Yuan, X.; Xiong, T.; Wang, H.; Jiang, L. Bioremediation of co-contaminated soil with heavy metals and pesticides: Influence factors, mechanisms and evaluation methods. Chem. Eng. J. 2020, 398, 125657. [Google Scholar] [CrossRef]
- Pande, V.; Pandey, S.C.; Sati, D.; Bhatt, P.; Samant, M. Microbial Interventions in Bioremediation of Heavy Metal Contaminants in Agroecosystem. Front. Microbiol. 2022, 13, 824084. [Google Scholar] [CrossRef]
- Chau, T.P.; Bulgariu, L.; Saravanan, M.; Rajkumar, R.; Chinnathambi, A.; Salmen, S.H.; Jhanani, G. Bioremediation efficiency of free and immobilized form of Aspergillus niger and Aspergillus tubigenesis biomass on tannery effluent. Environ. Res. 2023, 231, 116275. [Google Scholar] [CrossRef]
- Cao, R.; Zhang, Y.; Ju, Y.; Wang, W.; Zhao, Y.; Liu, N.; Zhang, G.; Wang, X.; Xie, X.; Dai, C.; et al. Exopolysaccharide-producing bacteria enhanced Pb immobilization and influenced the microbiome composition in rhizosphere soil of pakchoi (Brassica chinensis L.). Front. Microbiol. 2023, 14, 1117312. [Google Scholar] [CrossRef]
- Igiri, B.E.; Okoduwa, S.I.R.; Idoko, G.O.; Akabuogu, E.P.; Adeyi, A.O.; Ejiogu, I.K. Toxicity and Bioremediation of Heavy Metals Contaminated Ecosystem from Tannery Wastewater: A Review. J. Toxicol. 2018, 2018, 2568038. [Google Scholar] [CrossRef]
- Mathivanan, K.; Chandirika, J.U.; Vinothkanna, A.; Yin, H.; Liu, X.; Meng, D. Bacterial adaptive strategies to cope with metal toxicity in the contaminated environment—A review. Ecotoxicol. Environ. Saf. 2021, 226, 112863. [Google Scholar] [CrossRef]
- Velásquez, A.; Quintero, M.; Jíménez-Vergar, E.; Blandón, L.; Gómez-León, J. Microorganismos marinos extremófilos con potencial interes en estudios de bioprospección. Rev. Fac. Cienc. 2018, 7, 1–40. [Google Scholar]
- Anaya, J.A.; Gutiérrez-Vélez, V.H.; Pacheco-Pascagaza, A.M.; Palomino-Ángel, S.; Han, N.; Balzter, H. Drivers of Forest Loss in a Megadiverse Hotspot on the Pacific Coast of Colombia. Remote Sens. 2020, 12, 1235. [Google Scholar] [CrossRef]
- Luisa, F.; Espinosa, Paola Obando y Ostin Garcés (Eds.) INVEMAR, 2020, Diagnóstico y Evaluación de la Calidad de las Aguas Marinas y Costeras en el Caribe y Pacífico Colombianos. Red de Vigilancia Para la Conservación y Protección de las Aguas Marinas y costeras de Colombia−REDCAM: INVEMAR, MinAmbiente, CORALINA, CORPOGUAJIRA, CORPAMAG, CRA, CARDIQUE, CARSUCRE, CVS, CORPOURABÁ, CODECHOCÓ, CVC, CRC, and CORPONARIÑO). Informe técnico 2019. Serie de Publicaciones Periódicas No. 4 del INVEMAR, Santa Marta. 171p. Available online: https://siam.invemar.org.co/documentos-detalle/15654 (accessed on 8 September 2023).
- Rodríguez-Aparicio, J.; Vergara-Buitrago, P.A. Environmental analysis of coal mining in the strategic ecosystem of paramo (Boyacá, Colombia). Sci. Tech. 2021, 26, 398–405. [Google Scholar]
- Mohammad, A.M.; Salah Eldin, T.A.; Hassan, M.A.; El-Anadouli, B.E. Efficient treatment of lead-containing wastewater by hydroxyapatite/chitosan nanostructures. Arab. J. Chem. 2017, 10, 683–690. [Google Scholar] [CrossRef]
- Ramirez-Rodriguez, J.C. Tratamiento de aguas residuales y problemáticas ambientales del sector textil en Colombia: Una revisión. Inf. Téc. 2023, 87, 82–106. [Google Scholar] [CrossRef]
- Mahecha-Pulido, J.D.; Trujillo-González, J.M.; Torres-Mora, M.A.; Mahecha-Pulido, J.D.; Trujillo-González, J.M.; Torres-Mora, M.A. Analysis of Studies in Heavy Metals in Agricultural Areas of Colombia. ORINOQUIA 2017, 21, 83–93. [Google Scholar]
- Sánchez Pinzón, M.S. Contaminación por Metales Pesados en el Botadero de Basuras de Moravia en Medellín Transferencia a Flora y Fauna y Evaluación del Potencial Fitorremediador de Especies Nativas e Introducidas. 2010. Available online: http://repository.javeriana.edu.co/handle/10554/837 (accessed on 19 October 2023).
