Monophasic Variant of Salmonella Typhimurium Infection Affects the Serum Metabolome in Swine
Abstract
1. Importance
2. Introduction
3. Material and Methods
3.1. Animal Procedures
3.2. Metabolomic Analysis
3.2.1. Samples Preparation
3.2.2. LC–MS
3.2.3. Data Extraction and Metabolic Signals Identification
3.3. Microbiota Analysis
Samples Preparation
3.4. Statistical Analysis
3.4.1. Serum Metabolome Analysis
3.4.2. Fecal Microbiota Analysis
3.4.3. Link between Fecal Microbiota Composition and Serum Metabolome Composition
3.5. Data Availability
4. Results
4.1. Serum Metabolome Analysis
4.2. Comparison of Infected Pigs to Noninfected Animals
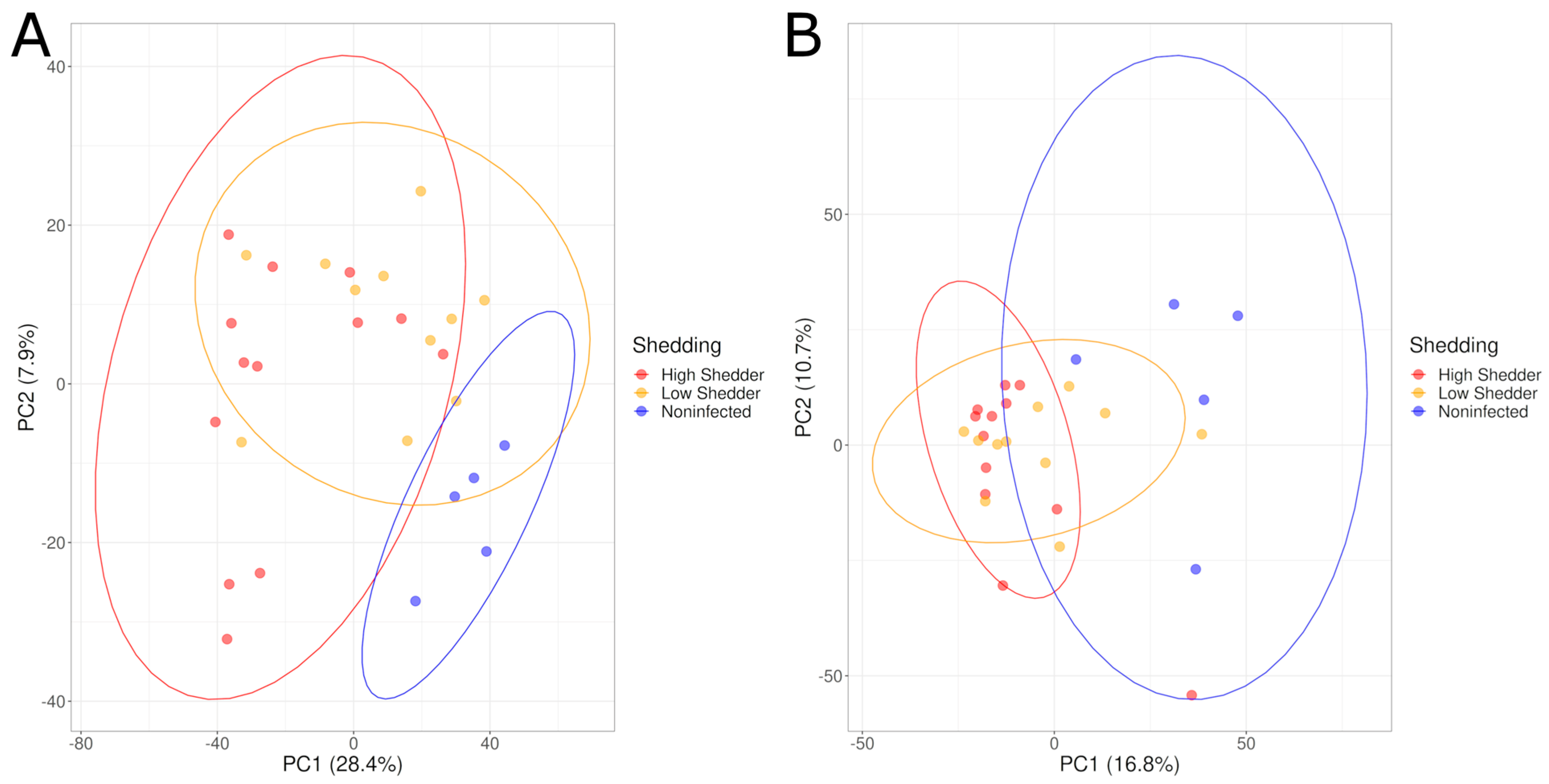
4.3. Comparison According to the Shedding Levels of SALMONELLA
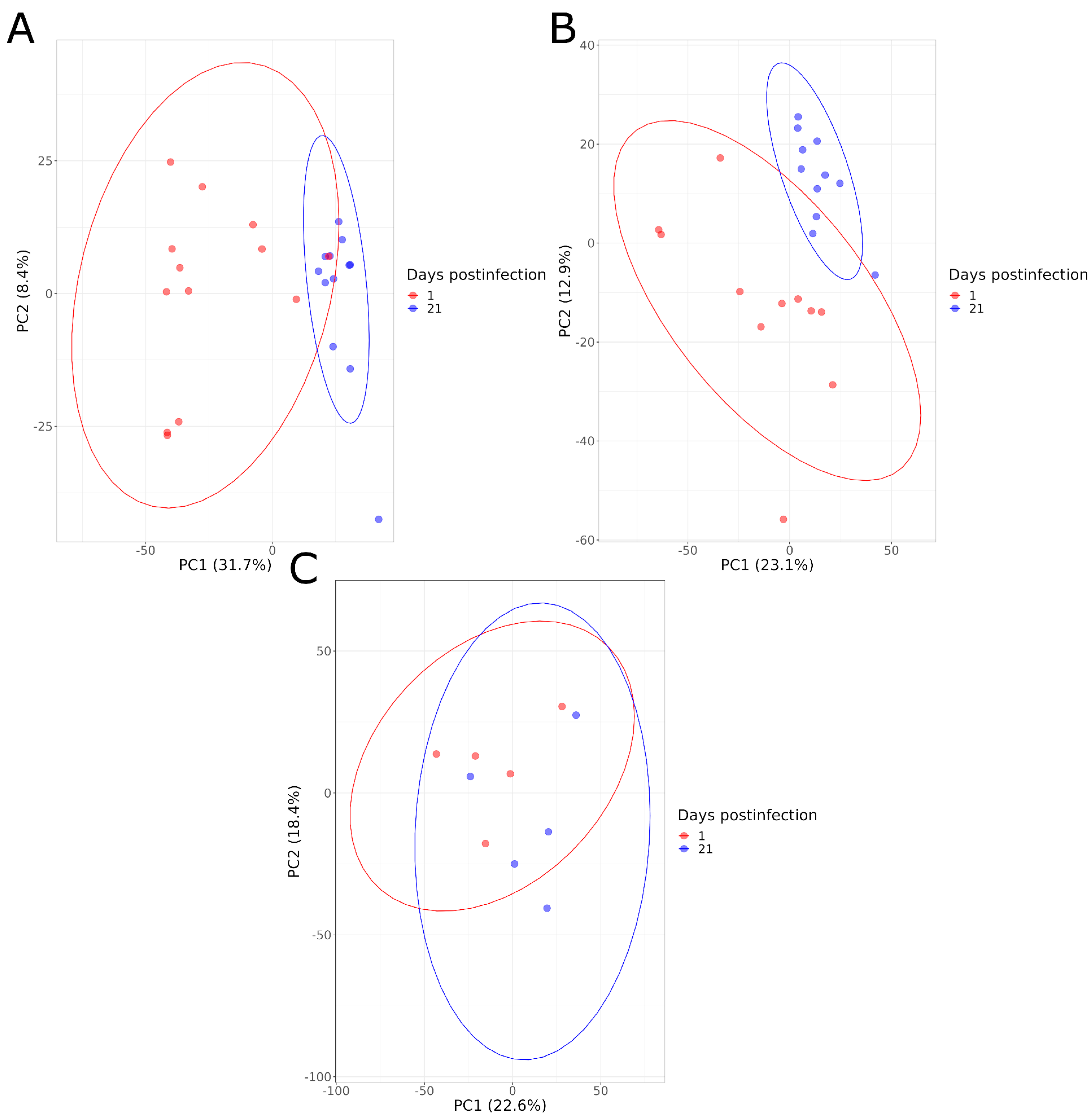
4.4. Comparison According to Time Postinfection
4.5. Comparison According to the Salmonella Seropositivity of the Animals
4.6. Fecal Microbiota Analysis
