Abstract
The monophasic variant of S. Typhimurium 4,[5],12:i:- (MVST) is the third most commonly reported Salmonella serovar involved in human infections (8.8%) in the EU and ranks after S. Enteritidis (54.6%) and S. Typhimurium (11.4%). In Italy, in contrast, the MVST has achieved peculiar epidemiological and ecological success which has allowed it to be, since 2011, the serovar most frequently isolated from humans. In the summer of 2022, a foodborne outbreak of the MVST involving 63 people occurred in the Marche Region (Central Italy). A common food exposure source among some human cases was a roasted, ready-to-eat (RTE) pork product, porchetta, which is a typical product of Central Italy. This paper describes the results of investigations conducted to clarify this outbreak. The porchetta was produced by a local manufacturing plant and distributed to at least two local retail stores, one of which was the retail outlet for the manufacturing plant. The MVST was isolated from surface samples collected at the porchetta manufacturing plant and at both local retail stores via bacterial analysis, and the porchetta sampled at one store contained the MVST. These data confirm this type of RTE pork product can be a source of Salmonella infection in humans.
1. Introduction
Salmonellosis is the second most commonly reported gastrointestinal infection in humans after campylobacteriosis and is a major cause of foodborne outbreaks in the EU/EEA. The past three decades have seen the rapid, worldwide emergence of a new Salmonella serovar, namely the monophasic variant of S. Typhimurium (MVST), with the antigenic formula 4,[5],12:i:- [1]. This serovar was first identified in chicken carcasses in Portugal in the 1980s [2] and has gradually become prevalent in the swine chain and therefore in pork products, especially in Europe and in the United States [3]. This link to swine, pork, and pork products caused infections with the MVST to become a global public health emergency with regard to human infections [4].
In recent years, the MVST has overtaken S. Typhimurium (ST) in some countries including Italy, where it ranked as the top serovar isolated from humans in 2021 (1125 reported cases of the MVST against 360 ST cases) [4] and from food samples in 2019 (206 strains of the MVST against 62 strains of ST) [5]. In Italy, the MVST has spread so successfully that is has overtaken S. Enteritidis (SE), which is still the serovar most frequently identified as responsible for human and veterinary infections in the majority of European countries, according to the EFSA-ECDC One Health Zoonoses report from 2021 [6].
This rapid diffusion suggests that the MVST has a competitive advantage over ST. For example, according to D’Incau et al. [7], when considering the association between the clinical signs of salmonellosis in pigs and the serovar, the clinical signs are more associated with ST than with the MVST, rendering the infections caused by the MVST more difficult to recognize and thus less likely to be controlled.
Heavy metal and antibiotic resistance might be further reasons explaining the spread of the MVST. The presence of heavy metal resistance genes (HMRGs) could favor the MVST, meaning it better escapes the metal-mediated antimicrobial response of human macrophages [8], which are able to poison bacteria in the phagosome with large amounts of copper and zinc [9]. According to Mastrorilli et al. [10], among 50 epidemiologically unrelated isolates of the MVST found in Italy from 2010 to 2016, the main shared genetic trait was the presence of HMTGs encoding efflux systems involved in silver and copper tolerance as a consequence of the usual practice, especially in swine husbandry, of supplementing swine feed with copper and zinc to promote animal growth [11].
The antibiotic resistance patterns in the MVST also represent a gain of function for the evolutionary success of this serovar and its clones [3]. The first antibiotic resistance pattern associated with swine and pork products dates from 1997, namely the so-called Spanish clone [12], which showed plasmid-mediated resistance toward seven antimicrobial drugs, ampicillin, chloramphenicol, gentamicin, streptomycin/spectinomycin, sulfonamides, tetracyclines, and trimethoprim (ACGSSuTTmp type), and it was ascribed to sequence type (ST) 19. The European clone, ascribed to ST34, is characterized by chromosomally encoded resistance to ampicillin, streptomycin, sulphonamides, and tetracycline (ASSuT type), and has, over the years, progressively overtaken the Spanish and the United States clones. This latter clone, on the contrary, rarely shows multidrug resistance (MDR) patterns [3].
According to data from the European database of sales of veterinary antimicrobial agents [13], in swine breeding, the most commonly used antibiotics are tetracyclines, penicillins, and sulfonamides. This evidence could explain the specific antibiotic resistance pattern in the clones circulating in Europe. However, in combination with the usage of antimicrobials in animal husbandry and human medicine, the versatility of plasmids may have largely contributed to the spread of antimicrobial resistance in Salmonella enterica [14].
An additional favorable factor for the spread of the MVST is the presence in the MVST genome of type II toxin–antitoxin (TA) cassettes involved in persistence phenomena, as demonstrated by Mastrorilli et al. [10].
Many outbreaks due to the MVST have been reported to date in Europe and in the United States, and most of them have been linked to RTE food products. For example, two outbreaks were linked to the consumption of salami and pork products in 2009 [15] and 2010 [16], respectively, in North Italy; three different outbreaks were linked to the same product, dried pork sausage, in France [17,18] and in Spain [19] in 2011 and again in France in 2020 [20]; one outbreak was associated with roast pork in Spain in 2016 [21]; and a multi-state outbreak was linked to pork products in Washington State in 2015 [22].
Although the MVST is especially associated with swine products [23], over the years, several outbreaks of the MVST in Europe have been linked to different sources, such as pets in Italy in 2021 [24], chocolate products in 12 countries belonging to the European Union’s European Economic Area and the UK in 2022 [25,26], and tomatoes in Sweden in 2019 [27].
In the present study, we describe an outbreak of the MVST that occurred in the period of July-September 2022 in the Marche region (Central Italy) and involved 63 people. Epidemiological investigations carried out by the Prevention Department of the Marche Sanitary Local Health Authority in the territorial areas involved identified an RTE roast pork product, namely porchetta, as the most probable source of the outbreak when some patients declared that they had consumed this product and had purchased the porchetta at the same retail stores.
In this outbreak report, we present the results of a microbiological investigation conducted at the two retail stores (RS(A) and RS(B)) and at a food-processing plant (FPP) in order to trace the source of the infection.
2. Materials and Methods
2.1. The Collection of Human, Food, and Environmental Strains and an Epidemiological Investigation
Between 14 July and 7 September, 2022, the Regional Reference Centre for Pathogenic Enterobacteria (CRRPE) of the Marche region (Central Italy) of the Istituto Zooprofilattico Sperimentale of Umbria and Marche, Peripheral Health Structure of Tolentino (IZSUM), received an unusual number (n = 102) of human strains of Salmonella submitted for serotyping from the regional hospital analysis laboratories and the private analysis laboratories participating in human surveillance (Enter-Net Italia) for the Marche region.
The epidemiological investigations for the salmonellosis cases were carried out by the Prevention Department of the Local Health Authority (LHA) and were shared with the CRRPE with respect for anonymity, accordingly, as part of an IZSUM research project funded by the Ministry of Health titled, “Implementation of an integrated system for the management of apparently sporadic cases of salmonellosis through the use of the latest generation molecular techniques”. The LHA arranged interviews with the case patients using a standard epidemiological questionnaire. The cases were described according to demographics (area of residence, age, and gender), the date of onset of symptoms, clinical illness (the type of symptoms, hospitalization, and the duration of illness), and food consumption over a three-day period before the onset of illness.
