Profile of Pancreatic and Ileal Microbiota in Experimental Acute Pancreatitis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice Experimental Design
2.2. Histopathology
2.3. Serum Biochemistry
2.4. Western Blotting
2.5. DNA Extraction PCR Amplification and Sequencing of Bacteria in Pancreas and Ileum
2.6. Bioinformatics Analysis of Sequencing Data
2.7. Statistical Analysis
3. Results
3.1. Caerulein, Caerulein+LPS, and L-Arginine Induced AP in Mice
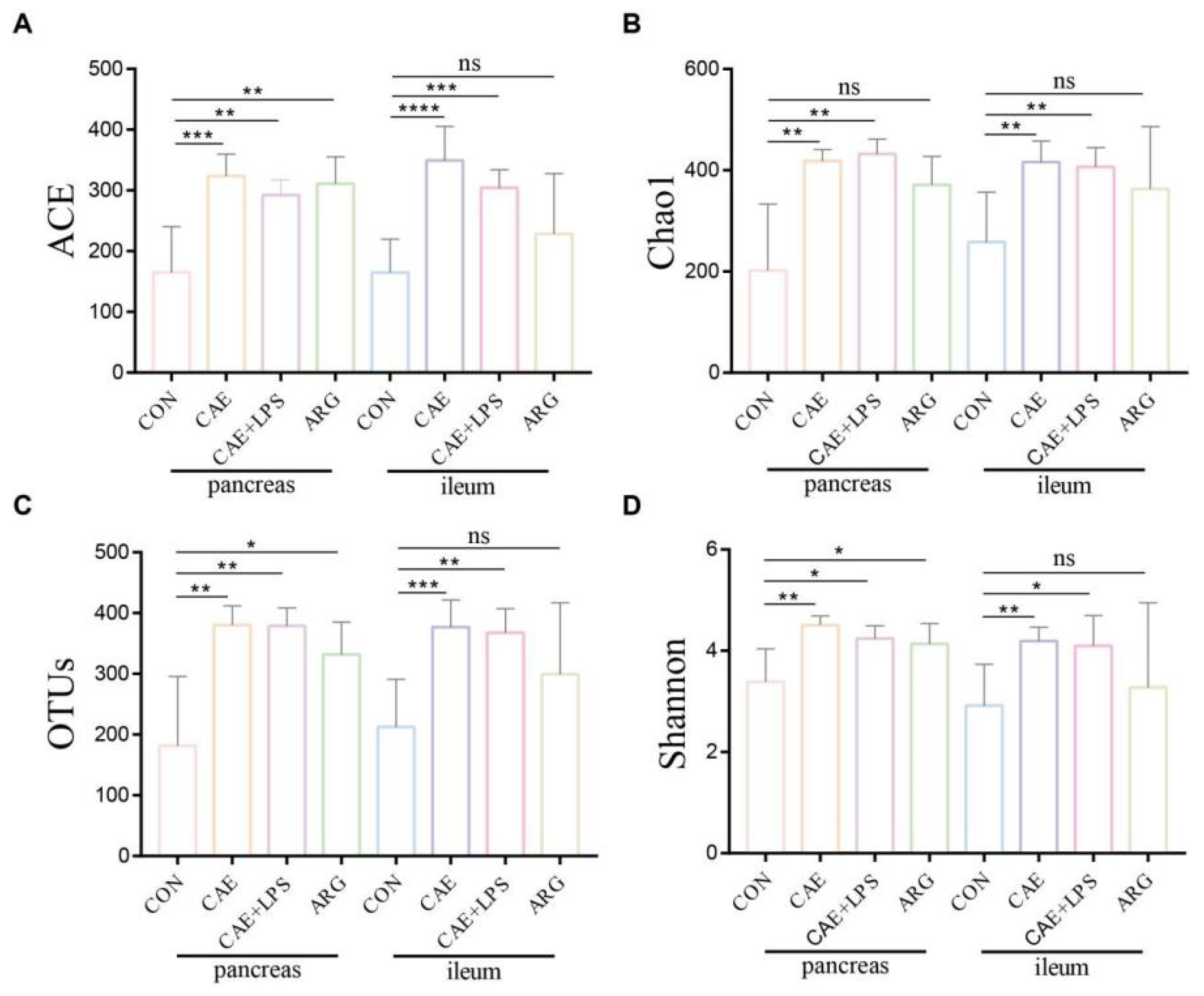
3.2. AP Mice Had a More Abundant Microbiota in the Pancreas and Ileum
3.3. AP Changed Pancreatic Microbiota
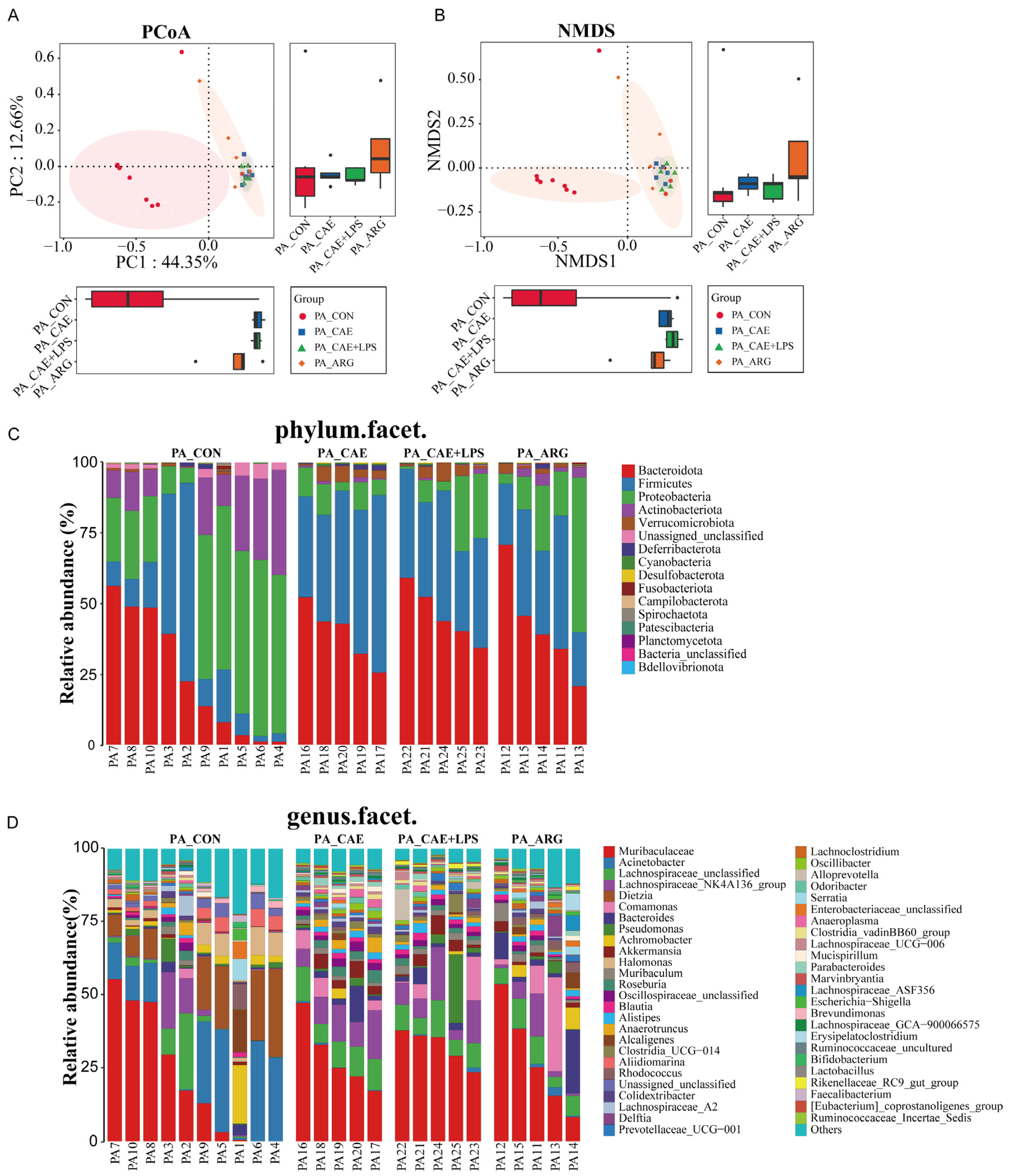
3.3.1. Differences in Microbiome Compositions in the Pancreas
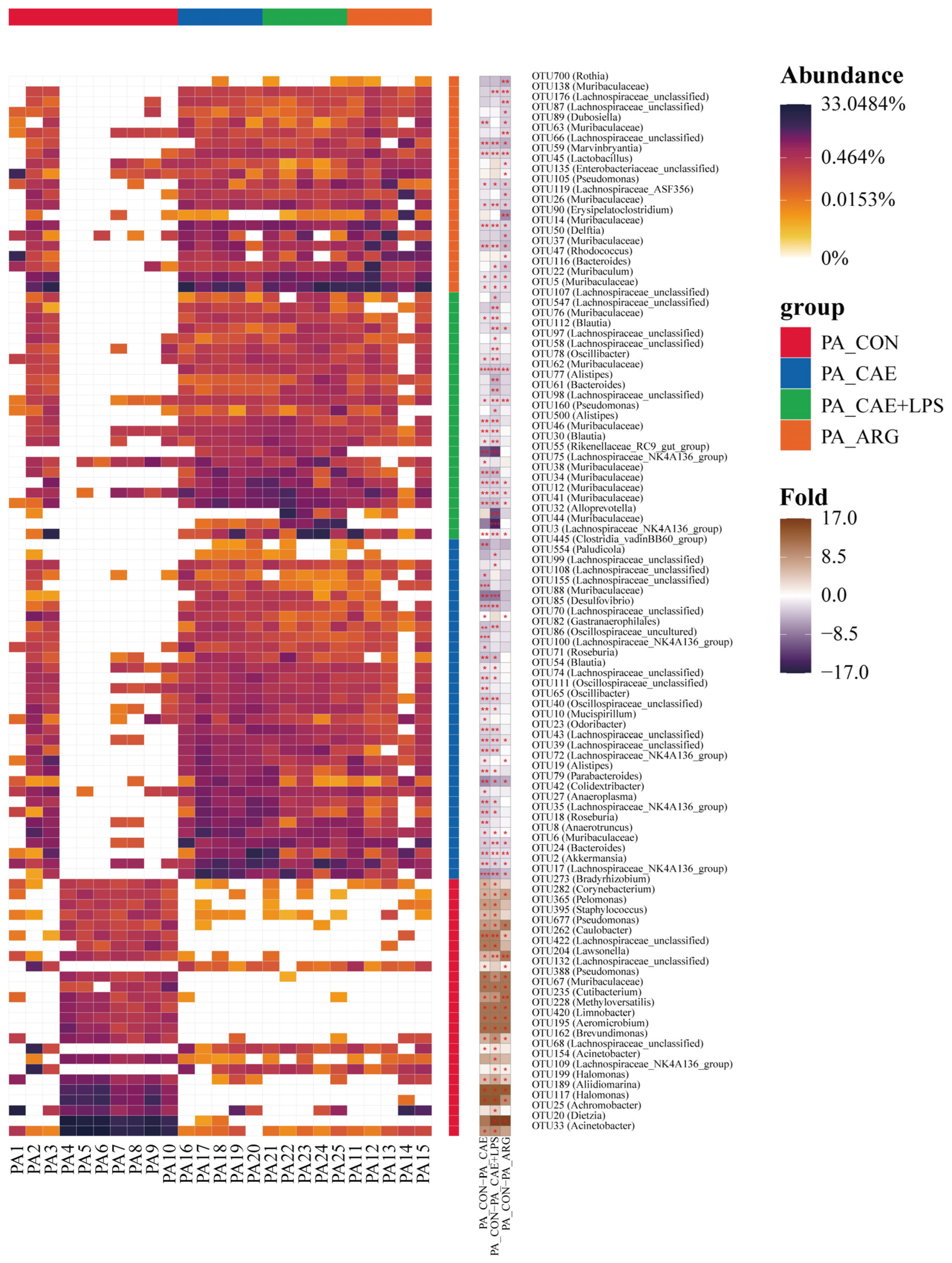
3.3.2. Differences in the Microbiota of Three AP Groups in the Pancreas
3.3.3. Functional Changes of Three AP Groups in the Pancreas
3.4. AP Changed Ileal Microbiota
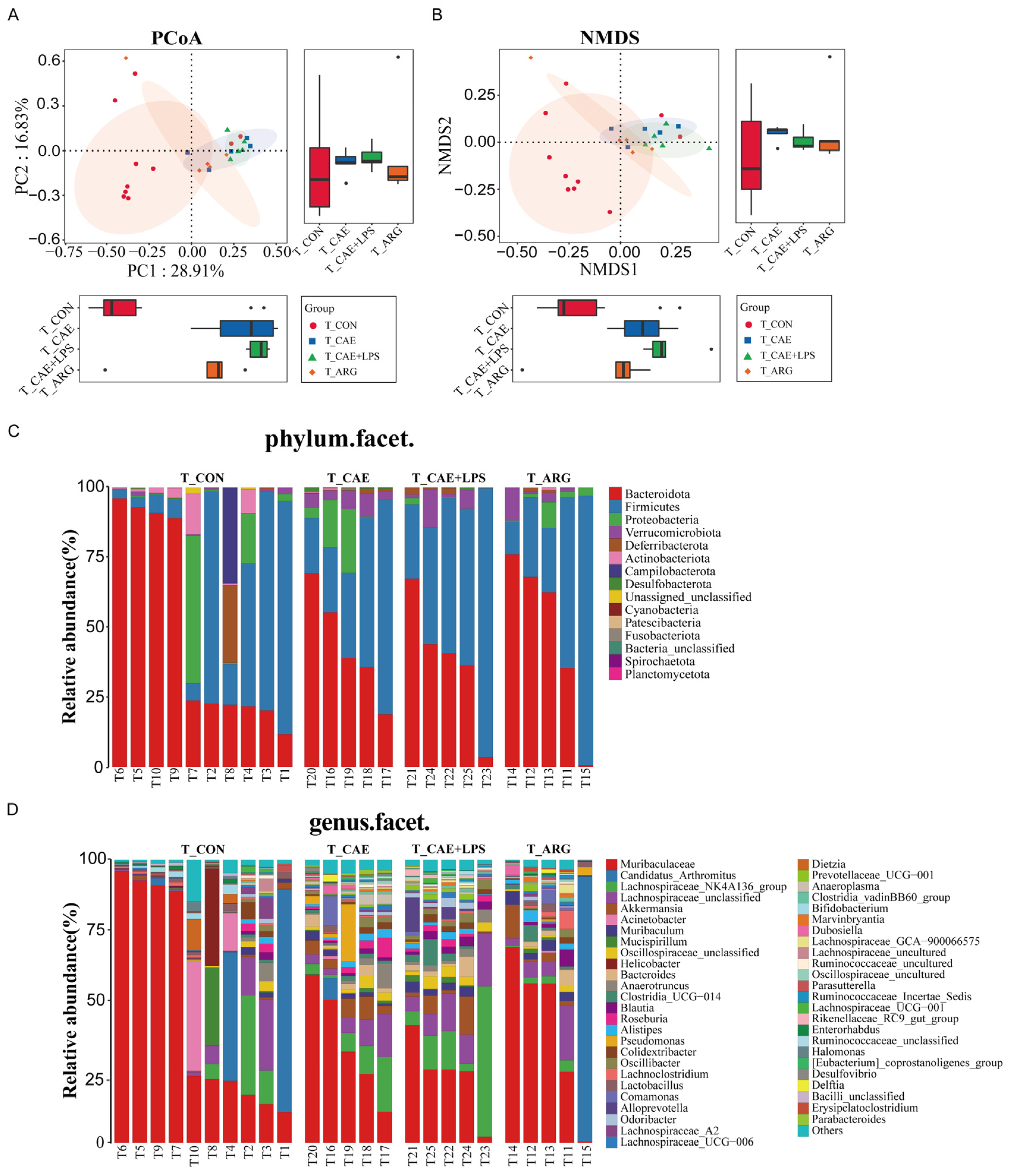
3.4.1. Differences in Microbiome Compositions in the Ileum
3.4.2. Differences in the Microbiota of Three AP Groups in the Ileum
3.4.3. Functional Changes of Three AP Groups in the Ileum
3.5. The Three AP Mice Models Had Distinct Pancreatic and Ileal Microbiota
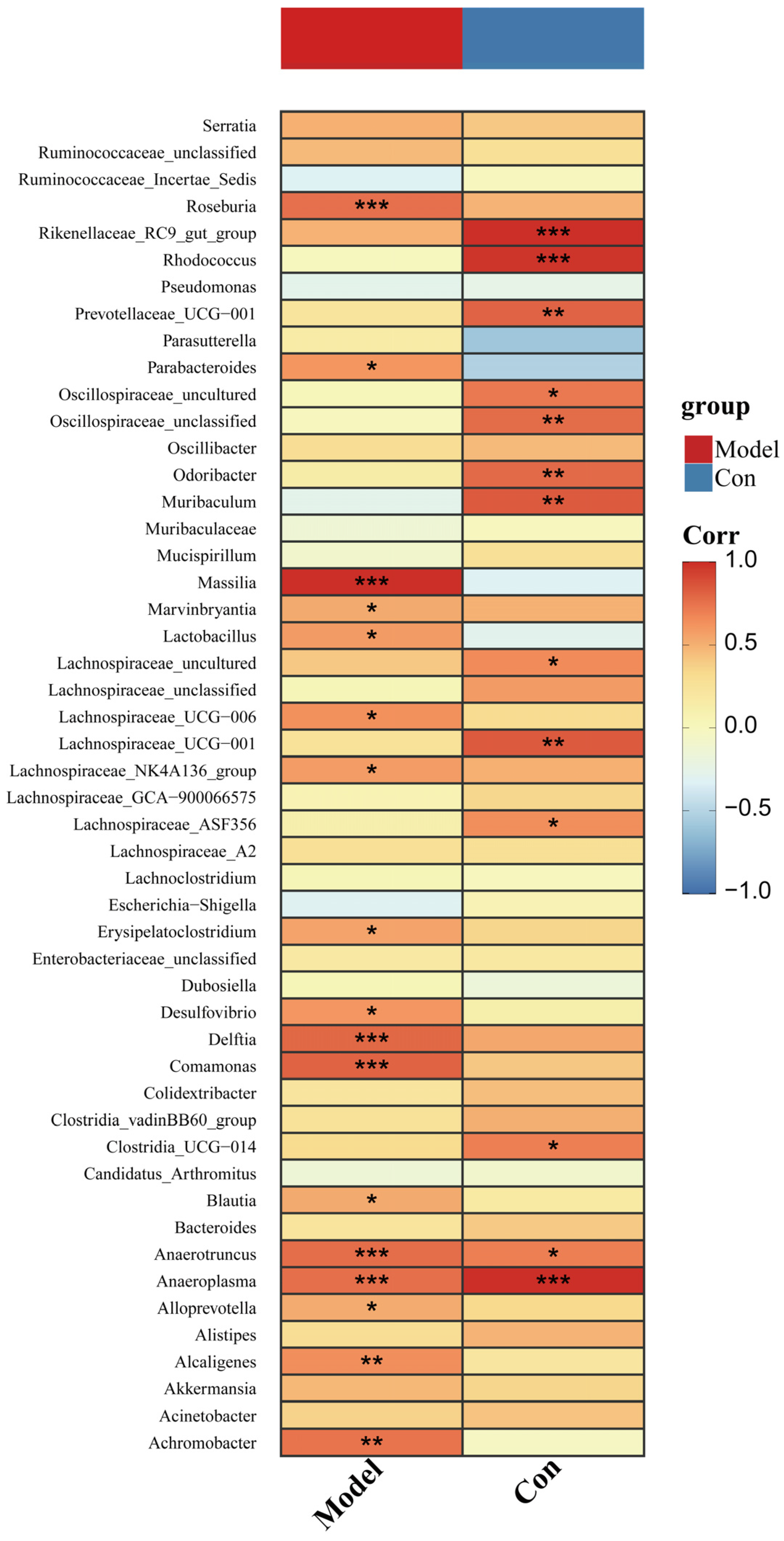
3.6. Changed Pancreatic Microbiota Was Associated with the Ileal Microbiota
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sendler, M.; Maertin, S.; John, D.; Persike, M.; Weiss, F.U.; Krüger, B.; Wartmann, T.; Wagh, P.; Halangk, W.; Schaschke, N.; et al. Cathepsin B Activity Initiates Apoptosis via Digestive Protease Activation in Pancreatic Acinar Cells and Experimental Pancreatitis. J. Biol. Chem. 2016, 291, 14717–14731. [Google Scholar] [CrossRef] [PubMed]
