Exploring the Association between Gut Microbiota and Inflammatory Skin Diseases: A Two-Sample Mendelian Randomization Analysis
Abstract
1. Introduction
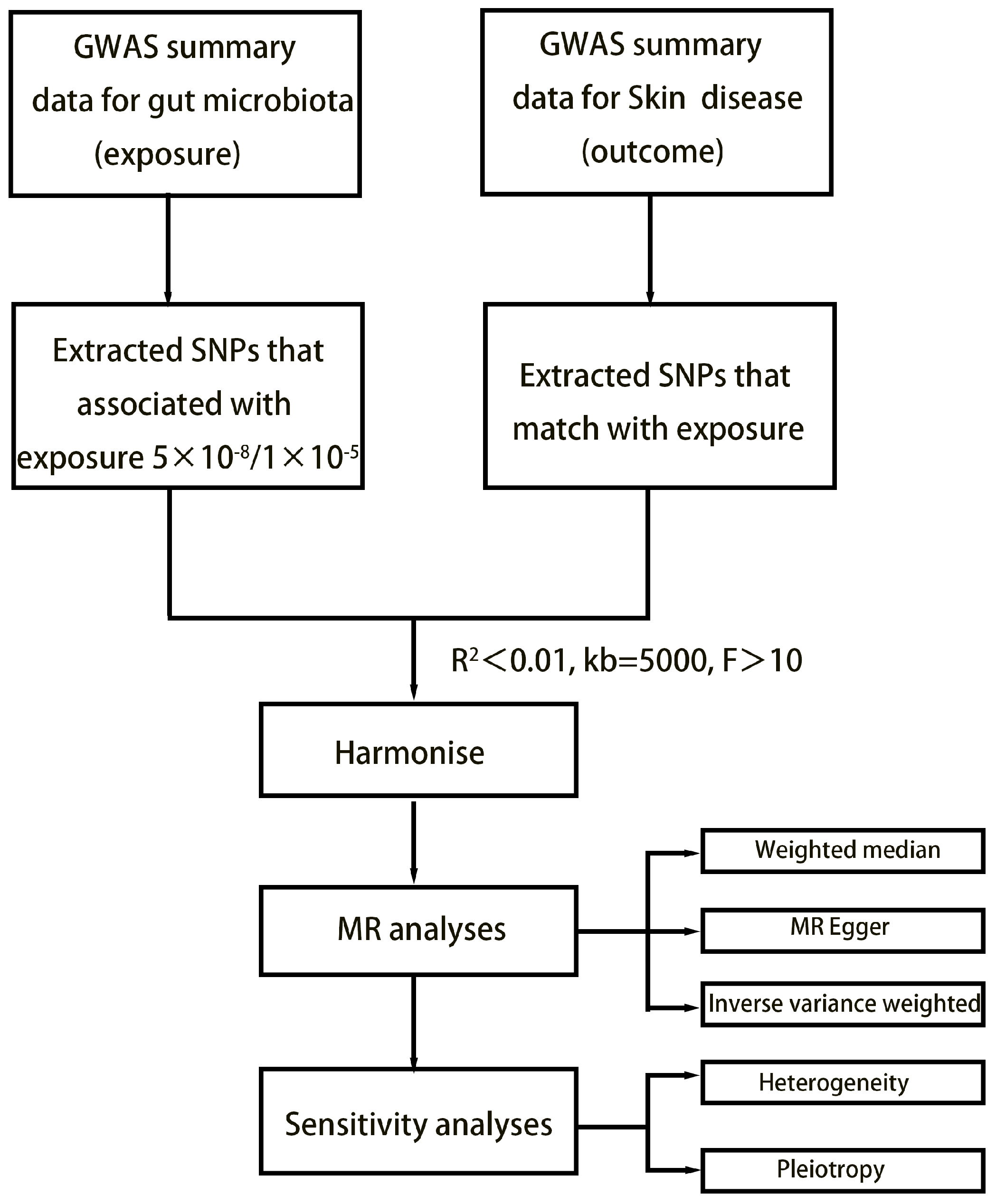
2. Method
2.1. Exposure Data
2.2. Outcome Data
2.3. Selection of Instrumental Variables
2.4. Statistical Analysis
3. Results
3.1. SNP Selection
3.2. Results of the TSMR Analysis (Genome-Wide Statistical Significance Threshold, p < 5 × 10−8)
3.3. Results of the TSMR Analysis (Gene Locus Range Significance Levels, p < 1 × 10−5)
3.4. Sensitivity Analyses
3.5. Reverse TSMR Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| GWAS | Genome-wide association study |
| MR | Mendelian randomization |
| GSA | Gut–skin axis |
| SCFAs | Short-chain fatty acids |
| IVs | Instrumental variables |
| SNP | Single-nucleotide polymorphism |
| IVW | Inverse variance weighted |
References
- Karimkhani, C.; Dellavalle, R.P.; Coffeng, L.E.; Flohr, C.; Hay, R.J.; Langan, S.M.; Naghavi, M. Global Skin Disease Morbidity and Mortality: An Update from the Global Burden of Disease Study 2013. JAMA Dermatol. 2017, 153, 406–412. [Google Scholar] [CrossRef] [PubMed]
- Salem, I.; Ramser, A.; Isham, N.; Ghannoum, M.A. The Gut Microbiome as a Major Regulator of the Gut-Skin Axis. Front. Microbiol. 2018, 9, 1459. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, M.R.; Akter, S.; Tamanna, S.K.; Mazumder, L.; Esti, I.Z.; Banerjee, S.; Pirttilä, A.M. Impact of gut microbiome on skin health: Gut-skin axis observed through the lenses of therapeutics and skin diseases. Gut Microbes 2022, 14, 2096995. [Google Scholar] [CrossRef]
- Willemsen, L.E.M.; Koetsier, M.A.; van Deventer, S.J.H.; Van Tol, E.A.F. Short chain fatty acids stimulate epithelial mucin 2 expression through differential effects on prostaglandin E1 and E2 production by intestinal myofibroblasts. Gut 2003, 52, 1442–1447. [Google Scholar] [CrossRef] [PubMed]
- Samuelson, D.R.; Welsh, D.A.; Shellito, J.E. Regulation of lung immunity and host defense by the intestinal microbiota. Front. Microbiol. 2015, 6, 1085. [Google Scholar] [CrossRef] [PubMed]