It was therefore possible to identify two retail stores, RS(A) and RS(B), both located in the province of Fermo, where some patients had purchased the same type of food a few days prior to the onset of symptoms. Moreover, the food-producing plant (FPP) for this commonly mentioned food, also located in Fermo province, was identified. The potential source of infection was identified as an RTE roasted pork product, porchetta, which is a typical product of Central Italy.
In the framework of the outbreak investigation, the LHA inspected RS(A), RS(B), and the FPP, performing controls on traceability, the labeling of the porchetta, and the hygiene conditions at the premises. Furthermore, at the FPP, the procedures for good manufacturing and hygiene practices and those based on a hazard analysis and critical control points (HACCP) principles were assessed. Furthermore, the LHA gained access to the FPP’s self-assessment to verify the compliance of the food business operator (FBOp) with the microbiological criteria established by Regulation (EC) No 2073/2005.
During the inspections, the LHA performed food (porchetta) and environmental sampling at RS(A) and RS(B) on 17 August and 23 August, respectively, and at the FPP on 24 August.
Environmental samples were collected aseptically from food contact surfaces (FCSs) and non-food contact surfaces (NFCSs). The sampling was performed using sterile sponges (Whirl-Pak Speci-Sponge Bags) which were pre-hydrated with Dey-Engley neutralizing buffer.
At RS(A), 10 environmental samples were collected (via the sponge swab method) from unsanitized surfaces, i.e., the anteroom access door handle, a porchetta knife, a porchetta-picking scoop, a Teflon chopping board used to support and cut porchetta, a Teflon cutting board from the butcher’s area, a Teflon worktop in the butcher’s area, a Teflon cutting board for the white meat counter, a scale keyboard from the fresh meat area keyboard, a fresh meat slicer, and a Teflon cutting board from behind the counter.
At RS(B), the LHA collected one sample of porchetta and five environmental samples (these latter samples were collected via the sponge swab method) from unsanitized surfaces, i.e., a wooden chopping board for porchetta, a porchetta knife, a porchetta spatula, a Teflon cutting board to the right of the wooden cutting board, and a steel table to the left of the wooden chopping board.
Finally, at the FPP, LHA staff collected one sample of porchetta and six environmental samples from unsanitized surfaces in the meat-cooking areas, i.e., a knife, a refrigerator grill, a refrigerator wall, a transporting board for cooked porchetta, a refrigerator bottom, and a refrigerator handle, and two samples from sanitized surfaces in the raw-meat-processing areas, i.e., a raw-porchetta-handling table and a knife.
2.2. Microbiological Analysis and the Serotyping of Salmonella Strains
Food and environmental samples were analyzed to detect Salmonella at the IZSUM Laboratory according to UNI EN ISO 6579-1:2020 [28].
All Salmonella strains from humans and the food-related and environmental samples were serotyped by the CRRPE according to ISO/TR 6579-3:2014 [29].
Phenotypically, the MVST strains were also tested using a multiplex PCR with specific oligonucleotides for the presence of the gene fljb (flagellar antigen H:2) [30,31].
2.3. Antimicrobial Susceptibility Testing
The antimicrobial susceptibility of each of the MVST strains was determined via the disk diffusion method, according to the Clinical and Laboratory Standards Institute guidelines (CLSI, 2023), using ampicillin (AMP, 10 µg), amoxicillin–clavulanic acid (AMC, 30 µg), cefotaxime (CTX, 30 µg), ceftazidime (CAZ, 30 µg), cefoxitin (FOX, 30 µg), chloramphenicol (C, 30 µg), gentamicin (CN, 10 µg), meropenem (MEM, 10 µg), tetracycline (TE, 30 µg), trimethoprim–sulfamethoxazole (SXT, 23.75/1.25 µg), trimethoprim (TMP, 5 µg), sulfisoxazole (ST, 300 µg), pefloxacin (PEF, 5 µg), and streptomycin (S 10, µg). The reference strain Escherichia coli ATCC 25922 was used for antimicrobial susceptibility. The CLSI interpretive criteria for the disk diffusion susceptibility testing of Salmonella (CLSI, 2023) were used [32].
2.4. Multiple Locus Variable Number Tandem Repeats Analysis (MLVA)
All the MVST strains were tested via a multiple-locus variable-number tandem repeat analysis (MLVA) carried out by the Department of Infectious Diseases of Istituto Superiore di Sanità and the National Reference Centre for Salmonellosis of Istituto Zooprofilattico Sperimentale delle Venezie for human and for food and environmental samples, respectively.
The MLVA was performed following the laboratory SOP by the ECDC [33], and the MLVA profiles were each reported as a string of five characters (STTR9-STTR5-STTR6-STTR10-STTR3) representing the number of repeats at the corresponding locus.
2.5. Whole Genome Sequencing (WGS)
Whole genome sequencing (WGS) was performed on a representative selection of 30 strains based on the MLVA results and epidemiological information.
The DNA of all the 30 Salmonella strains was extracted using a commercial column-based protocol (QIAamp DNA Mini, QIAGEN, Valencia, CA, USA), and purified gDNA was quantified using a Qubit 3.0 Fluorometer (Life Technologies, Carlsbad, CA, USA). Libraries for whole genome sequencing were prepared using a Nextera XT DNA sample preparation kit (Illumina, San Diego, CA, USA). High-throughput sequencing was performed using a MiSeq Reagent kit v3, resulting in paired-end reads that were 301 bp’s long.
For the analysis of the WGS data, an in-house pipeline was used which included steps for the trimming and quality control check of the reads (Fastp) [34]. A genome assembly of the paired-end reads was performed using Shovill (https://github.com/tseemann/shovill; accessed on 21 August 2023) with the default parameters.
2.5.1. In Silico Multilocus Sequence Typing (MLST)
The multilocus sequence typing (MLST) scheme used to characterize the Salmonella strains was based on a sequence analysis of the following seven housekeeping genes: chorismate synthase (aroC), β sliding clamp (dnaN), uroporphyrinogen-III synthase (hemD), histidinol dehydrogenase (hisD), N5-carboxyaminoimidazole ribonucleotide mutase (purE), 2-oxoglutarate decarboxylase (sucA), and fused aspartate kinase/homoserine dehydrogenase 1 (thrA).
The seven genes of the MLST scheme (ST) were deducted in silico using the program mlst v2.23.0 (https://github.com/tseemann/mlst) (https://pubmlst.org/) (accessed on 21 August 2023).
2.5.2. Core Genome MLST
For a cluster analysis of the strains, core genome MLST (cgMLST), was performed according to an INNUENDO scheme of 3255 target loci (https://zenodo.org/record/1323684; accessed on 21 August 2023), using the chewBBACA v 3.0.0 allele calling algorithm [35]. Using the software GrapeTree v.1.5.0 [36], a minimum spanning tree (MSTreeV2) showing the relationships among the strains in terms of allelic mismatches was created.
Strains presenting seven or fewer allelic differences were considered to belong to the same cgMLST cluster.