4.6.1. Comparison According to the Shedding Levels of Salmonella
4.6.2. Comparison According to Time Postinfection
4.6.3. Comparison According to the Salmonella Seropositivity of the Animals
4.7. Link between Fecal Microbiota Composition and Serum Metabolome Composition
5. Discussion
5.1. Comparison of Infected Pigs to Noninfected Animals
5.2. Comparison According to the Shedding Levels of Salmonella
5.3. Comparison According to Time Postinfection
5.4. Comparison According to the Salmonella Seropositivity of the Animals
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- EFSA. The European Union One Health 2021 Zoonoses Report. EFSA J. 2022, 20, e07666. [Google Scholar]
- De Knegt, L.V.; Pires, S.M.; Hald, T. Attributing foodborne salmonellosis in humans to animal reservoirs in the European Union using a multi-country stochastic model. Epidemiol. Infect. 2015, 143, 1175–1186. [Google Scholar] [CrossRef]
- Griffith, R.W.; Carlson, S.A.; Krull, A.C. Salmonellosis. In Diseases of Swine; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2019; pp. 912–925. [Google Scholar]
- EFSA. Report of the Task Force on Zoonoses Data Collection on the Analysis of the baseline survey on the prevalence of Salmonella in slaughter pigs, in the EU, 2006–2007—Part A: Salmonella prevalence estimates. EFSA J. 2008, 6, 135r. [Google Scholar] [CrossRef]
- EFSA. Analysis of the baseline survey on the prevalence of Salmonella in holdings with breeding pigs in the EU, 2008—Part A: Salmonella prevalence estimates. EFSA J. 2009, 7, 1377. [Google Scholar]
- Arguello, H.; Rubio, P.; Carvajal, A.; Arguello, H.; Rubio, P.; Carvajal, A. Salmonella Control Measures at Farm in Swine Production. Salmonella—Distribution, Adaptation, Control Measures and Molecular Technologies. IntechOpen 2012. Available online: https://www.intechopen.com/chapters/37795 (accessed on 10 October 2022).