- Banks, P.A.; Bollen, T.L.; Dervenis, C.; Gooszen, H.G.; Johnson, C.D.; Sarr, M.G.; Tsiotos, G.G.; Vege, S.S. Classification of acute pancreatitis—2012: Revision of the Atlanta classification and definitions by international consensus. Gut 2013, 62, 102–111. [Google Scholar] [CrossRef] [PubMed]
- Capurso, G.; Zerboni, G.; Signoretti, M.; Valente, R.; Stigliano, S.; Piciucchi, M.; Delle, F.G. Role of the gut barrier in acute pancreatitis. J. Clin. Gastroenterol. 2012, 46, S46–S51. [Google Scholar] [CrossRef]
- Ojima, M.; Motooka, D.; Shimizu, K.; Gotoh, K.; Shintani, A.; Yoshiya, K.; Nakamura, S.; Ogura, H.; Iida, T.; Shimazu, T. Metagenomic Analysis Reveals Dynamic Changes of Whole Gut Microbiota in the Acute Phase of Intensive Care Unit Patients. Dig. Dis. Sci. 2016, 61, 1628–1634. [Google Scholar] [CrossRef]
- Tan, C.; Ling, Z.; Huang, Y.; Cao, Y.; Liu, Q.; Cai, T.; Yuan, H.; Liu, C.; Li, Y.; Xu, K. Dysbiosis of Intestinal Microbiota Associated with Inflammation Involved in the Progression of Acute Pancreatitis. Pancreas 2015, 44, 868–875. [Google Scholar] [CrossRef]
- Zhu, Y.; He, C.; Li, X.; Cai, Y.; Hu, J.; Liao, Y.; Zhao, J.; Xia, L.; He, W.; Liu, L.; et al. Gut microbiota dysbiosis worsens the severity of acute pancreatitis in patients and mice. J. Gastroenterol. 2019, 54, 347–358. [Google Scholar] [CrossRef]
- Fritz, S.; Hackert, T.; Hartwig, W.; Rossmanith, F.; Strobel, O.; Schneider, L.; Will-Schweiger, K.; Kommerell, M.; Büchler, M.W.; Werner, J. Bacterial translocation and infected pancreatic necrosis in acute necrotizing pancreatitis derives from small bowel rather than from colon. Am. J. Surg. 2010, 200, 111–117. [Google Scholar] [CrossRef]