3. Results
From 14 July to 7 September 2022, as part of Enter-Net Italia surveillance, an increase in the number of clinical strains of Salmonella submitted to the CRRPE of the Marche region was observed. In fact, the total number of strains received in the considered period (n = 102) was double the number collected in the same period over the two previous years, 48 in 2021 and 55 in 2020, suggesting the onset of an outbreak of salmonellosis.
3.1. Clinical Strains
3.1.1. Serotyping and PCR Analysis
Seventy-eight of 102 Salmonella strains were serotyped and confirmed via a PCR as the MVST. The remaining 24 strains were other serovars.
3.1.2. Antimicrobial Susceptibility Testing
Fifty-seven of the seventy-eight strains of the MVST showed resistance to the following antibiotics: ampicillin (A), chloramphenicol (C), streptomycin (S), sulfisoxazole (Su), gentamicin (Gm), trimethoprim (Tmp), and trimethoprim–sulfamethoxazole (Sxt) (ACSSu+Gm+Tmp+Sxt, i.e., these 57 strains had one “cluster strain” antibiotic resistance type). The peculiar antibiotic resistance to Gm, which is unusual for the main MVST clone circulating in Italy, allowed us to make a first definition in this study of a cluster on the basis of phenotype. Therefore, at first, this cluster strain’s antibiotic-resistance type was used to define the control strain with respect to the outbreak investigation against which all other potentially related isolates were compared to.
Furthermore, seven strains of the MVST had different partial types of antibiotic resistance with respect to the type of cluster strain: three strains (ACSSu+Tmp+Sxt), one strain (ACSSu), one strain (ACSSu+Gm), one strain (ACSSu+Amc+Gm+Tmp+Sxt), and one strain (CSSu+Gm+Tmp+Sxt).
The remaining 14 strains showed the following antibiotic resistance types: ASSuT (n = 8), ASSu (n = 2), ASFox (n = 1), ASSuFox (n = 1), ASSuGm (n = 1), and ASSuTPef (n = 1) (Table 1).

Table 1.
Antimicrobial susceptibility testing, multiple-locus variable-number tandem repeat analysis (MLVA) typing, and inclusiveness in the cluster of human strains of the MVST (n = 78). The following are the criteria of inclusiveness: the “cluster strain” antibiotic resistance profile ACSSu+Gm+Tmp+Sxt and the MLVA profile 3-14-10-na-211; the “cluster strain” antibiotic resistance profile ACSSu+Gm+Tmp+Sxt and an MLVA profile different for just one tandem repeat in one locus compared to the MLVA profile 3-14-10-na-211; different partial types of antibiotic resistance with respect to the type of cluster strain (at least four antimicrobials shared) and the MLVA profile 3-14-10-na-211.
3.1.3. Multiple-Locus Variable-Number Tandem Repeats Analysis (MLVA)
Sixty-three of the sixty-four strains of the MVST strains (fifty-seven with the cluster-strain antibiotic resistance type and seven with antimicrobial resistance types close to the cluster-strain type) showed the same MLVA profile corresponding to 3-14-10-na-211, while the remaining strains showed a closely related MLVA profile corresponding to 3-13-10-na-211. The other 14 MVST strains had different MLVA profiles.
Therefore, a total of 57 isolates from cases of human salmonellosis were identified as belonging to the same cluster of the MVST (antibiotic resistance type ACSSu+Gm+Tmp+Sxt and MLVA 3-14-10-na-211 or 3-13-10-na-211). Additionally, seven isolates from human cases were defined as being closely related to this cluster of the MVST (Table 1).
Therefore, from this point, either the cluster-strain antibiotic resistance type ACSSu+Gm+Tmp+Sxt or the MLVA profile 3-14-10-na-211 were considered reference characteristics defining the control strain with respect to the outbreak investigation.
3.1.4. Epidemiological Investigations and Inquires in Neighboring Regions
The geographical distribution of the 64 human salmonellosis cases encompassed the entire Marche region and, therefore, all five local health authorities.
Thirty-three out of the sixty-four cases were male, and the most affected age group was 5–14 years old, with 26 cases. The cases were first reported between 14 July and 7 September 2022 and were distributed as follows: 22 in July, 37 in August, and 5 in September. The peak number of cases was recorded between 21 and 27 July (Figure 1). Twenty-nine patients of the sixty-four cases were hospitalized; for ten people out of the remaining thirty-five, information about their hospitalization was not available.
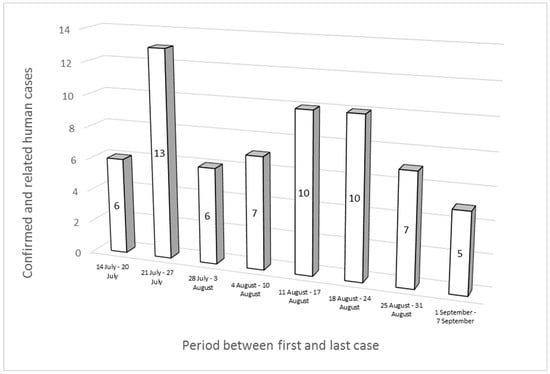
Figure 1.
Epidemic curve of “MVST clone” cases (n = 57) and “MVST clone—related” cases (n = 7).
In 10 of the 43 epidemiological investigations that were conducted, the consumption of a food, a roasted, RTE pork product, porchetta, was commonly reported, together with the names of the retail shops from which the porchetta had been purchased. In the remaining 33 epidemiological investigations, this information was not described.
This allowed us to determine that one brand of porchetta produced by one FPP and sold at two retail outlets could be of interest. A food safety inspection at the FPP revealed poor basic hygiene practices and poor maintenance of the facility, particularly in the cooking areas and with respect to the equipment used for the production of porchetta. Furthermore, inadequate procedures regarding the identification and management of critical control points (CCPs) were revealed, e.g., the cooking and rapid cooling of porchetta were not considered by the food business operator (FBO) to be CCPs but were just managed using good manufacturing practices. In addition, conflicting information about the actual production process compared to the process described in the self-monitoring manual in relation to the temperatures used to cook and blast chill the pork meat product was provided by the FBO during an interview.
Following the verification by the LHA of the FPP’s self-assessment, no positivity for Salmonella spp. for the ready-to-eat products was found; only carcass swabs carried out for compliance with hygiene process criteria at the slaughterhouse of the FPP were positive. No serotyping data were available for these isolates.
Based on the results of the food safety inspections, the LHA suspended porchetta production at the FPP beginning on 31 August 2022. Furthermore, an information notification for attention (https://webgate.ec.europa.eu/irasff/notification/view/; notification number: 568075; accessed on 2 September 2023) was sent to the Rapid Alert System for Food and Feed. In addition, the LHA ordered extraordinary cleaning, disinfection, and maintenance processes to be carried out at the FPP, along with an expert review of permanent procedures based on HACCP principles.
Following the isolation of the cluster strain of the MVST from environmental samples obtained at RS(A) and RS(B), the LHA also prescribed the extraordinary sanitation of all the surfaces (whether they tested positive or not) and of all the equipment in all food-related areas in these retail shops. The training of the workers at all three locations (the FPP, RS(A), and RS(B)) with respect to the management of cooked products in order to prevent further recontamination was also prescribed.