- Pothmann, A.; Illing, T.; Wiegand, C.; Hartmann, A.A.; Elsner, P. The Microbiome and Atopic Dermatitis: A Review. Am. J. Clin. Dermatol. 2019, 20, 749–761. [Google Scholar] [CrossRef]
- Lee, Y.B.; Byun, E.J.; Kim, H.S. Potential Role of the Microbiome in Acne: A Comprehensive Review. J. Clin. Med. 2019, 8, 987. [Google Scholar] [CrossRef]
- Yan, D.; Issa, N.; Afifi, L.; Jeon, C.; Chang, H.-W.; Liao, W. The Role of the Skin and Gut Microbiome in Psoriatic Disease. Curr. Dermatol. Rep. 2017, 6, 94–103. [Google Scholar] [CrossRef]
- Chen, Y.-J.; Lee, W.-H.; Ho, H.J.; Tseng, C.-H.; Wu, C.-Y. An altered fecal microbial profiling in rosacea patients compared to matched controls. J. Formos. Med. Assoc. 2020, 120 Pt 1, 256–264. [Google Scholar] [CrossRef]
- Dan, Y.-L.; Wang, P.; Cheng, Z.; Wu, Q.; Wang, X.-R.; Wang, D.-G.; Pan, H.-F. Circulating adiponectin levels and systemic lupus erythematosus: A two-sample Mendelian randomization study. Rheumatology 2020, 60, 940–946. [Google Scholar] [CrossRef]
- Kurilshikov, A.; Medina-Gomez, C.; Bacigalupe, R.; Radjabzadeh, D.; Wang, J.; Demirkan, A.; Le Roy, C.I.; Garay, J.A.R.; Finnicum, C.T.; Liu, X.; et al. Large-scale association analyses identify host factors influencing human gut microbiome composition. Nat. Genet. 2021, 53, 156–165. [Google Scholar] [CrossRef]
- Burgess, S.; Thompson, S.G.; CRP CHD Genetics Collaboration. Avoiding bias from weak instruments in Mendelian randomization studies. Int. J. Epidemiol. 2011, 40, 755–764. [Google Scholar] [CrossRef]
- Burgess, S.; Butterworth, A.; Thompson, S.G. Mendelian Randomization Analysis with Multiple Genetic Variants Using Summarized Data. Genet. Epidemiol. 2013, 37, 658–665. [Google Scholar] [CrossRef] [PubMed]
- Verbanck, M.; Chen, C.-Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef]
- Bowden, J.; Smith, G.D.; Haycock, P.C.; Burgess, S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet. Epidemiol. 2016, 40, 304–314. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Dudbridge, F.; Thompson, S.G. Combining information on multiple instrumental variables in Mendelian randomization: Comparison of allele score and summarized data methods. Stat. Med. 2016, 35, 1880–1906. [Google Scholar] [CrossRef] [PubMed]