- Letellier, A.; Beauchamp guy Guevrement, E.; D’allaire, S.; Hurnik, D.; Quessy, S. Risk Factors at Slaughter Associated with Presence of Salmonella on Hog Carcasses in Canada. J. Food Prot. 2009, 72, 2326–2331. [Google Scholar] [PubMed]
- Nair, S.; Farzan, A.; Poljak, Z.; Friendship, R. Identifying Active Salmonella Infections in Swine Nurseries Using Serology and Bacterial Culture and Evaluating Associated Risk Factors. Animals 2020, 10, 1517. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar]
- Gika, H.; Virgiliou, C.; Theodoridis, G.; Plumb, R.S.; Wilson, I.D. Untargeted LC/MS-based metabolic phenotyping (metabonomics/metabolomics): The state of the art. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2019, 1117, 136–147. [Google Scholar]
- Want, E.J. LC-MS Untargeted Analysis. Methods Protoc. 2018, 1738, 99–116. [Google Scholar]
- McCreath, G.; Whitfield, P.D.; Roe, A.J.; Watson, M.J.; Sim, M.A.B. A Metabolomics approach for the diagnosis Of SecondAry InfeCtions in COVID-19 (MOSAIC): A study protocol. BMC Infect. Dis. 2021, 21, 1204. [Google Scholar] [CrossRef]
- Tounta, V.; Liu, Y.; Cheyne, A.; Larrouy-Maumus, G. Metabolomics in infectious diseases and drug discovery. Mol. Omics 2021, 17, 376–393. [Google Scholar] [CrossRef] [PubMed]
- Isa, F.; Collins, S.; Lee, M.H.; Decome, D.; Dorvil, N.; Joseph, P.; Smith, L.; Salerno, S.; Wells, M.T.; Fischer, S.; et al. Mass Spectrometric Identification of Urinary Biomarkers of Pulmonary Tuberculosis. EBioMedicine 2018, 31, 157–165. [Google Scholar] [CrossRef] [PubMed]
- Khan, I.; Nam, M.; Kwon, M.; Seo, S.; Jung, S.; Han, J.S.; Hwang, G.-S.; Kim, M.K. LC/MS-Based Polar Metabolite Profiling Identified Unique Biomarker Signatures for Cervical Cancer and Cervical Intraepithelial Neoplasia Using Global and Targeted Metabolomics. Cancers 2019, 11, 511. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Milani, C.; de Giori, G.S.; Sesma, F.; van Sinderen, D.; Ventura, M. Bacteria as vitamin suppliers to their host: A gut microbiota perspective. Curr. Opin. Biotechnol. 2013, 24, 160–168. [Google Scholar] [CrossRef]
- Neuman, H.; Debelius, J.W.; Knight, R.; Koren, O. Microbial endocrinology: The interplay between the microbiota and the endocrine system. FEMS Microbiol. Rev. 2015, 39, 509–521. [Google Scholar] [CrossRef]
- Silva, Y.P.; Bernardi, A.; Frozza, R.L. The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Front. Endocrinol. 2020, 11, 25. [Google Scholar] [CrossRef] [PubMed]
- Claus, S.P.; Tsang, T.M.; Wang, Y.; Cloarec, O.; Skordi, E.; Martin, F.-P.; Rezzi, S.; Ross, A.; Kochhar, S.; Holmes, E.; et al. Systemic multicompartmental effects of the gut microbiome on mouse metabolic phenotypes. Mol. Syst. Biol. 2008, 4, 219. [Google Scholar] [CrossRef] [PubMed]
- Lee-Sarwar, K.A.; Lasky-Su, J.; Kelly, R.S.; Litonjua, A.A.; Weiss, S.T. Metabolome–Microbiome Crosstalk and Human Disease. Metabolites 2020, 10, 181. [Google Scholar] [CrossRef] [PubMed]
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703. [Google Scholar] [CrossRef]