- Hernández Rodríguez, C.; Gutierrez-Malaxechebarria, A.; Zafra Mejía, C. Reported Lead Levels in Different Environmental Matrices in Colombia. Ing. Univ. 2021, 25, 1–29. [Google Scholar] [CrossRef]
- Samimi, M.; Shahriari-Moghadam, M. Isolation and identification of Delftia lacustris Strain-MS3 as a novel and efficient adsorbent for lead biosorption: Kinetics and thermodynamic studies, optimization of operating variables. Biochem. Eng. J. 2021, 173, 108091. [Google Scholar] [CrossRef]
- Palacios-Torres, Y.; Caballero-Gallardo, K.; Olivero-Verbel, J. Mercury pollution by gold mining in a global biodiversity hotspot, the Choco biogeographic region, Colombia. Chemosphere 2018, 193, 421–430. [Google Scholar] [CrossRef]
- Gutiérrez-Mosquera, H.; Marrugo-Negrete, J.; Díez, S.; Morales-Mira, G.; Montoya-Jaramillo, L.J.; Jonathan, M.P. Distribution of chemical forms of mercury in sediments from abandoned ponds created during former gold mining operations in Colombia. Chemosphere 2020, 258, 127319. [Google Scholar] [CrossRef]
- Córdoba-Tovar, L.; Marrugo-Negrete, J.; Ramos Barón, P.A.; Calao-Ramos, C.R.; Díez, S. Toxic metal(loids) levels in the aquatic environment and nuclear alterations in fish in a tropical river impacted by gold mining. Environ. Res. 2023, 224, 115517. [Google Scholar] [CrossRef]
- Miller, J.H. Experiments in Molecular Genetics; Cold Spring Harbor Laboratory: New York, NY, USA, 1972; Volume xvi, 466p. [Google Scholar]
- Abu-Dieyeh, M.H.; Alduroobi, H.M.; Al-Ghouti, M.A. Potential of mercury-tolerant bacteria for bio-uptake of mercury leached from discarded fluorescent lamps. J. Environ. Manag. 2019, 237, 217–227. [Google Scholar] [CrossRef] [PubMed]
- Ezeobiora, C.; Igbokwe, N.; Hatem, D.; Enwuru, N.; Okpalanwa, C.; Mendie, U. Uncovering the biodiversity and biosynthetic potentials of rare actinomycetes. Future J. Pharm. Sci. 2022, 8, 23. [Google Scholar] [CrossRef]
- Bedoya Vélez, J.M.; Castaño, G.; Ochoa Agudelo, S.; Bedoya Vélez, J.M.; Castaño, G.; Ochoa Agudelo, S. Tolerancia al plomo de aislamientos nativos de Pseudomonas spp. de aguas residuales del Valle de Aburrá. Rev. Colomb. Biotecnol. 2019, 21, 135–143. [Google Scholar]
- Lane, D. 16S/23S rRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematics; Stackebrandt, E., Goodfellow, M., Eds.; Wiley: Chichester, UK, 1991; pp. 115–175. [Google Scholar]
- Weisburg, W.G.; Barns, S.M.; Pelletier, D.A.; Lane, D.J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 1991, 173, 697–703. [Google Scholar] [CrossRef] [PubMed]
- Chakansin, C.; Yostaworakul, J.; Warin, C.; Kulthong, K.; Boonrungsiman, S. Resazurin rapid screening for antibacterial activities of organic and inorganic nanoparticles: Potential, limitations and precautions. Anal. Biochem. 2022, 637, 114449. [Google Scholar] [CrossRef] [PubMed]
- Costa, P.; TPCGomes, A.; Braz, M.; Pereira, C. Application of the Resazurin Cell Viability Assay to Monitor Escherichia coli and Salmonella Typhimurium Inactivation Mediated by Phages. Antibiotics 2021, 10, 974. [Google Scholar] [CrossRef]
- Mohapatra, R.K.; Parhi, P.K.; Pandey, S.; Bindhani, B.K.; Thatoi, H.; Panda, C.R. Active and passive biosorption of Pb(II)using live and dead biomass of marine bacterium Bacillus xiamenensis PbRPSD202: Kinetics and isotherm studies. J. Environ. Manag. 2019, 247, 121–134. [Google Scholar] [CrossRef]
- American Public Health Association, 2017, Standard Methods for the Examination of Water and Wastewater. Available online: https://www.standardmethods.org/ (accessed on 31 August 2023).