- Van Minnen, L.P.; Timmerman, H.M.; Lutgendorff, F.; Verheem, A.; Harmsen, W.; Konstantinov, S.R.; Smidt, H.; Visser, M.R.; Rijkers, G.T.; Gooszen, H.G.; et al. Modification of intestinal microbiota with multispecies probiotics reduces bacterial translocation and improves clinical course in a rat model of acute pancreatitis. Surgery 2007, 141, 470–480. [Google Scholar] [CrossRef]
- Bedirli, A.; Gokahmetoglu, S.; Sakrak, O.; Soyuer, I.; Ince, O.; Sozuer, E. Beneficial effects of recombinant platelet-activating factor acetylhydrolase and BN 52021 on bacterial translocation in cerulein-induced pancreatitis. Eur. Surg. Res. 2004, 36, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, B.; Wu, J.; Wang, G. Beneficial effects of growth hormone on bacterial translocation during the course of acute necrotizing pancreatitis in rats. Pancreas 2001, 23, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Das, N.K.; Schwartz, A.J.; Barthel, G.; Inohara, N.; Liu, Q.; Sankar, A.; Hill, D.R.; Ma, X.; Lamberg, O.; Schnizlein, M.K.; et al. Microbial Metabolite Signaling Is Required for Systemic Iron Homeostasis. Cell Metab. 2020, 31, 115–130. [Google Scholar] [CrossRef]
- Tang, D.; Chen, X.; Kang, R.; Kroemer, G. Ferroptosis: Molecular mechanisms and health implications. Cell Res. 2021, 31, 107–125. [Google Scholar] [CrossRef]
- Han, X.; Li, B.; Ye, X.; Mulatibieke, T.; Wu, J.; Dai, J.; Wu, D.; Ni, J.; Zhang, R.; Xue, J.; et al. Dopamine D2 receptor signalling controls inflammation in acute pancreatitis via a PP2A-dependent Akt/NF-kappaB signalling pathway. Br. J. Pharmacol. 2017, 174, 4751–4770. [Google Scholar] [CrossRef]
- Schmidt, J.; Rattner, D.W.; Lewandrowski, K.; Compton, C.C.; Mandavilli, U.; Knoefel, W.T.; Warshaw, A.L. A better model of acute pancreatitis for evaluating therapy. Ann. Surg. 1992, 215, 44–56. [Google Scholar] [CrossRef]