- Argüello, H.; Estellé, J.; Zaldívar-López, S.; Jiménez-Marín, Á.; Carvajal, A.; López-Bascón, M.A.; Crispie, F.; O’Sullivan, O.; Cotter, P.D.; Priego-Capote, F.; et al. Early Salmonella Typhimurium infection in pigs disrupts Microbiome composition and functionality principally at the ileum mucosa. Sci. Rep. 2018, 8, 7788. [Google Scholar] [CrossRef]
- Kempf, F.; Cordoni, G.; Chaussé, A.-M.; Drumo, R.; Brown, H.; Horton, D.L.; Paboeuf, F.; Denis, M.; Velge, P.; La Ragione, R.; et al. Inflammatory Responses Induced by the Monophasic Variant of Salmonella Typhimurium in Pigs Play a Role in the High Shedder Phenotype and Fecal Microbiota Composition. mSystems 2023, 8, e0085222. [Google Scholar] [CrossRef]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Giacomoni, F.; Le Corguillé, G.; Monsoor, M.; Landi, M.; Pericard, P.; Pétéra, M.; Duperier, C.; Tremblay-Franco, M.; Martin, J.-F.; Jacob, D.; et al. Workflow4Metabolomics: A collaborative research infrastructure for computational metabolomics. Bioinformatics 2015, 31, 1493–1495. [Google Scholar] [CrossRef] [PubMed]
- van der Kloet, F.M.; Bobeldijk, I.; Verheij, E.R.; Jellema, R.H. Analytical error reduction using single point calibration for accurate and precise metabolomic phenotyping. J. Proteome Res. 2009, 8, 5132–5141. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef]
- Larivière-Gauthier, G.; Thibodeau, A.; Letellier, A.; Yergeau, É.; Fravalo, P. Reduction of Salmonella Shedding by Sows during Gestation in Relation to Its Fecal Microbiome. Front. Microbiol. 2017, 8, 2219. [Google Scholar] [CrossRef] [PubMed]
- Thévenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the Human Adult Urinary Metabolome Variations with Age, Body Mass Index, and Gender by Implementing a Comprehensive Workflow for Univariate and OPLS Statistical Analyses. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e0061217. [Google Scholar] [CrossRef] [PubMed]
- Oksanen, J.; Simpson, G.L.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’Hara, R.B.; Solymos, P.; Stevens, M.H.H.; Szoecs, E.; et al. vegan: Community Ecology Package, Version 2.6-2; GitHub, Inc.: San Francisco, CA, USA, 2022. [Google Scholar]
- Rohart, F.; Gautier, B.; Singh, A.; Cao, K.-A.L. mixOmics: An R package for ‘omics feature selection and multiple data integration. PLoS Comput. Biol. 2017, 13, e1005752. [Google Scholar] [CrossRef]
- Mon, K.K.Z.; Zhu, Y.; Chanthavixay, G.; Kern, C.; Zhou, H. Integrative analysis of gut microbiome and metabolites revealed novel mechanisms of intestinal Salmonella carriage in chicken. Sci. Rep. 2020, 10, 4809. [Google Scholar] [CrossRef]
- Kogut, M.H.; Arsenault, R.J. Immunometabolic Phenotype Alterations Associated with the Induction of Disease Tolerance and Persistent Asymptomatic Infection of Salmonella in the Chicken Intestine. Front. Immunol. 2017, 8, 372. [Google Scholar] [CrossRef] [PubMed]
- Arsenault, R.J.; Napper, S.; Kogut, M.H. Salmonella enterica Typhimurium infection causes metabolic changes in chicken muscle involving AMPK, fatty acid and insulin/mTOR signaling. Vet. Res. 2013, 44, 35. [Google Scholar] [CrossRef]