- Pagoni, P.; Dimou, N.L.; Murphy, N.; Stergiakouli, E. Using Mendelian randomisation to assess causality in observational studies. BMJ Ment. Health 2019, 22, 67–71. [Google Scholar] [CrossRef]
- Bowden, J.; Hemani, G.; Smith, G.D. Invited Commentary: Detecting Individual and Global Horizontal Pleiotropy in Mendelian Randomization—A Job for the Humble Heterogeneity Statistic? Am. J. Epidemiol. 2018, 187, 2681–2685. [Google Scholar] [CrossRef]
- Kwon, M.-S.; Lim, S.K.; Jang, J.-Y.; Lee, J.; Park, H.K.; Kim, N.; Yun, M.; Shin, M.-Y.; Jo, H.E.; Oh, Y.J.; et al. Lactobacillus sakei WIKIM30 Ameliorates Atopic Dermatitis-Like Skin Lesions by Inducing Regulatory T Cells and Altering Gut Microbiota Structure in Mice. Front. Immunol. 2018, 9, 1905. [Google Scholar] [CrossRef]
- Morotomi, M.; Nagai, F.; Sakon, H.; Tanaka, R. Dialister succinatiphilus sp. nov. and Barnesiella intestinihominis sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2008, 58 Pt 12, 2716–2720. [Google Scholar] [CrossRef]
- Arpaia, N.; Campbell, C.; Fan, X.; Dikiy, S.; Van Der Veeken, J.; DeRoos, P.; Liu, H.; Cross, J.R.; Pfeffer, K.; Coffer, P.J.; et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 2013, 504, 451–455. [Google Scholar] [CrossRef]
- Ohira, H.; Tsutsui, W.; Fujioka, Y. Are Short Chain Fatty Acids in Gut Microbiota Defensive Players for Inflammation and Atherosclerosis? J. Atheroscler. Thromb. 2017, 24, 660–672. [Google Scholar] [CrossRef]
- Zhuge, A.; Li, B.; Yuan, Y.; Lv, L.; Li, Y.; Wu, J.; Yang, L.; Bian, X.; Wang, K.; Wang, Q.; et al. Lactobacillus salivarius LI01 encapsulated in alginate-pectin microgels ameliorates d-galactosamine-induced acute liver injury in rats. Appl. Microbiol. Biotechnol. 2020, 104, 7437–7455. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yang, L.; Zhao, L.; Bai, F.; Liu, X. Comparison of Intestinal Microbes in Noninfectious Anterior Scleritis Patients with and Without Rheumatoid Arthritis. Front. Microbiol. 2022, 13, 925929. [Google Scholar] [CrossRef]