- Chiu, C.J.; McArdle, A.H.; Brown, R.; Scott, H.J.; Gurd, F.N. Intestinal mucosal lesion in low-flow states. I. A morphological, hemodynamic, and metabolic reappraisal. Arch. Surg. 1970, 101, 478–483. [Google Scholar] [CrossRef] [PubMed]
- Tan, B.; Chen, M.J.; Guo, Q.; Tang, H.; Li, Y.; Jia, X.M.; Xu, Y.; Zhu, L.; Wang, M.Z.; Qian, J.M. Clinical-radiological characteristics and intestinal microbiota in patients with pancreatic immune-related adverse events. Thorac. Cancer 2021, 12, 1814–1823. [Google Scholar] [CrossRef]
- Huang, C.; Chen, J.; Wang, J.; Zhou, H.; Lu, Y.; Lou, L.; Zheng, J.; Tian, L.; Wang, X.; Cao, Z.; et al. Dysbiosis of Intestinal Microbiota and Decreased Antimicrobial Peptide Level in Paneth Cells during Hypertriglyceridemia-Related Acute Necrotizing Pancreatitis in Rats. Front. Microbiol. 2017, 8, 776. [Google Scholar] [CrossRef]
- Chen, J.; Huang, C.; Wang, J.; Zhou, H.; Lu, Y.; Lou, L.; Zheng, J.; Tian, L.; Wang, X.; Cao, Z.; et al. Dysbiosis of intestinal microbiota and decrease in paneth cell antimicrobial peptide level during acute necrotizing pancreatitis in rats. PLoS ONE 2017, 12, e0176583. [Google Scholar] [CrossRef] [PubMed]
- Smith, B.J.; Miller, R.A.; Schmidt, T.M. Muribaculaceae Genomes Assembled from Metagenomes Suggest Genetic Drivers of Differential Response to Acarbose Treatment in Mice. mSphere 2021, 6, e0085121. [Google Scholar] [CrossRef] [PubMed]
- Liao, J.; Li, Q.; Lei, C.; Yu, W.; Deng, J.; Guo, J.; Han, Q.; Hu, L.; Li, Y.; Pan, J.; et al. Toxic effects of copper on the jejunum and colon of pigs: Mechanisms related to gut barrier dysfunction and inflammation influenced by the gut microbiota. Food Funct. 2021, 12, 9642–9657. [Google Scholar] [CrossRef] [PubMed]
- Wong, E.O.; Brownlie, E.J.E.; Ng, K.M.; Kathirgamanathan, S.; Yu, F.B.; Merrill, B.D.; Huang, K.C.; Martin, A.; Tropini, C.; Navarre, W.W. The CIAMIB: A Large and Metabolically Diverse Collection of Inflammation-Associated Bacteria from the Murine Gut. mBio 2022, 13, e0294921. [Google Scholar] [CrossRef]