- Wu, X.-M.; Yang, X.; Fan, X.-C.; Chen, X.; Wang, Y.-X.; Zhang, L.-X.; Song, J.-K.; Zhao, G.-H. Serum metabolomics in chickens infected with Cryptosporidium baileyi. Parasit. Vectors 2021, 14, 336. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Dai, X.; Zhou, C.-C.; Li, K.-X.; Zhang, Y.-J.; Lou, X.-Y.; Zhu, Y.-M.; Sun, Y.-L.; Peng, B.-X.; Cui, W. Integrated analysis of the faecal metagenome and serum metabolome reveals the role of gut microbiome-associated metabolites in the detection of colorectal cancer and adenoma. Gut 2022, 71, 1315–1325. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Lai, J.; Zhang, P.; Ding, J.; Jiang, J.; Liu, C.; Huang, H.; Zhen, H.; Xi, C.; Sun, Y.; et al. Multi-omics analyses of serum metabolome, gut microbiome and brain function reveal dysregulated microbiota-gut-brain axis in bipolar depression. Mol. Psychiatry 2022, 27, 4123–4135. [Google Scholar] [CrossRef]
- Zhao, F.; An, R.; Wang, L.; Shan, J.; Wang, X. Specific Gut Microbiome and Serum Metabolome Changes in Lung Cancer Patients. Front. Cell Infect. Microbiol. 2021, 11, 725284. [Google Scholar] [CrossRef]
- Bearson, S.M.D.; Allen, H.K.; Bearson, B.L.; Looft, T.; Brunelle, B.W.; Kich, J.D.; Tuggle, C.K.; Bayles, D.O.; Alt, D.; Levine, U.Y.; et al. Profiling the gastrointestinal microbiota in response to Salmonella: Low versus high Salmonella shedding in the natural porcine host. Infect. Genet. Evol. 2013, 16, 330–340. [Google Scholar] [CrossRef]
- Kim, H.B.; Isaacson, R.E. Salmonella in Swine: Microbiota Interactions. Annu. Rev. Anim. Biosci. 2017, 5, 43–63. [Google Scholar] [CrossRef]
- Surendran Nair, M.; Yao, D.; Chen, C.; Pieters, M. Serum metabolite markers of early Mycoplasma hyopneumoniae infection in pigs. Vet. Res. 2019, 50, 98. [Google Scholar] [CrossRef]
- Jurburg, S.D.; Bossers, A. Age Matters: Community Assembly in the Pig Fecal Microbiome in the First Month of Life. Front. Microbiol. 2021, 12, 564408. [Google Scholar] [CrossRef]
- Ke, S.; Fang, S.; He, M.; Huang, X.; Yang, H.; Yang, B.; Chen, C.; Huang, L. Age-based dynamic changes of phylogenetic composition and interaction networks of health pig gut microbiome feeding in a uniformed condition. BMC Vet. Res. 2019, 15, 172. [Google Scholar] [CrossRef] [PubMed]
- Beloeil, P.-A.; Chauvin, C.; Proux, K.; Fablet, C.; Madec, F.; Alioum, A. Risk factors for Salmonella seroconversion of fattening pigs in farrow-to-finish herds. Vet. Res. 2007, 38, 835–848. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cevallos-Almeida, M.; Fablet, C.; Houdayer, C.; Dorenlor, V.; Eono, F.; Denis, M.; Kerouanton, A. Longitudinal study describing time to Salmonella seroconversion in piglets on three farrow-to-finish farms. Vet. Rec. Open 2019, 6, e000287. [Google Scholar] [CrossRef] [PubMed]
- Lo Fo Wong, D.M.A.; Dahl, J.; Stege, H.; van der Wolf, P.J.; Leontides, L.; von Altrock, A.; Thorberg, B.M. Herd-level risk factors for subclinical Salmonella infection in European finishing-pig herds. Prev. Vet. Med. 2004, 62, 253–266. [Google Scholar] [CrossRef] [PubMed]
- Cevallos-Almeida, M.; Martin, L.; Houdayer, C.; Rose, V.; Guionnet, J.M.; Paboeuf, F.; Denis, M. Kerouanton. Experimental infection of pigs by Salmonella Derby, S. Typhimurium and monophasic variant of S. Typhimurium: Comparison of colonization and serology. Vet. Microbiol. 2019, 231, 147–153. [Google Scholar] [CrossRef] [PubMed]