- Bowe, W.P.; Logan, A.C. Acne vulgaris, probiotics and the gut-brain-skin axis-back to the future? Gut Pathog. 2011, 3, 1. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, I.; Layton, A.M.; Ogawa, R. Updated Treatment for Acne: Targeted Therapy Based on Pathogenesis. Dermatol. Ther. 2021, 11, 1129–1139. [Google Scholar] [CrossRef] [PubMed]
- Salamon, M.; Sysa-Jedrzejowska, A.; Lukamowicz, J.; Lukamowicz, M.; Swiatkowska, E.; Wozniacka, A. Concentration of selected cytokines in serum of patients with acne rosacea. Przegl. Lek. 2008, 65, 371–374. [Google Scholar]
- Zafar, H.; Saier, M.H., Jr. Gut Bacteroides species in health and disease. Gut Microbes 2021, 13, 1848158. [Google Scholar] [CrossRef]
- Kim, J.M.; Cho, S.J.; Oh, Y.; Jung, H.; Kim, Y.; Kim, N. Nuclear factor-kappa B activation pathway in intestinal epithelial cells is a major regulator of chemokine gene expression and neutrophil migration induced by Bacteroides fragilis enterotoxin. Clin. Exp. Immunol. 2002, 130, 59–66. [Google Scholar] [CrossRef]
- Takeshita, K.; Mizuno, S.; Mikami, Y.; Sujino, T.; Saigusa, K.; Matsuoka, K.; Naganuma, M.; Sato, T.; Takada, T.; Tsuji, H.; et al. A Single Species of Clostridium Subcluster XIVa Decreased in Ulcerative Colitis Patients. Inflamm. Bowel Dis. 2016, 22, 2802–2810. [Google Scholar] [CrossRef]
- Caillon, F.; O’connell, M.; Eady, E.; Jenkins, G.; Cove, J.; Layton, A.; Mountford, A. Interleukin-10 secretion from CD14+ peripheral blood mononuclear cells is downregulated in patients with acne vulgaris. Br. J. Dermatol. 2010, 162, 296–303. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fabbrocini, G.; Bertona, M.; Picazo, Ó.; Pareja-Galeano, H.; Monfrecola, G.; Emanuele, E. Supplementation with Lactobacillus rhamnosus SP1 normalises skin expression of genes implicated in insulin signalling and improves adult acne. Benef. Microbes 2016, 7, 625–630. [Google Scholar] [CrossRef] [PubMed]
- Sikora, M.; Stec, A.; Chrabaszcz, M.; Knot, A.; Waskiel-Burnat, A.; Rakowska, A.; Olszewska, M.; Rudnicka, L. Gut Microbiome in Psoriasis: An Updated Review. Pathogens 2020, 9, 463. [Google Scholar] [CrossRef]
- Chen, Y.; Ho, H.J.; Tseng, C.; Lai, Z.; Shieh, J.; Wu, C. Intestinal microbiota profiling and predicted metabolic dysregulation in psoriasis patients. Exp. Dermatol. 2018, 27, 1336–1343. [Google Scholar] [CrossRef]