- Chen, J.; Ding, X.; Wu, R.; Tong, B.; Zhao, L.; Lv, H.; Meng, X.; Liu, Y.; Ren, B.; Li, J.; et al. Novel Sesquiterpene Glycoside from Loquat Leaf Alleviates Type 2 Diabetes Mellitus Combined with Nonalcoholic Fatty Liver Disease by Improving Insulin Resistance, Oxidative Stress, Inflammation, and Gut Microbiota Composition. J. Agric. Food Chem. 2021, 69, 14176–14191. [Google Scholar] [CrossRef] [PubMed]
- Valle, M.; Mitchell, P.L.; Pilon, G.; Varin, T.; Hénault, L.; Rolin, J.; McLeod, R.; Gill, T.; Richard, D.; Vohl, M.C.; et al. Salmon peptides limit obesity-associated metabolic disorders by modulating a gut-liver axis in vitamin D-deficient mice. Obes. Silver Spring 2021, 29, 1635–1649. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Li, M.; Wang, Y.; Wu, X.; Shen, J.; Xiao, Z.; Zhao, Y.; Du, F.; Chen, Y.; Wu, Z.; et al. Excessive Intake of Longan Arillus Alters gut Homeostasis and Aggravates Colitis in Mice. Front. Pharmacol. 2021, 12, 640417. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Wang, Z.; Fan, B.; Wang, L.; Liu, M.; An, Z.; Zhao, X. A comparative study of the effects of different fucoidans on cefoperazone-induced gut microbiota disturbance and intestinal inflammation. Food Funct. 2021, 12, 9087–9097. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Wang, N.; Tan, H.Y.; Li, S.; Zhang, C.; Feng, Y. Function of Akkermansia muciniphila in Obesity: Interactions with Lipid Metabolism, Immune Response and Gut Systems. Front. Microbiol. 2020, 11, 219. [Google Scholar] [CrossRef]
- Zhai, Q.; Feng, S.; Arjan, N.; Chen, W. A next generation probiotic, Akkermansia muciniphila. Crit. Rev. Food Sci. Nutr. 2019, 59, 3227–3236. [Google Scholar] [CrossRef]