- Ameen, F.A.; Hamdan, A.M.; El-Naggar, M.Y. Assessment of the heavy metal bioremediation efficiency of the novel marine lactic acid bacterium, Lactobacillus plantarum MF042018. Sci. Rep. 2020, 10, 314. [Google Scholar] [CrossRef]
- Toledo, A.G.R.; Andrade, J.C.R.; Palmieri, M.C.; Bevilaqua, D.; Sponchiado, S.R.P. Innovative method for encapsulating highly pigmented biomass from Aspergillus nidulans mutant for copper ions removal and recovery. PLoS ONE 2021, 16, e0259315. [Google Scholar]
- Cardona, G.I.; Escobar, M.C.; Acosta-González, A.; Marín, P.; Marqués, S. Highly mercury-resistant strains from different Colombian Amazon ecosystems affected by artisanal gold mining activities. Appl. Microbiol. Biotechnol. 2022, 106, 2775–2793. [Google Scholar] [CrossRef]
- Zar, J.H. Análisis Bioestadístico, 5th ed.; Prentice-Hall/Pearson: Upper Saddle River, NJ, USA, 2010. [Google Scholar]
- Knox, G.A. The Ecology of Seashores; CRS Press: Boca Raton, FL, USA, 2001. [Google Scholar]
- Gupta, P.; Diwan, B. Bacterial Exopolysaccharide mediated heavy metal removal: A Review on biosynthesis, mechanism and remediation strategies. Biotechnol. Rep. 2017, 13, 58–71. [Google Scholar] [CrossRef] [PubMed]
- Oren, A.; Garrity, G.M. Valid publication of the names of forty-two phyla of prokaryotes. Int. J. Syst. Evol. Microbiol. 2021, 71, 005056. [Google Scholar] [CrossRef]
- George, F.; Mahieux, S.; Daniel, C.; Titécat, M.; Beauval, N.; Houcke, I.; Neut, C.; Allorge, D.; Borges, F.; Jan, G.; et al. Assessment of Pb(II), Cd(II), and Al(III) Removal Capacity of Bacteria from Food and Gut Ecological Niches: Insights into Biodiversity to Limit Intestinal Biodisponibility of Toxic Metals. Microorganisms 2021, 9, 456. [Google Scholar] [CrossRef] [PubMed]
- El-Kady, A.A.; Abdel-Wahhab, M.A. Occurrence of trace metals in foodstuffs and their health impact. Trends Food Sci. Technol. 2018, 75, 36–45. [Google Scholar] [CrossRef]
- Cai, J.; Gao, J.; Zhou, Y. CRISPR-mediated genome editing in non-conventional yeasts for biotechnological applications. Microb. Cell Factories 2019, 18, 63. [Google Scholar] [CrossRef] [PubMed]
- Lan, M.M.; Liu, C.; Liu, S.J.; Qiu, R.L.; Tang, Y.T. Phytostabilization of Cd and Pb in Highly Polluted Farmland Soils Using Ramie and Amendments. Int. J. Environ. Res. Public Health 2020, 17, 1661. [Google Scholar] [CrossRef]
- Oh, S.E.; Hassan, S.; Joo, J. Biosorption of Heavy Metals by Lyophilized Cells of Pseudomonas stutzeri. World J. Microbiol. Biotechnol. 2009, 25, 1771–1778. [Google Scholar] [CrossRef]
- Naik, M.M.; Dubey, S.K. Lead resistant bacteria: Lead resistance mechanisms, their applications in lead bioremediation and biomonitoring. Ecotoxicol. Environ. Saf. 2013, 98, 1–7. [Google Scholar] [CrossRef]
- De la Cruz, C.E.B.; Rafael, F.A.M.; Moreno, S.M. G Cadmium (II) and Lead (II) bioadsorption ability by using dead bacterial biomass in aqueous solutions. Theorēma Lima Segunda Época En Línea 2015, 2, 95–106. [Google Scholar]
- Tur-Naranjo, E.; de los Orberá-Ratón T., M.; Romagosa-Álvare, Y.; Pérez-Silva, R.M. Bioadsorción de plomo (II) por biomasa microbiana seca: Efecto del pH. Rev. Cuba. Quím. 2013, 25, 75–81. [Google Scholar]