- Kim, J.; Moreno, A.; Krueger, J.G. The imbalance between Type 17 T-cells and regulatory immune cell subsets in psoriasis vulgaris. Front. Immunol. 2022, 13, 1005115. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Liu, Q.; Wu, H.; Tang, T.; Zhao, T.; Li, Z. The dysbiosis gut microbiota induces the alternation of metabolism and imbalance of Th17/Treg in OSA patients. Arch. Microbiol. 2022, 204, 217. [Google Scholar]
- Louis, P.; Scott, K.P.; Duncan, S.H.; Flint, H.J. Understanding the effects of diet on bacterial metabolism in the large intestine. J. Appl. Microbiol. 2007, 102, 1197–1208. [Google Scholar] [CrossRef]
- Chang, P.V.; Hao, L.; Offermanns, S.; Medzhitov, R. The microbial metabolite butyrate regulates intestinal macrophage function via histone deacetylase inhibition. Proc. Natl. Acad. Sci. USA 2014, 111, 2247–2252. [Google Scholar] [CrossRef]
- Schwarz, A.; Philippsen, R.; Schwarz, T. Induction of Regulatory T Cells and Correction of Cytokine Disbalance by Short-Chain Fatty Acids: Implications for Psoriasis Therapy. J. Investig. Dermatol. 2020, 141, 95–104.e2. [Google Scholar] [CrossRef]
- Salonen, A.; Lahti, L.; Salojärvi, J.; Holtrop, G.; Korpela, K.; Duncan, S.H.; Date, P.; Farquharson, F.; Johnstone, A.M.; Lobley, G.E.; et al. Impact of diet and individual variation on intestinal microbiota composition and fermentation products in obese men. ISME J. 2014, 8, 2218–2230. [Google Scholar] [CrossRef] [PubMed]
- Chiodini, R.J.; Dowd, S.E.; Chamberlin, W.M.; Galandiuk, S.; Davis, B.; Glassing, A. Microbial Population Differentials between Mucosal and Submucosal Intestinal Tissues in Advanced Crohn’s Disease of the Ileum. PLoS ONE 2015, 10, e0134382. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.D.; Spoendlin, J.; Chien, A.L.; Baldwin, H.; Chang, A.L.S. Evidence-based update on rosacea comorbidities and their common physiologic pathways. J. Am. Acad. Dermatol. 2018, 78, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Zhen, Z.; Xia, L.; You, H.; Jingwei, Z.; Shasha, Y.; Xinyi, W.; Wenjing, L.; Xin, Z.; Chaomei, F. An Integrated Gut Microbiota and Network Pharmacology Study on Fuzi-Lizhong Pill for Treating Diarrhea-Predominant Irritable Bowel Syndrome. Front. Pharmacol. 