The traceback carried out by the LHA allowed us to identify the suppliers of the pork meat used by the FPP located in the Abruzzo and Umbria regions. To determine whether the cluster strain was identified from clinical or food samples in the same period in the Abruzzo and Umbria regions, the peculiar feature of the circulating clone (e.g., its antibiotic resistance type) was shared with the CRRPE of Perugia (Umbria) and the CRRPE of Teramo (Abruzzo).
During the period of interest, no strains of the MVST which were of human or animal/food origin and characterized by the ACSSu+Gm+Tmp+Sxt antibiotic resistance type were identified by the CRRPE of Abruzzo. In the period of interest in Umbria, three strains of the MVST were isolated from pig carcasses at a slaughterhouse that was a supplier of pork meat for the FPP, but these strains had MLVA profiles unrelated to the MVST cluster.
In the same period, two clinical strains of the MVST isolated from two different Umbrian hospitals showed the same antibiotic resistance pattern as the cluster clone. No MLVA data were available for these strains.
3.2. Food-Related and Environmental Strains
3.2.1. Microbiological, Serotyping, and PCR Analyses
Six of the twenty-five food and environmental samples tested positive for Salmonella, and the isolated strains were serotyped as the MVST (n = 5) and S. Infantis (n = 1) (Table 2). All the MVST strains were confirmed via a multiplex PCR. More details are reported in Table S1 (Supplementary Materials).

Table 2.
Antimicrobial susceptibility testing and the MLVA typing of food and environmental strains of the MVST (n = 5).
The five samples which were positive for the MVST were a Teflon chopping board for supporting and cutting porchetta (unsanitized surface) at RS(A); a wooden chopping board for porchetta, a porchetta knife (unsanitized surface), and a porchetta sample at RS(B), and a transporting board for cooked porchetta (unsanitized surface) at the FPP.
3.2.2. Antimicrobial Susceptibility and MLVA
All five food/environmental strains of the MVST had the antibiotic resistance type ACSSu+Gm+Tmp+Sxt and the MLVA profile 3-14-10-na-211, corresponding to those of the human cluster strains (Table 2).
3.3. Whole Genome Sequencing Analysis of the Clinical, Food-Related, and Environmental Strains
For the 30 genomes analyzed (25 clinical strains out of 64 total belonging to the MVST cluster on the basis of the MLVA results and all five food/environmental strains of the MVST) (Table 3), we obtained sequence data in accordance with the quality-control thresholds recommended for Salmonella as Q30 > 70%, average coverage ≥ 30×, a de novo assembly seq. length between 4.3 and 5.3 Mb, and the number of contigs ≤ 300 [37].

Table 3.
List of strains analyzed using WGS and epidemiological and analytic data.
Furthermore, the MLST analysis confirmed that all 30 genomes belonged to ST34.
Cluster Analysis
The cgMLST analysis showed that no significant allelic distance was highlighted between the clinical, food-related, and environmental strains except for one clinical strain (Figure 2).
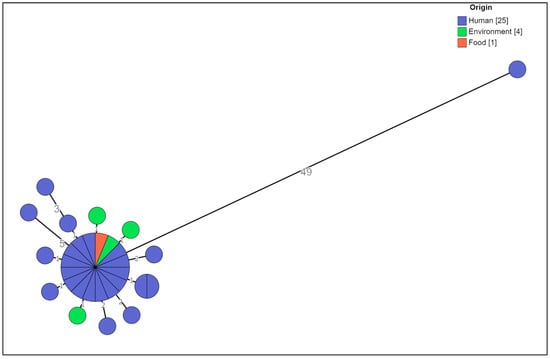
Figure 2.
Cluster analysis of a selection of the MVST strains colored by origin. The numbers reported in the branches indicate the allelic differences existing between the strains.
The central core included strains (n = 16) sharing the same allelic profile isolated from patients who consumed porchetta (n = 9) or from patients for whom this information was not available (n = 5), from porchetta (n = 1), and from the transporting board for cooked porchetta (n = 1). They showed the ACSSu+Gm+Tmp+Sxt “cluster strain” antibiotic resistance type (n = 15) and the ACSSu+Amc+Gm+Tmp+Sxt (n = 1) antibiotic resistance type and 3-14-10-na-211 MLVA profile (n = 16) (Figure 3).
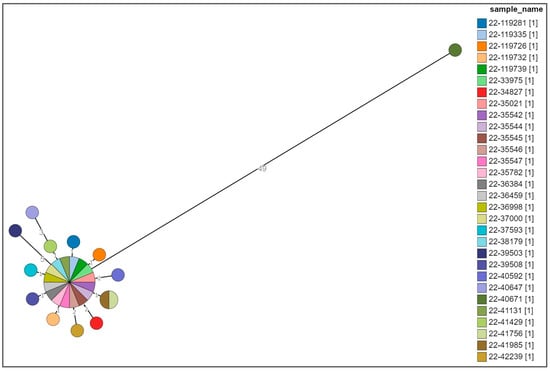
Figure 3.
Cluster analysis of a selection of the MVST strains. Different colors are representative of the identification detail of each strain.
Distances ranging from one to five alleles from the main group were seen for thirteen isolates, including strains from patients for whom information about the consumption of porchetta was not available (n = 10), from the Teflon chopping board used for supporting and cutting porchetta (n = 1), from the wooden chopping board for porchetta (n = 1), and from the porchetta knife (n = 1), with the ACSSu+Gm+Tmp+Sxt “cluster strain” antibiotic resistance type (n = 8) and different partial types of antibiotic resistance with respect to the type of cluster strain (n = 5) and the 3-14-10-na-211 (n = 12) and and 3-13-10-na-211 (n = 1) MLVA profiles, (Figure 3).
Only one strain, 22-40671, which was previously defined as being closely related to this MVST cluster on the basis of the MLVA profile 3-14-10-na-211 and the partial type of antibiotic resistance with respect to the type of cluster strain, i.e., ACSSu, was shown to be 49 alleles distant from the main group; therefore, it was considered a different clone (Figure 3).
4. Discussion
Our study provided evidence of the MVST contamination of both a porchetta sample and environmental samples from equipment used for handling this typical Italian food. The contaminated porchetta was the source of a foodborne outbreak involving 63 people.
Throughout the 1990s, pork production increased worldwide, resulting in consequent rapid globalization, and through the years, consensus on the close link between the MVST and the swine food chain became stronger, supported by the increasing evidence of human infections traced back to swine and pork products, especially RTE products [38].
In Italy, pork products are widely consumed [16]. In addition to production at the national level, there are many local traditional products such as porchetta, which is typical of Central Italy. Porchetta is an RTE product made from boned, seasoned, and roasted swine carcass meat. Roasting lasts from five to eight hours, depending on the size of the animal (a maximum weight of one quintal), followed by blast chilling down to 4 °C, which is measured in the thermal center of the product.
This type of pork product has already been identified as a source of Salmonella infections [21,39]. It is a food at risk of potentially spreading Salmonella due to the heterogeneous composition of its ingredients, which determines the variable pH and water activity levels depending on the sampling site.