- Sar, P.; Kazy, S.K.; Asthana, R.K.; Singh, S.P. Metal adsorption and desorption by lyophilized Pseudomonas aeruginosa. Int. Biodeterior. Biodegrad. 1999, 44, 101–110. [Google Scholar] [CrossRef]
- Palacios-Torres, Y.; de la Rosa, J.D.; Olivero-Verbel, J. Trace elements in sediments and fish from Atrato River: An ecosystem with legal rights impacted by gold mining at the Colombian Pacific. Environ. Pollut. 2020, 256, 113290. [Google Scholar] [CrossRef] [PubMed]
- Caicedo-Rivas, G.; Salas-Moreno, M.; Marrugo-Negrete, J. Health Risk Assessment for Human Exposure to Heavy Metals via Food Consumption in Inhabitants of Middle Basin of the Atrato River in the Colombian Pacific. Int. J. Environ. Res. Public Health 2022, 20, 435. [Google Scholar] [CrossRef] [PubMed]
- Feria, J.J.; Marrugo, J.L.; González, H. Heavy metals in Sinú river, department of Córdoba, Colombia, South America. Rev. Fac. Ing. Univ. Antioquia 2010, 5, 35–44. [Google Scholar]
- Decree 1076 of 2015 [with force of law]. Por Medio del Cual se Expide el Código de Recursos Naturales Renovables y de Protección al Medio Ambiente; Bogotá D.C, Colombia. 27 January 1974. D.O. No. 34243. Available online: https://www.funcionpublica.gov.co/eva/gestornormativo/norma.php?i=78153 (accessed on 8 September 2023).
- Xu, W.; Qiao, Y.; Wei, J.; Jiang, Q.; Xue, J. Bacterial Communities and Culturable Petroleum Hydrocarbon Degrading Bacteria in Marine Sediments in the Northeastern South China Sea. Front. Environ. Sci. 2022, 10, 865636. [Google Scholar] [CrossRef]
- Dell’anno, F.; Rastelli, E.; Tangherlini, M.; Corinaldesi, C.; Sansone, C.; Brunet, C.; Balzano, S.; Ianora, A.; Musco, L.; Montereali, M.R.; et al. Highly Contaminated Marine Sediments Can Host Rare Bacterial Taxa Potentially Useful for Bioremediation. Front. Microbiol. 2021, 12, 584850. [Google Scholar] [CrossRef]
- Bubnova, E.N.; Georgieva, M.L.; Grum-Grzhimailo, O.A. Method for Isolation and Enumeration of Fungi Developing in Marine Sediments. Microbiology 2018, 87, 777–782. [Google Scholar] [CrossRef]
- Da Silva, M.; Umbuzeiro, G.A.; Pfenning, L.H.; Canhos, V.P.; Esposito, E. Filamentous Fungi Isolated from Estuarine Sediments Contaminated with Industrial Discharges. Soil. Sediment. Contam. Int. J. 2003, 12, 345–356. [Google Scholar] [CrossRef]
- Bazzicalupo, A. Local adaptation in fungi. FEMS Microbiol. Rev. 2022, 46, fuac026. [Google Scholar] [CrossRef]
- Lai, J.; Cheah, W.; Palaniveloo, K.; Suwa, R.; Sharma, S. A Systematic Review of the Physicochemical and Microbial Diversity of Well-Preserved, Restored, and Disturbed Mangrove Forests: What Is Known and What Is the Way Forward? Forests 2022, 13, 2160. [Google Scholar] [CrossRef]
- Palit, K.; Rath, S.; Chatterjee, S.; Das, S. Microbial diversity and ecological interactions of microorganisms in the mangrove ecosystem: Threats, vulnerability, and adaptations. Environ. Sci. Pollut. Res. 2022, 29, 32467–32512. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Zhang, H.; Ma, Y.; Zhou, J.; Zhang, Y. Bio-removal of cadmium by growing deep-sea bacterium Pseudoalteromonas sp. SCSE709-6. Extremophiles 2013, 17, 723–731. [Google Scholar] [CrossRef] [PubMed]