- Wang, K.; Liao, M.; Zhou, N.; Bao, L.; Ma, K.; Zheng, Z.; Wang, Y.; Liu, C.; Wang, W.; Wang, J.; et al. Parabacteroides distasonis Alleviates Obesity and Metabolic Dysfunctions via Production of Succinate and Secondary Bile Acids. Cell Rep. 2019, 26, 222–235.e5. [Google Scholar] [CrossRef]
- Lei, Y.; Tang, L.; Liu, S.; Hu, S.; Wu, L.; Liu, Y.; Yang, M.; Huang, S.; Tang, X.; Tang, T.; et al. Parabacteroides produces acetate to alleviate heparanase-exacerbated acute pancreatitis through reducing neutrophil infiltration. Microbiome 2021, 9, 115. [Google Scholar] [CrossRef] [PubMed]
- Nasri Nasrabadi, M.R.; Razavi, S.H. Use of response surface methodology in a fed-batch process for optimization of tricarboxylic acid cycle intermediates to achieve high levels of canthaxanthin from Dietzia natronolimnaea HS-1. J. Biosci. Bioeng. 2010, 109, 361–368. [Google Scholar] [CrossRef]
- Nie, Y.; Liang, J.; Fang, H.; Tang, Y.Q.; Wu, X.L. Two novel alkane hydroxylase-rubredoxin fusion genes isolated from a Dietzia bacterium and the functions of fused rubredoxin domains in long-chain n-alkane degradation. Appl. Environ. Microbiol. 2011, 77, 7279–7288. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Xu, J.B.; Nie, Y.; Wu, X.L. Pan-genomic analysis reveals that the evolution of Dietzia species depends on their living habitats. Environ. Microbiol. 2021, 23, 861–877. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Xu, H.; Chen, Y.; Wu, J.; Jin, F.; Wu, Q.; Yao, X.M. Therapeutic effect of Bifidobacterium combined with early enteral nutrition in the treatment of severe acute pancreatitis: A pilot study. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 4018–4024. [Google Scholar]