2021, 12, 746923. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.D.; Steinhoff, M. Integrative concepts of rosacea pathophysiology, clinical presentation and new therapeutics. Exp. Dermatol. 2016, 26, 659–667. [Google Scholar] [CrossRef]
- Saber, W.I.A.; Ghoniem, A.A.; Al-Otibi, F.O.; El-Hersh, M.S.; Eldadamony, N.M.; Menaa, F.; Elattar, K.M. A comparative study using response surface methodology and artificial neural network towards optimized production of melanin by Aureobasidium pullulans AKW. Sci. Rep. 2023, 13, 13545. [Google Scholar] [CrossRef]
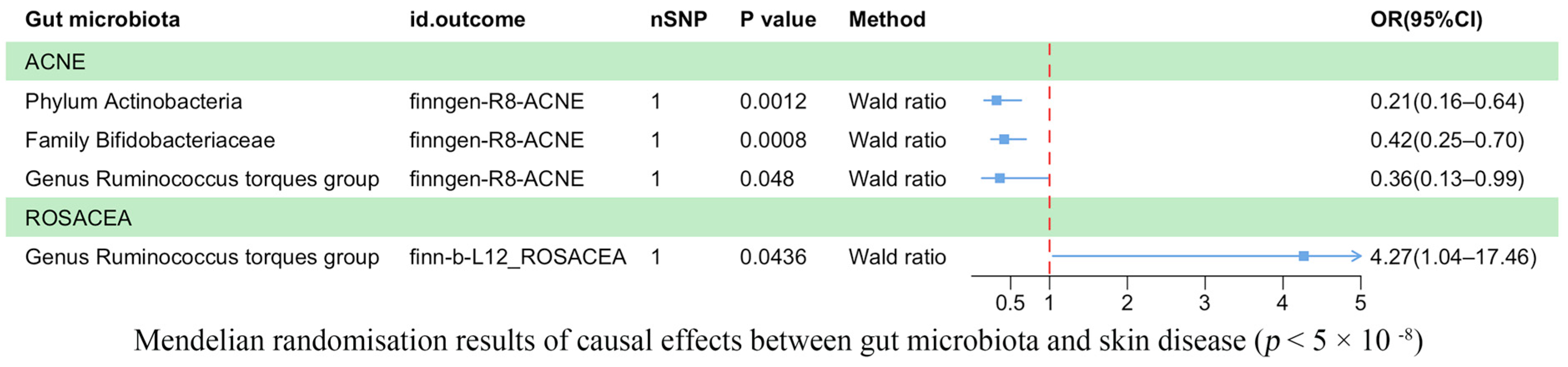
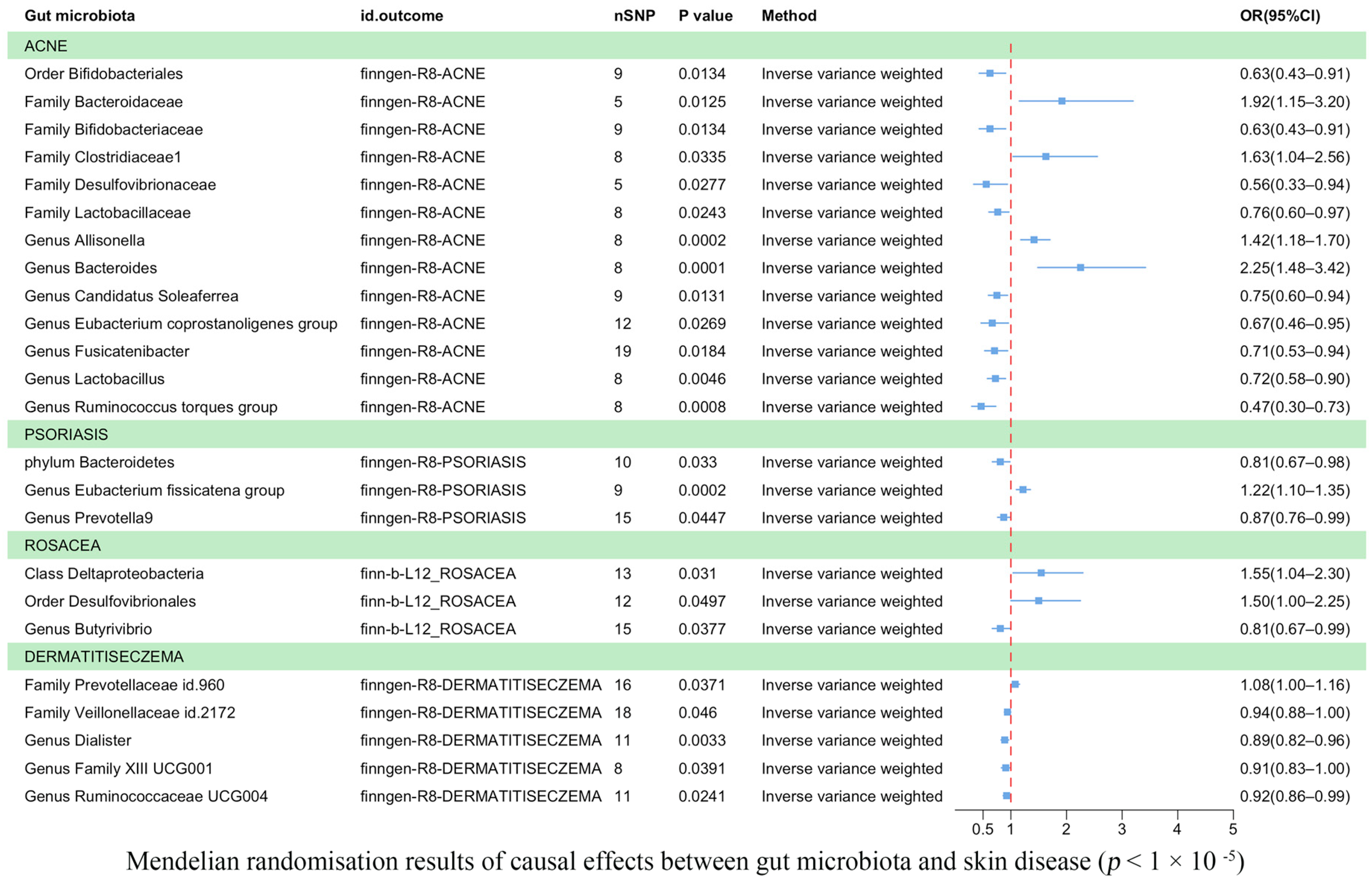

| Threshold/Method | Outcome | Id. Exposure | Id. Outcome | Nsnp | Beta | Se | p-Value | Trait |
|---|---|---|---|---|---|---|---|---|
| 5 × 10−8/Wald ratio | ACNE | ebi-a-GCST90017110 | finngen-R8-ACNE | 1 | −1.14272 | 0.351571 | 0.001153 | Phylum Actinobacteria id.400 |
| 1 × 10−5/IVW | ACNE | ebi-a-GCST90017093 | finngen-R8-ACNE | 9 | −0.46978 | 0.190061 | 0.013446 | Order Bifidobacteriales id.432 |
| 5 × 10−8/Wald ratio | ACNE | ebi-a-GCST90016929 | finngen-R8-ACNE | 1 | −0.875 | 0.26169 | 0.000827 | Family Bifidobacteriaceae id.433 |
| 1 × 10−5/IVW | ACNE | ebi-a-GCST90016927 | finngen-R8-ACNE | 5 | 0.651397 | 0.260806 | 0.012503 | Family Bacteroidaceae id.917 |
| 1 × 10−5/IVW | ACNE | ebi-a-GCST90016929 | finngen-R8-ACNE | 9 | −0.46978 | 0.190061 | 0.013446 | Family Bifidobacteriaceae id.433 |
| 1 × 10−5/IVW | ACNE | ebi-a-GCST90016931 | finngen-R8-ACNE | 8 | 0.488064 | 0.229627 | 0.033548 | Family Clostridiaceae1 id.1869 |
| 1 × 10−5/IVW | ACNE | ebi-a-GCST90016935 | finngen-R8-ACNE | 5 | −0.58438 | 0.265392 | 0.027668 | Family Desulfovibrionaceae id.3169 |
| 1 × 10−5/IVW | ACNE | ebi-a-GCST90016941 | finngen-R8-ACNE | 8 | −0.26929 | 0.119557 | 0.024295 | Family Lactobacillaceae id.1836 |
| 5 × 10−8/Wald ratio | ACNE | ebi-a-GCST90017066 | finngen-R8-ACNE | 1 | −1.0184 | 0.515063 | 0.048014 | Genus Ruminococcus torques group id.14377 |