Based on the porchetta production process, Salmonella can occur in this cooked, RTE pork product via two main routes. The first route is related to the use of Salmonella-positive carcasses or Salmonella-positive dissected raw meat; in this case, the pathogen is a consequence of meat contamination during slaughter or during the further handling procedures of boning, seasoning, and assembly [40].
According to the EFSA-ECDC EU One Health Zoonoses Report 2021 [6], considering all process hygiene criteria (PHC) monitoring data obtained from pig carcasses collected at slaughterhouses after dressing but before chilling, according to Regulation (EC) 2073/2005 and as reported by 23 EU member states (MSs), the overall proportion of Salmonella-positive samples based on official controls was 1.7% (N = 24,802), and it was significantly higher than that based on the FBOps’ own checks (1.4%, N = 103,270).
Regarding the Italian situation and bearing in mind that Italy is an MS that reports data collected by both the competent authority (CA) and the FBOps, 174 of 5147 samples reported by the CA (3.4%) and 107 of 11,494 samples reported by FBOps (0.93%) tested positive for Salmonella. Moreover, considering Salmonella along the swine production chain [41], for primary production, the most common Salmonella serovars causing human infections at the national level (i.e., the MVST and ST) are strictly associated with swine sources. To date, however, no control programs aimed at reducing the prevalence of Salmonella in swine farms have been implemented in Italy.
Furthermore, one of the main risks concerning carcass contamination is the persistence of Salmonella strains in slaughter or processing environments. Subsequent improper cooking procedures, such as inadequate thermal treatments, might not be sufficient to kill all present bacteria and can thus lead to the proliferation of Salmonella in the final product.
The second route for porchetta contamination occurs after cooking, whereby Salmonella originates either from the environment, through contaminated equipment or surfaces, or from healthy human carriers. The absence of competitive bacteria in RTE, cooked products like porchetta is another factor favoring the growth of Salmonella in a contaminated product if it is stored in unsuitable conditions.
The isolation of the MVST cluster clone at the FPP from a sponge swab on a transporting board used for cooked porchetta allowed us to exclude the post-cooking contamination of the porchetta at the two retail stores. At these locations, a Teflon chopping board used for supporting and cutting porchetta (RS(A)) and a wooden chopping board also used for porchetta, a porchetta knife (unsanitized surface), and porchetta (RS(B)) harbored the MVST cluster clone.
Nonetheless, it was not possible to establish the ultimate origin of the contamination at the FPP, nor it was possible to clarify whether the cluster clone originally contaminated the swine meat at the slaughterhouse level (i.e., the level of the meat suppliers) or was initially an environmental contaminant of the final product at the production level.
The evidence that no strain of the MVST was isolated from the swine carcasses at the Abruzzo slaughterhouse and that strains of the MVST with MLVA allelic profiles unrelated to the MVST cluster were isolated from the swine carcasses at the Umbrian slaughterhouse, both of which were suppliers of pork meat to the FPP, allows us to hypothesize that it is more probable that either the environmental contamination of the pork associated with some weaknesses in the porchetta production process or the contamination of the final product at the FPP occurred. The lack of evidence of Salmonella detected on a raw porchetta-manipulating table cannot exclude the potential environmental contamination of the meat since the table surface was sanitized prior to sampling.
The environmental contamination of the final product at the FPP also appears probable due to the detection of Salmonella on the transporting board used for cooked porchetta on this premises. The post-cooking contamination of the porchetta at the FPP could be the result of the persistence of the MVST strain in the FPP environment as a consequence of equipment being shared between raw- and cooked-meat-processing areas and the lack of sanitation procedures.
Following the positivity for Salmonella of the sampled porchetta, the Local Health Authority forced the FPP to cease production. At that point, the batch of porchetta that tested positive was no longer on the market as it had been sold and potentially already consumed due to its shelf-life.
Production activity at the FPP resumed on 12 October, following a favorable inspection carried out by the competent authority and the verification of compliance with the instructions previously given, i.e., by then, separate equipment was being used in the raw- and cooked-meat-processing areas, and CCPs were properly implemented for cooking and blast-chilling management.
The peculiar antibiotic resistance to gentamicin of the outbreak strain, which is unusual for the main MVST clone circulating in Italy, allowed us to obtain a first definition of the outbreak cluster on a phenotypic basis, resulting in the timely initiation of the investigation. This evidence underlines the importance of testing antibiotic resistance susceptibility as a preliminary screening method of characterizing Salmonella strains.
The identification of the unique MLVA profile of the investigated isolates of the MVST established, at first, the epidemiological relationship between the human cases and porchetta consumption and, therefore, the porchetta as the probable source of human infection.
This MLVA profile (3-14-10-na-211) is infrequent in the Italian human database, Enter-Net Italia (there were eight strains of the MVST of human origin with this profile from 2019 to date) and in the food–veterinary database, Enter-Vet (28 isolates from 2019 to date: 8 in 2019, 3 in 2020, 3 in 2021 and 14 in 2022; the isolates were of swine origin, with the exception of the 2022 isolates, which were all of bovine origin and were transmitted from a single laboratory).
The cgMLST analysis performed on a selection of 30 isolates of the MVST confirmed the belonging of clinical (n = 24), food-related (porchetta) (n = 1), and environmental (n = 4) strains to the same cluster except for one clinical strain that was shown to be 49 alleles distant from the main group and was therefore considered a different clone. Regarding this strain, it was at first considered closely related to the MVST cluster on the basis of the partial type of antibiotic resistance (ACSSu) with respect to the type of cluster strain (ACSSu+Gm+Tmp+Sxt) and of the MLVA profile (3-14-10-na-211). The epidemiological investigation surrounding this case was not carried out due to a failure to report to the Prevention Department of the Marche Sanitary Local Health Authority. Therefore, it is plausible that the clinical case in question never bought nor consumed porchetta.
Regarding the remaining 40 clinical strains which were not analyzed via WGS, since they showed the cluster-strain antibiotic resistance type ACSSu+Gm+Tmp+Sxt and the MLVA cluster strain profile 3-14-10-na-211, we included them in the cluster which, therefore, involved 63 people in total.
Regarding the evidence that the 30 genomes were ascribed to ST34, this sequence type is typical of the European clone [42], which, has gradually overtaken the Spanish and U.S. clones to become the most prevalent clone worldwide at present [43]. However, the isolates belonging to ST34 show the typical ASSuT phenotype. The antibiotic resistance type of this cluster strain, ACSSu+Gm+Tmp+Sxt, could be the result of a selective pressure imposed by the use of antimicrobials in the farms of origin of the pork used later for the production of porchetta. Following the WGS evidence of the inclusiveness in the cluster of six out of seven strains showing partial types of antibiotic resistance with respect to the type of cluster strain, the resistance pattern ACSSu+Gm+Tmp+Sxt could also be explained by the European clone’s plasmid-mediated acquisition of antimicrobial resistance genes and their subsequent loss.
Further phylogenetic analyses of this clone will be needed to better understand its origin and its evolution inside the ST34 population. The presence of specific genes of antimicrobial and heavy metal resistance, as well genes against biocides that could have favored the persistence of this clone of the MVST in the FPP environment, will be also investigated.