- Liu, Y.; Liu, H.; Rong, Y.; Shi, Q.; Yang, Q.; Li, H.; Zhang, Z.; Tao, J. Alterations of oral microbiota are associated with the development and severity of acute pancreatitis. J. Oral Microbiol. 2023, 15, 2264619. [Google Scholar] [CrossRef] [PubMed]
- Farrell, J.J.; Zhang, L.; Zhou, H.; Chia, D.; Elashoff, D.; Akin, D.; Paster, B.J.; Joshipura, K.; Wong, D.T. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 2012, 61, 582–588. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Ni, H.M.; Chao, X.; Wang, H.; Bridges, B.; Kumer, S.; Schmitt, T.; Mareninova, O.; Gukovskaya, A.; De Lisle, R.C.; et al. Impaired TFEB-mediated lysosomal biogenesis promotes the development of pancreatitis in mice and is associated with human pancreatitis. Autophagy 2019, 15, 1954–1969. [Google Scholar] [CrossRef]
- Zhang, X.; Jin, T.; Shi, N.; Yao, L.; Yang, X.; Han, C.; Wen, L.; Du, D.; Szatmary, P.; Mukherjee, R.; et al. Mechanisms of Pancreatic Injury Induced by Basic Amino Acids Differ Between L-Arginine, L-Ornithine, and L-Histidine. Front. Physiol. 2018, 9, 1922. [Google Scholar] [CrossRef]
- Nie, K.; Ma, K.; Luo, W.; Shen, Z.; Yang, Z.; Xiao, M.; Tong, T.; Yang, Y.; Wang, X. Roseburia intestinalis: A Beneficial Gut Organism from the Discoveries in Genus and Species. Front. Cell Infect. Microbiol. 2021, 11, 757718. [Google Scholar]
- McKee, L.S.; La Rosa, S.L.; Westereng, B.; Eijsink, V.G.; Pope, P.B.; Larsbrink, J. Polysaccharide degradation by the Bacteroidetes: Mechanisms and nomenclature. Environ. Microbiol. Rep. 2021, 13, 559–581. [Google Scholar] [CrossRef]
- Zheng, J.; Lou, L.; Fan, J.; Huang, C.; Mei, Q.; Wu, J.; Guo, Y.; Lu, Y.; Wang, X.; Zeng, Y. Commensal Escherichia coli Aggravates Acute Necrotizing Pancreatitis through Targeting of Intestinal Epithelial Cells. Appl. Environ. Microbiol. 2019, 85, e00059-19. [Google Scholar] [CrossRef]