- Kalkan, S. Heavy metal resistance of marine bacteria on the sediments of the Black Sea. Mar. Pollut. Bull. 2022, 179, 113652. [Google Scholar] [CrossRef] [PubMed]
- Gillard, B.; Chatzievangelou, D.; Thomsen, L.; Ullrich, M.S. Heavy-Metal-Resistant Microorganisms in Deep-Sea Sediments Disturbed by Mining Activity: An Application Toward the Development of Experimental in vitro Systems. Front. Mar. Sci. 2019, 6, 462. [Google Scholar] [CrossRef]
- Ma, B.; Song, W.; Zhang, X.; Chen, M.; Li, J.; Yang, X.; Zhang, L. Potential application of novel cadmium-tolerant bacteria in bioremediation of Cd-contaminated soil. Ecotoxicol. Environ. Saf. 2023, 255, 114766. [Google Scholar] [CrossRef]
- Chakraborty, J.; Das, S. Characterization and cadmium-resistant gene expression of biofilm-forming marine bacterium Pseudomonas aeruginosa JP-11. Environ. Sci. Pollut. Res. Int. 2014, 21, 14188–14201. [Google Scholar] [CrossRef]
- Touahir, N.; Alouache, S.; Dehane, D. Assessment and characterization of heavy metals resistance bacteria isolated in Southwestern Mediterranean coastal waters (Bou-Ismail Bay): Impacts of anthropogenic activities. Mar. Pollut. Bull. 2023, 192, 115085. [Google Scholar] [CrossRef]
- Pal, A.; Paul, A.K. Microbial extracellular polymeric substances: Central elements in heavy metal bioremediation. Indian. J. Microbiol. 2008, 48, 49–64. [Google Scholar] [CrossRef]
- Morillo, J.A.; Aguilera, M.; Ramos-Cormenzana, A.; Monteoliva-Sánchez, M. Production of a metal-binding exopolysaccharide by Paenibacillus jamilae using two-phase olive-mill waste as fermentation substrate. Curr. Microbiol. 2006, 53, 189–193. [Google Scholar] [CrossRef]
- Franco, N.R.; Giraldo, M.; López-Alvarez, D.; Gallo-Franco, J.J.; Dueñas, L.F.; Puentes, V.; Castillo, A. Bacterial Composition and Diversity in Deep-Sea Sediments from the Southern Colombian Caribbean Sea. Diversity 2021, 13, 10. [Google Scholar] [CrossRef]
- Blandón, L.M.; Marín, M.A.; Quintero, M.; Jutinico-Shubach, L.M.; Montoya-Giraldo, M.; Santos-Acevedo, M.; Gómez-León, J. Diversity of cultivable bacteria from deep-sea sediments of the Colombian Caribbean and their potential in bioremediation. Antonie Van Leeuwenhoek. 2022, 115, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Edoamodu, C.E.; Nwodo, U.U. Marine sediment derived bacteria Enterobacter asburiae ES1 and Enterobacter sp. Kamsi produce laccase with high dephenolisation potentials. Prep. Biochem. Biotechnol. 2022, 52, 748–761. [Google Scholar] [CrossRef] [PubMed]
- Lo Giudice, A.; Poli, A.; Finore, I.; Rizzo, C. Peculiarities of extracellular polymeric substances produced by Antarctic bacteria and their possible applications. Appl. Microbiol. Biotechnol. 2020, 104, 2923–2934. [Google Scholar] [CrossRef] [PubMed]
- Pages, D.; Rose, J.; Conrod, S.; Cuine, S.; Carrier, P.; Heulin, T.; Achouak, W. Heavy Metal Tolerance in Stenotrophomonas maltophilia. PLoS ONE 2023, 3, 1539. [Google Scholar] [CrossRef]
- Bautista-Hernández, D.A.; Ramírez-Burgos, L.I.; Duran-Páramo, E.; Fernández-Linares, L. Zinc and Lead Biosorption by Delftia tsuruhatensis: A Bacterial Strain Resistant to Metals Isolated from Mine Tailings. J. Water Resour. Prot. 2012, 4, 207–216. [Google Scholar] [CrossRef]