5. Conclusions
According to the Food and Agriculture Organization of United Nations, global pork production is expected to increase by 13.1% by 2030 and account for 34% of global meat consumption [44].
If the routes through which Salmonella can enter the swine chain generally remain unclear [45], it is equally true that a structured control plan, as has been put in place for the poultry industry, could allow for the identification and therefore the control of Salmonella along the chain, with the consequent containment of its spread to humans via food.
The key factor in the management of this investigation was, indeed, the intersectoral collaboration among microbiologists, veterinarians, and health and food safety authorities at national, regional, and local levels.
The laboratory-based surveillance network and consolidated system for contact and collaboration, like the one implemented in Italy’s Marche region, was essential in detecting the outbreak in a timely manner and for conducting a further investigation to identify the source of the outbreak clone.
Therefore, the content of this manuscript may be especially relevant for food and public health laboratories and epidemiologists.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11102567/s1, Table S1: Total number (n = 25) of food (n = 2) and environmental (n = 23) samples at RS(A) (n = 10), RS(B) (n = 6) and the FPP (n = 9); * FCS: surface in contact with food, ** NFCS: surface not in contact with food.
Author Contributions
Conceptualization, M.N., L.V., L.B. and G.B. (Giuliana Blasi); methodology, M.N., L.V., L.B. and G.B. (Giuliana Blasi); formal analysis, A.T., G.B. (Giulia Baggio) and A.M.D.; investigation, M.N., A.A., E.F., S.R., M.S., E.R., V.S. and G.B. (Giuliana Blasi); resources, A.A., E.F. and S.R.; data curation, M.N.; writing—original draft preparation, M.N.; writing—review and editing, M.N., L.V., L.B., C.L., A.T., E.F., B.M. and G.B. (Giuliana Blasi); visualization, M.N.; supervision, A.A., E.F., S.R. and G.B. (Giuliana Blasi); project administration, A.A., E.F., E.R. and G.B. (Giuliana Blasi); funding acquisition, M.N. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Italian Ministry of Health: Progetto di Ricerca Corrente IZSUM 07/21 RC.
Data Availability Statement
The data presented in this study are available on request from the corresponding author. The data are not publicly available due to privacy.
Acknowledgments
We gratefully acknowledge the contribution of the Enter-Net Peripheral Laboratories Referents for the Marche region who provided the Salmonella human strains: Paola Notaris, AST Pesaro-Urbino—Marche, UOC Patologia Clinica, Stabilimento Santa Maria della Misericordia, Urbino; Francesca Orecchioni, AOU Ospedali Riuniti di Ancona, SOD Medicina di Laboratorio, Ancona; Francesca Brecciaroli, AST Ancona—Marche, UOC Patologia Clinica, Presidio Ospedaliero Carlo Urbani, Jesi (Ancona); Stefano Lodolini, AST Ancona—Marche, UOSD Laboratorio Analisi e Biologia Molecolare HPV, Presidio Ospedaliero Principe di Piemonte, Senigallia (Ancona); Luciana Gironacci, AST Macerata—Marche, Laboratorio Analisi Presidio Ospedaliero di Civitanova Marche (Macerata); Emidio Alesiani, Laboratorio Analisi ClinicaLab, Civitanova Marche (Macerata); Maurizio Giorgi, Laboratorio Analisi Clinica Villa dei Pini, Civitanova Marche (Macerata); Maria Silvia Lancellotti, AST Fermo—Marche, UOC Patologia Clinica, Presidio Ospedaliero Augusto Murri, Fermo; Daniela Bracciani, AST Ascoli Piceno—Marche, UOC Patologia Clinica, Presidio Ospedaliero C. e G. Mazzoni, Ascoli Piceno; Franco Vagnarelli, Qualis Lab Ormodiagnostica, Grottammare (Ascoli Piceno). We gratefully acknowledge the contribution of staff from the Prevention Departments of the Sani-tary Territorial Health Authority of Marche region, who provided Salmonella epidemiological investigations: Augusto Liverani, AST Pesaro-Urbino—Marche, UOC Igiene, Sanità Pubblica e Prevenzione Malattie Infettive; Daniela Cimini, AST Ancona—Marche, UOC Igiene, Sanità Pubblica, Sorveglianza e Prevenzione delle Malattie Infettive e Cronico Degenerative; Franca Laici, AST Macerata—Marche, UOC Igiene e Sanità Pubblica—Prevenzione malattie infettive e cronico-degenerative; Giuseppe Ciarrocchi, AST Fermo—Marche, UOC Igiene e Sanità Pubblica; and Claudio Angelini, AST Ascoli Piceno—Marche, UOC Igiene, Epidemiologia e Sanità Pubblica. We gratefully acknowledge the contribution of the technical personnel of Istituto Zooprofilattico Sperimentale dell’Umbria e delle Marche “Togo Rosati”, UOC Controllo Alimenti Marche, Laboratorio Alimenti di Fermo, who analyzed the food and environmental samples collected at RS(A), RS(B), and the FPP.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Crayford, G.; Coombes, J.L.; Humphrey, T.J.; Wigley, P. Monophasic Expression of FliC by Salmonella 4,[5],12:I:- DT193 Does Not Alter Its Pathogenicity during Infection of Porcine Intestinal Epithelial Cells. Microbiology 2014, 160, 2507–2516. [Google Scholar] [CrossRef]
- Machado, J.; Bernardo, F. Prevalence of Salmonella in Chicken Carcasses in Portugal. J. Appl. Bacteriol. 1990, 69, 477–480. [Google Scholar] [CrossRef]
- Sun, H.; Wan, Y.; Du, P.; Bai, L. The Epidemiology of Monophasic Salmonella Typhimurium. Foodborne Pathog. Dis. 2020, 17, 87–97. [Google Scholar] [CrossRef]
- Survellaince Atlas of Infectious Diseases. Available online: https://atlas.ecdc.europa.eu/public/index.aspx (accessed on 20 April 2023).