| OTU | Rho | p_Value | |
|---|---|---|---|
| 1 | OTU44 (Muribaculaceae) | 0.934755574 | <0.001 |
| 2 | OTU117 (Halomonas) | 0.903396145 | <0.001 |
| 3 | OTU27 (Anaeroplasma) | 0.894354839 | <0.001 |
| 4 | OTU8 (Anaerotruncus) | 0.890898251 | <0.001 |
| 5 | OTU67 (Muribaculaceae) | 0.884818826 | <0.001 |
| 6 | OTU3 (Lachnospiraceae_NK4A136_group) | 0.88152809 | <0.001 |
| 7 | OTU55 (Rikenellaceae_RC9_gut_group) | 0.875320249 | <0.001 |
| 8 | OTU41 (Muribaculaceae) | 0.870283019 | <0.001 |
| 9 | OTU35(Lachnospiraceae_NK4A136_group) | 0.845194922 | <0.001 |
| 10 | OTU20 (Dietzia) | 0.831554236 | <0.001 |
| 11 | OTU189 (Aliidiomarina) | 0.825593033 | <0.001 |
| 12 | OTU50 (Delftia) | 0.809621838 | <0.001 |
| 13 | OTU43 (Lachnospiraceae_unclassified) | 0.804835266 | <0.001 |
| 14 | OTU88 (Muribaculaceae) | 0.802774593 | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, M.; Cui, M.; Jiang, Q.; Wang, J.; Lu, Y. Profile of Pancreatic and Ileal Microbiota in Experimental Acute Pancreatitis. Microorganisms 2023, 11, 2707. https://doi.org/10.3390/microorganisms11112707
Zhao M, Cui M, Jiang Q, Wang J, Lu Y. Profile of Pancreatic and Ileal Microbiota in Experimental Acute Pancreatitis. Microorganisms. 2023; 11(11):2707. https://doi.org/10.3390/microorganisms11112707
Chicago/Turabian StyleZhao, Mengqi, Mengyan Cui, Qiaoli Jiang, Jingjing Wang, and Yingying Lu. 2023. "Profile of Pancreatic and Ileal Microbiota in Experimental Acute Pancreatitis" Microorganisms 11, no. 11: 2707. https://doi.org/10.3390/microorganisms11112707
APA StyleZhao, M., Cui, M., Jiang, Q., Wang, J., & Lu, Y. (2023). Profile of Pancreatic and Ileal Microbiota in Experimental Acute Pancreatitis. Microorganisms, 11(11), 2707. https://doi.org/10.3390/microorganisms11112707