| 1 × 10−5/IVW | ACNE | ebi-a-GCST90016963 | finngen-R8-ACNE | 8 | 0.347787 | 0.094021 | 0.000216 | Genus Allisonella id.2174 |
| 1 × 10−5/IVW | ACNE | ebi-a-GCST90016968 | finngen-R8-ACNE | 8 | 0.812298 | 0.213034 | 0.000137 | Genus Bacteroides id.918 |
| 1 × 10−5/IVW | ACNE | ebi-a-GCST90016976 | finngen-R8-ACNE | 9 | −0.28914 | 0.116587 | 0.013137 | Genus Candidatus Soleaferrea id.11350 |
| 1 × 10−5/IVW | ACNE | ebi-a-GCST90016997 | finngen-R8-ACNE | 12 | −0.40734 | 0.184114 | 0.026938 | Genus Eubacterium coprostanoligenes group id.11375 |
| 1 × 10−5/IVW | ACNE | ebi-a-GCST90017011 | finngen-R8-ACNE | 19 | −0.34742 | 0.147413 | 0.018436 | Genus Fusicatenibacter id.11305 |
| 1 × 10−5/IVW | ACNE | ebi-a-GCST90017030 | finngen-R8-ACNE | 8 | −0.32463 | 0.11442 | 0.004552 | Genus Lactobacillus id.1837 |
| 1 × 10−5/IVW | ACNE | ebi-a-GCST90017066 | finngen-R8-ACNE | 8 | −0.76493 | 0.229111 | 0.000842 | Genus Ruminococcus torques group id.14377 |
| 1 × 10−5/IVW | PSORIASIS | ebi-a-GCST90017111 | finngen-R8-PSORIASIS | 10 | −0.21072 | 0.098831 | 0.032997 | Phylum Bacteroidetes id.905 |
| 1 × 10−5/IVW | PSORIASIS | ebi-a-GCST90016999 | finngen-R8-PSORIASIS | 9 | 0.197321 | 0.052692 | 0.000181 | Genus Eubacterium fissicatena group id.14373 |
| 1 × 10−5/IVW | PSORIASIS | ebi-a-GCST90017045 | finngen-R8-PSORIASIS | 15 | −0.13764 | 0.068571 | 0.044716 | Genus Prevotella 9 id.11183 |
| 1 × 10−5/IVW | ROSACEA | ebi-a-GCST90016915 | finn-b-L12_ROSACEA | 13 | 0.435394 | 0.201843 | 0.030998 | Class Deltaproteobacteria id.3087 |
| 1 × 10−5/IVW | ROSACEA | ebi-a-GCST90017097 | finn-b-L12_ROSACEA | 12 | 0.405676 | 0.2067 | 0.049689 | Order Desulfovibrionales id.3156 |
| 5 × 10−8/Wald ratio | ROSACEA | ebi-a-GCST90017066 | finn-b-L12_ROSACEA | 1 | 1.450923 | 0.718926 | 0.043572 | Genus Ruminococcus torques group id.14377 |
| 1 × 10−5/IVW | ROSACEA | ebi-a-GCST90016975 | finn-b-L12_ROSACEA | 15 | −0.20966 | 0.100904 | 0.037723 | Genus Butyrivibrio id.1993 |
| 1 × 10−5/IVW | DERMATITISECZEMA | ebi-a-GCST90016948 | finngen-R8-DERMATITISECZEMA | 16 | 0.074607 | 0.035786 | 0.037084 | Family Prevotellaceae id.960 |
| 1 × 10−5/IVW | DERMATITISECZEMA | ebi-a-GCST90016956 | finngen-R8-DERMATITISECZEMA | 18 | −0.06238 | 0.031264 | 0.046024 | Family Veillonellaceae id.2172 |