- Leati, M.; Cibin, V.; Mancin, M.; Pestelli, P.; Barco, L. Enter-Vet Report Dati 2019, Centro Di Referenza Nazionale per Le Salmonellosi; Istituto Zooprofiliattico Sperimentale Delle Venezi: Legnaro, Italy, 2019. [Google Scholar]
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union One Health 2021 Zoonoses Report. EFSA J. 2022, 20, e07666. [Google Scholar] [CrossRef]
- D’Incau, M.; Salogni, C.; Giovannini, S.; Ruggeri, J.; Scali, F.; Tonni, M.; Formenti, N.; Guarneri, F.; Pasquali, P.; Alborali, G.L. Occurrence of Salmonella Typhimurium and Its Monophasic Variant (4,[5],12:I:-) In Healthy and Clinically Ill Pigs in Northern Italy. Porc. Health Manag. 2021, 7, 34. [Google Scholar] [CrossRef] [PubMed]
- Kapetanovic, R.; Bokil, N.J.; Achard, M.E.S.; Ong, C.Y.; Peters, K.M.; Stocks, C.J.; Phan, M.; Monteleone, M.; Schroder, K.; Irvine, K.M.; et al. Salmonella Employs Multiple Mechanisms to Subvert the TLR-inducible Zinc-mediated Antimicrobial Response of Human Macrophages. FASEB J. 2016, 30, 1901–1912. [Google Scholar] [CrossRef] [PubMed]
- Neyrolles, O.; Wolschendorf, F.; Mitra, A.; Niederweis, M. Mycobacteria, Metals, and the Macrophage. Immunol. Rev. 2015, 264, 249–263. [Google Scholar] [CrossRef] [PubMed]
- Mastrorilli, E.; Pietrucci, D.; Barco, L.; Ammendola, S.; Petrin, S.; Longo, A.; Mantovani, C.; Battistoni, A.; Ricci, A.; Desideri, A.; et al. A Comparative Genomic Analysis Provides Novel Insights Into the Ecological Success of the Monophasic Salmonella Serovar 4,[5],12:I:-. Front. Microbiol. 2018, 9, 715. [Google Scholar] [CrossRef]
- Medardus, J.J.; Molla, B.Z.; Nicol, M.; Morrow, W.M.; Rajala-Schultz, P.J.; Kazwala, R.; Gebreyes, W.A. In-Feed Use of Heavy Metal Micronutrients in U.S. Swine Production Systems and Its Role in Persistence of Multidrug-Resistant Salmonellae. Appl. Environ. Microbiol. 2014, 80, 2317–2325. [Google Scholar] [CrossRef]
- Mourão, J.; Machado, J.; Novais, C.; Antunes, P.; Peixe, L. Characterization of the Emerging Clinically-Relevant Multidrug-Resistant Salmonella Enterica Serotype 4,[5],12:I:-(Monophasic Variant of S. Typhimurium) Clones. Eur. J. Clin. Microbiol. Infect. Dis. 2014, 33, 2249–2257. [Google Scholar] [CrossRef]
- European Medicines Agency. Sales of Veterinary Antimicrobial Agents in 31 European Countries in 2021: Trends from 2010 to 2021: Twelfth ESVAC Report; Publications Office: Luxembourg, 2022. [Google Scholar]
- Plasmid-Mediated. Antimicrobial Resistance in Salmonella Enterica. Curr. Issues Mol. Biol. 2003, 5, 113–122. [Google Scholar] [CrossRef]
- Barco, L.; Ramon, E.; Cortini, E.; Longo, A.; Dalla Pozza, M.C.; Lettini, A.A.; Dionisi, A.M.; Olsen, J.E.; Ricci, A. Molecular Characterization of Salmonella enterica Serovar 4,[5],12:I:-DT193 ASSuT Strains from Two Outbreaks in Italy. Foodborne Pathog. Dis. 2014, 11, 138–144. [Google Scholar] [CrossRef]
- Andreoli, G.; Merla, C.; Valle, C.D.; Corpus, F.; Morganti, M.; D’incau, M.; Colmegna, S.; Marone, P.; Fabbi, M.; Barco, L.; et al. Foodborne Salmonellosis in Italy: Characterization of Salmonella enterica Serovar Typhimurium and Monophasic Variant 4,[5],12:I−Isolated from Salami and Human Patients. J. Food Prot. 2017, 80, 632–639. [Google Scholar] [CrossRef] [PubMed]
- Gossner, C.M.; van Cauteren, D.; Le Hello, S.; Weill, F.X.; Terrien, E.; Tessier, S.; Janin, C.; Brisabois, A.; Dusch, V.; Vaillant, V.; et al. Nationwide Outbreak of Salmonella enterica Serotype 4,[5],12:I:-Infection Associated with Consumption of Dried Pork Sausage, France, November to December 2011. Eurosurveillance 2012, 17, 20071. [Google Scholar] [CrossRef] [PubMed]
- Raguenaud, M.E.; Le Hello, S.; Salah, S.; Weill, F.X.; Brisabois, A.; Delmas, G.; Germonneau, P. Epidemiological and Microbiological Investigation of a Large Outbreak of Monophasic Salmonella Typhimurium 4,5,12:I:-In Schools Associated with Imported Beef in Poitiers, France, October 2010. EuroSurveill 2012, 17, 20289. [Google Scholar] [CrossRef]
- Arnedo-Pena, A.; Sabater-Vidal, S.; Herrera-León, S.; Bellido-Blasco, J.B.; Silvestre-Silvestre, E.; Meseguer-Ferrer, N.; Yague-Muñoz, A.; Gil-Fortuño, M.; Romeu-García, A.; Moreno-Muñoz, R. An Outbreak of Monophasic and Biphasic Salmonella Typhimurium, and Salmonella Derby Associated with the Consumption of Dried Pork Sausage in Castellon (Spain). Enfermedades Infecc. Y Microbiol. Clínica 2016, 34, 544–550. [Google Scholar] [CrossRef]
- Pardos De La Gandara, M.; Fournet, N.; Bonifait, L.; Lefèvre, S.; Chemaly, M.; Grastilleur, C.; Cadel-Six, S.; Fach, P.; Pignault, A.; Brisabois, A.; et al. Countrywide Multi-Serotype Outbreak of Salmonella Bovismorbificans ST142 and Monophasic Salmonella Typhimurium ST34 Associated with Dried Pork Sausages in France, September 2020* to January 2021. Eurosurveillance 2023, 28, 2200123. [Google Scholar] [CrossRef]
- de Frutos, M.; López-Urrutia, L.; Berbel, C.; Allue, M.; Herrera, S.; Azcona, J.M.; Beristaín, X.; Aznar, E.; Albert, M.; Ruiz, C.; et al. [Monophasic Salmonella Typhimurium outbreak due to the consumption of roast pork meat]. Rev. Esp. Quim. 2018, 31, 156–159. [Google Scholar]
- Kawakami, V.M.; Bottichio, L.; Angelo, K.; Linton, N.; Kissler, B.; Basler, C.; Lloyd, J.; Inouye, W.; Gonzales, E.; Rietberg, K.; et al. Notes from the Field: Outbreak of Multidrug-Resistant Salmonella Infections Linked to Pork—Washington, 2015. MMWR Morb. Mortal. Wkly. Rep. 2016, 65, 379–381. [Google Scholar] [CrossRef]
- Biswas, S.; Li, Y.; Elbediwi, M.; Yue, M. Emergence and Dissemination of Mcr-Carrying Clinically Relevant Salmonella Typhimurium Monophasic Clone ST34. Microorganisms 2019, 7, 298. [Google Scholar] [CrossRef]
- Russini, V.; Corradini, C.; Rasile, E.; Terracciano, G.; Senese, M.; Bellagamba, F.; Amoruso, R.; Bottoni, F.; De Santis, P.; Bilei, S.; et al. A Familiar Outbreak of Monophasic Salmonella Serovar Typhimurium (ST34) Involving Three Dogs and Their Owner’s Children. Pathogens 2022, 11, 1500. [Google Scholar] [CrossRef]
- Lund, S.; Tahir, M.; Vohra, L.I.; Hamdana, A.H.; Ahmad, S. Outbreak of Monophasic Salmonella Typhimurium Sequence Type 34 Linked to Chocolate Products. Ann. Med. Surg. 2022, 82, 104597. [Google Scholar] [CrossRef] [PubMed]
- Larkin, L.; Pardos De La Gandara, M.; Hoban, A.; Pulford, C.; Jourdan-Da Silva, N.; De Valk, H.; Browning, L.; Falkenhorst, G.; Simon, S.; Lachmann, R.; et al. Investigation of an International Outbreak of Multidrug-Resistant Monophasic Salmonella Typhimurium Associated with Chocolate Products, EU/EEA and United Kingdom, February to April 2022. Eurosurveillance 2022, 27, 2200314. [Google Scholar] [CrossRef] [PubMed]
- Colombe, S.; Jernberg, C.; Löf, E.; Angervall, A.L.; Mellström-Dahlgren, H.; Dotevall, L.; Bengnér, M.; Hall, I.; Sundqvist, L.; Kühlmann-Berenzon, S.; et al. Outbreak of Unusual H2S-Negative Monophasic Salmonella Typhimurium Strain Likely Associated with Small Tomatoes, Sweden, August to October 2019. Eurosurveillance 2019, 24, 1900643. [Google Scholar] [CrossRef]
- ISO 6579-1:2017/Amd 1:2020; Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 1: Detection of Salmonella spp.—Amendment 1: Broader Range of Incubation Temperatures, Amendment to the Status of Annex D, and Correction of the Composition of MSRV and SC. ISO: Geneva, Switzerland, 2020.