- Shamim, S. Chapter 2 Biosorption of Heavy Metals. In Biosorption; Derco, J., Vrana, B., Eds.; IntechOpen: Rijeka, Croacia, 2018; Volume 2, pp. 21–49. ISBN 978-1-78923-473-2. [Google Scholar]



| Locality | Sampling Site | Sampling Site Abbreviation | Latitude | Longitude | Depth (m) | Temperature (°C) | Salinity (ppt) | pH |
|---|---|---|---|---|---|---|---|---|
| Atrato river mouth | Margarita Atrato river mouth | MARM | N 8°8′47.052″ | W 76°50′8.337″ | 2.3 | 27.7 | 0.2 | 6.2 |
| Mangrove Paila Atrato river | MPAR | N 8°1′51″ | W 76°50′51″ | 1.1 | 28.0 | 7.5 | 6.5 | |
| Mangrove Roto Atrato river | MRAR | N 8°12′42.351″ | W 76°56′33.552″ | 3.2 | 27.6 | 0.0 | 6.2 | |
| Baudó river mouth | Mangrove via Usaraga Baudó river | MVUBR | N 4°56′38.4″ | W 77°21′48.6″ | 0.5 | 27.1 | 7.2 | 6.2 |
| Out to sea Baudó river | OBR | N 4°56′19.9″ | W 77°22′55.1″ | 7.6 | 29.7 | 6.5 | 6.4 | |
| San Juan river mouth | Mangrove Choncho San Juan river two | MCHSJ2 | N 4°2′56.2″ | W 77°27′03.1″ | 4.0 | 25.9 | 0.3 | 6.3 |
| Mangrove San Juan river two | MSJ2 | N 4°2′34.5″ | W 77°26′05.1″ | 4.0 | 26.2 | 3.1 | 6.4 | |
| Beach San Juan river mouth two | BSJM2 | N 4°1′31.2″ | W 77°26′24.1″ | 4.5 | 25.6 | 1.5 | 6.9 |
| Wavenumber (cm−1) | Group Functional | Wavenumber (cm−1) after Lead Removal | References |
|---|---|---|---|
| 3433 and 3294 | Hydroxyl groups | 3263–3140 | [54,55,56,57] |
| 1080 | Phosphate | 1118 | |
| 1780–1640 | (C=O) of amides | 1658–1539 | |
| 1543 | Carboxylate (COO-) | 1519 | |
| 1238 | SO3 | 1045 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alvarado-Campo, K.L.; Quintero, M.; Cuadrado-Cano, B.; Montoya-Giraldo, M.; Otero-Tejada, E.L.; Blandón, L.; Sánchez, O.; Zuleta-Correa, A.; Gómez-León, J. Heavy Metal Tolerance of Microorganisms Isolated from Coastal Marine Sediments and Their Lead Removal Potential. Microorganisms 2023, 11, 2708. https://doi.org/10.3390/microorganisms11112708
Alvarado-Campo KL, Quintero M, Cuadrado-Cano B, Montoya-Giraldo M, Otero-Tejada EL, Blandón L, Sánchez O, Zuleta-Correa A, Gómez-León J. Heavy Metal Tolerance of Microorganisms Isolated from Coastal Marine Sediments and Their Lead Removal Potential. Microorganisms. 2023; 11(11):2708. https://doi.org/10.3390/microorganisms11112708
Chicago/Turabian StyleAlvarado-Campo, Katleen L., Marynes Quintero, Bernarda Cuadrado-Cano, Manuela Montoya-Giraldo, Elver Luis Otero-Tejada, Lina Blandón, Olga Sánchez, Ana Zuleta-Correa, and Javier Gómez-León. 2023. "Heavy Metal Tolerance of Microorganisms Isolated from Coastal Marine Sediments and Their Lead Removal Potential" Microorganisms 11, no. 11: 2708. https://doi.org/10.3390/microorganisms11112708
APA StyleAlvarado-Campo, K. L., Quintero, M., Cuadrado-Cano, B., Montoya-Giraldo, M., Otero-Tejada, E. L., Blandón, L., Sánchez, O., Zuleta-Correa, A., & Gómez-León, J. (2023). Heavy Metal Tolerance of Microorganisms Isolated from Coastal Marine Sediments and Their Lead Removal Potential. Microorganisms, 11(11), 2708. https://doi.org/10.3390/microorganisms11112708