| 1 × 10−5/IVW | DERMATITISECZEMA | ebi-a-GCST90016988 | finngen-R8-DERMATITISECZEMA | 11 | −0.11711 | 0.039833 | 0.003282 | Genus Dialister id.2183 |
| 1 × 10−5/IVW | DERMATITISECZEMA | ebi-a-GCST90017009 | finngen-R8-DERMATITISECZEMA | 8 | −0.09576 | 0.046417 | 0.039115 | Genus Family XIII UCG001 id.11294 |
| 1 × 10−5/IVW | DERMATITISECZEMA | ebi-a-GCST90017055 | finngen-R8-DERMATITISECZEMA | 11 | −0.07922 | 0.035124 | 0.024099 | Genus Ruminococcaceae UCG004 id.11362 |
| Id. Outcome | Gut Microbiota (Outcome) | Exposure | Method | Number of Snps | Beta | Se | p-Value | Correct_Causal Direction | Steiger_Pval |
|---|---|---|---|---|---|---|---|---|---|
| ebi-a-GCST90017045 | genus Prevotella 9 | PSORIASIS | IVW | 75 | 0.04 | 0.02 | 0.02 | TRUE | 0.001 |
| ebi-a-GCST90017111 | phylum Bacteroidetes | PSORIASIS | IVW | 80 | 0.04 | 0.01 | 0.002 | TRUE | 0.0002 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Long, J.; Gu, J.; Yang, J.; Chen, P.; Dai, Y.; Lin, Y.; Wu, M.; Wu, Y. Exploring the Association between Gut Microbiota and Inflammatory Skin Diseases: A Two-Sample Mendelian Randomization Analysis. Microorganisms 2023, 11, 2586. https://doi.org/10.3390/microorganisms11102586
Long J, Gu J, Yang J, Chen P, Dai Y, Lin Y, Wu M, Wu Y. Exploring the Association between Gut Microbiota and Inflammatory Skin Diseases: A Two-Sample Mendelian Randomization Analysis. Microorganisms. 2023; 11(10):2586. https://doi.org/10.3390/microorganisms11102586
Chicago/Turabian StyleLong, Junhao, Jinglan Gu, Juexi Yang, Pu Chen, Yan Dai, Yun Lin, Ming Wu, and Yan Wu. 2023. "Exploring the Association between Gut Microbiota and Inflammatory Skin Diseases: A Two-Sample Mendelian Randomization Analysis" Microorganisms 11, no. 10: 2586. https://doi.org/10.3390/microorganisms11102586
APA StyleLong, J., Gu, J., Yang, J., Chen, P., Dai, Y., Lin, Y., Wu, M., & Wu, Y. (2023). Exploring the Association between Gut Microbiota and Inflammatory Skin Diseases: A Two-Sample Mendelian Randomization Analysis. Microorganisms, 11(10), 2586. https://doi.org/10.3390/microorganisms11102586


_Di_Marco.png)