- ISO/TR 6579-3:2014; Microbiology of the Food Chain—Horizontal Method for the Detection, Enumeration and Serotyping of Salmonella—Part 3: Guidelines for Serotyping of Salmonella spp. ISO: Geneva, Switzerland, 2014.
- Lim, Y.-H.; Hirose, K.; Izumiya, H.; Arakawa, E.; Takahashi, H.; Terajima, J.; Itoh, K.; Tamura, K.; Kim, S.-I.; Watanabe, H. Multiplex Polymerase Chain Reaction Assay for Selective Detection of Salmonella enterica Serovar Typhimurium. Jpn. J. Infect. Dis. 2003, 56, 151–155. [Google Scholar]
- Rahn, K.; De Grandis, S.A.; Clarke, R.C.; McEwen, S.A.; Galán, J.E.; Ginocchio, C.; Curtiss, R.; Gyles, C.L. Amplification of an invA Gene Sequence of Salmonella Typhimurium by Polymerase Chain Reaction as a Specific Method of Detection of Salmonella. Mol. Cell. Probes 1992, 6, 271–279. [Google Scholar] [CrossRef]
- Weinstein, M.P.; Clinical and Laboratory Standards Institute CLSI. Performance Standards for Antimicrobial Susceptibility Testing, 33rd ed.; CLSI Supplement M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2023. [Google Scholar]
- ECDC; Statens Serum Institut. Laboratory Standard Operating Procedure for MLVA of Salmonella Enterica Serotype Typhimurium; Publications Office: Luxembourg, 2011. [Google Scholar]
- Chen, S. Ultrafast One-pass FASTQ Data Preprocessing, Quality Control, and Deduplication Using Fastp. iMeta 2023, 2, e107. [Google Scholar] [CrossRef]
- Silva, M.; Machado, M.P.; Silva, D.N.; Rossi, M.; Moran-Gilad, J.; Santos, S.; Ramirez, M.; Carriço, J.A. chewBBACA: A Complete Suite for Gene-by-Gene Schema Creation and Strain Identification. Microb. Genom. 2018, 4, e000166. [Google Scholar] [CrossRef]
- Zhou, Z.; Alikhan, N.-F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carriço, J.A.; Achtman, M. GrapeTree: Visualization of Core Genomic Relationships among 100,000 Bacterial Pathogens. Genome Res. 2018, 28, 1395–1404. [Google Scholar] [CrossRef]
- European Food Safety Authority (EFSA); Costa, G.; Di Piazza, G.; Koevoets, P.; Iacono, G.; Liebana, E.; Pasinato, L.; Rizzi, V.; Rossi, M. Guidelines for Reporting Whole Genome Sequencing-based Typing Data through the EFSA One Health WGS System. EFS3 2022, 19, 7413E. [Google Scholar] [CrossRef]
- De la Torre, E.; Zapata, D.; Tello, M.; Mejía, W.; Frías, N.; García Peña, F.J.; Mateu, E.M.; Torre, E. Several Salmonella Enterica Subsp. Enterica Serotype 4,5,12:I:−Phage Types Isolated from Swine Samples Originate from Serotype Typhimurium DT U302. J. Clin. Microbiol. 2003, 41, 2395–2400. [Google Scholar] [CrossRef] [PubMed]
- Lettini, A.A.; Saccardin, C.; Ramon, E.; Longo, A.; Cortini, E.; Dalla Pozza, M.C.; Barco, L.; Guerra, B.; Luzzi, I.; Ricci, A. Characterization of an Unusual Salmonella Phage Type DT7a and Report of a Foodborne Outbreak of Salmonellosis. Int. J. Food Microbiol. 2014, 189, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Borch, E.; Nesbakken, T.; Christensen, H. Hazard Identification in Swine Slaughter with Respect to Foodborne Bacteria. Int. J. Food Microbiol. 1996, 30, 9–25. [Google Scholar] [CrossRef]
- Lauteri, C.; Festino, A.R.; Conter, M.; Vergara, A. Prevalence and Antimicrobial Resistance Profile in Salmonella Spp. Isolates from Swine Food Chain. Ital. J. Food Saf. 2022, 11, 9980. [Google Scholar] [CrossRef]
- Lucarelli, C.; Dionisi, A.M.; Torpdahl, M.; Villa, L.; Graziani, C.; Hopkins, K.; Threlfall, J.; Caprioli, A.; Luzzi, I. Evidence for a Second Genomic Island Conferring Multidrug Resistance in a Clonal Group of Strains of Salmonella Enterica Serovar Typhimurium and Its Monophasic Variant Circulating in Italy, Denmark, and the United Kingdom. J. Clin. Microbiol. 2010, 48, 2103–2109. [Google Scholar] [CrossRef]
- Elnekave, E.; Hong, S.; Mather, A.E.; Boxrud, D.; Taylor, A.J.; Lappi, V.; Johnson, T.J.; Vannucci, F.; Davies, P.; Hedberg, C.; et al. Salmonella enterica Serotype 4,[5],12:I:- In Swine in the United States Midwest: An Emerging Multidrug-Resistant Clade. Clin. Infect. Dis. 2018, 66, 877–885. [Google Scholar] [CrossRef] [PubMed]
- OECD; Food and Agriculture Organization of the United Nations. OECD-FAO Agricultural Outlook 2021–2030; OECD-FAO Agricultural Outlook; OECD: Paris, France, 2021; ISBN 978-92-64-43607-7. [Google Scholar]
- Harrison, O.L.; Rensing, S.; Jones, C.K.; Trinetta, V. Salmonella enterica 4,[5],12:I:-, an Emerging Threat for the Swine Feed and Pork Production Industry. J. Food Prot. 2022, 85, 660–663. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).