| Ionization Mode | Animal Shedding Status Comparison | DPI | R2Y | Q2 | pR2Y | pQ2 |
|---|---|---|---|---|---|---|
| Positive | Infected/Noninfected | 1 | 0.997 | 0.955 | 0.002 | 0.002 |
| 21 | 0.966 | 0.644 | 0.098 | 0.004 | ||
| High shedders/Low shedders/Noninfected | 1 | 0.989 | 0.755 | 0.002 | 0.002 | |
| 21 | 0.872 | 0.419 | 0.01 | 0.002 | ||
| High shedders/Noninfected | 1 | 0.99 | 0.891 | 0.004 | 0.002 | |
| 21 | 0.996 | 0.864 | 0.002 | 0.002 | ||
| Low shedders/Noninfected | 1 | 0.999 | 0.955 | 0.004 | 0.002 | |
| 21 | 1 | 0.719 | 0.002 | 0.062 | ||
| High shedders/Low Shedders | 1 | 0.99 | 0.696 | 0.116 | 0.02 | |
| 21 | 0.997 | 0.708 | 0.238 | 0.008 | ||
| Negative | Infected/Noninfected | 1 | 0.996 | 0.925 | 0.014 | 0.002 |
| 21 | 0.989 | 0.754 | 0.018 | 0.004 | ||
| High shedders/Low shedders/Noninfected | 1 | 0.98 | 0.757 | 0.002 | 0.002 | |
| 21 | 0.987 | 0.739 | 0.002 | 0.002 | ||
| High shedders/Noninfected | 1 | 0.992 | 0.916 | 0.012 | 0.002 | |
| 21 | 0.997 | 0.891 | 0.014 | 0.002 | ||
| Low shedders/Noninfected | 1 | 0.994 | 0.895 | 0.02 | 0.002 | |
| 21 | 0.998 | 0.724 | 0.016 | 0.044 | ||
| High shedders/Low Shedders | 1 | 0.995 | 0.823 | 0.084 | 0.002 | |
| 21 | 0.995 | 0.708 | 0.024 | 0.002 |
| Ionization Mode | Animal Shedding Status | R2Y | Q2 | pR2Y | pQ2 |
|---|---|---|---|---|---|
| Positive | Noninfected | 0.996 | 0.535 | 0.266 | 0.12 |
| High shedders | 0.997 | 0.939 | 0.002 | 0.002 | |
| Low shedders | 0.99 | 0.92 | 0.002 | 0.002 | |
| Negative | Noninfected | 0.989 | 0.562 | 0.162 | 0.062 |
| High shedders | 0.988 | 0.918 | 0.002 | 0.002 | |
| Low shedders | 0.993 | 0.931 | 0.002 | 0.002 |
| Ionisation Mode | Animal Shedding Status | DPI | R2Y | Q2 | pRY2 | pQ2 |
|---|---|---|---|---|---|---|
| Positive | All shedders | 1 | 0.993 | 0.802 | 0.022 | 0.002 |
| 21 | 0.996 | 0.633 | 0.43 | 0.026 | ||
| High shedders | 1 | 0.998 | 0.728 | 0.15 | 0.07 | |
| 21 | 0.995 | 0.489 | 0.272 | 0.152 | ||
| Low shedders | 1 | 0.965 | 0.245 | 0.614 | 0.682 | |
| 21 | 0.993 | 0.527 | 0.648 | 0.28 | ||
| Negative | All shedders | 1 | 0.897 | 0.463 | 0.094 | 0.026 |
| 21 | 0.944 | 0.371 | 0.556 | 0.088 | ||
| High shedders | 1 | 0.994 | 0.741 | 0.202 | 0.036 | |
| 21 | 0.989 | 0.518 | 0.132 | 0.176 | ||
| Low shedders | 1 | 0.921 | 0.311 | 0.492 | 0.548 | |
| 21 | 0.995 | 0.433 | 0.996 | 0.596 |
| Days Postinfection | All Positive | High Shedder | Low Shedder | Negative | |
|---|---|---|---|---|---|
| Observed OTUs | 1 DPI | 409 a | 433 | 381 b | 459 ab |
| 21 DPI | 620 a | 606 b | 635 | 665 ab | |
| Inverted Simpson’s | 1 DPI | 12.23 | 13.58 | 10.64 | 12.07 |
| 21 DPI | 31.974 | 32.91 | 30.95 | 28.34 | |
| Shannon evenness | 1 DPI | 0.635 | 0.645 | 0.623 | 0.6414 |
| 21 DPI | 0.732 | 0.728 | 0.736 | 0.737 |
| Animal Infection Status Comparison | DPI | PERMANOVA (p-Value) |
|---|---|---|
| Infected/Noninfected | 1 | 0.037 |
| 21 | 0.018 | |
| High shedders/Low shedders/noninfected | 1 | 0.119 |
| 21 | 0.060 | |
| High shedders/Noninfected | 1 | 0.071 |
| 21 | 0.048 | |
| Low shedders/Noninfected | 1 | 0.071 |
| 21 | 0.048 | |
| High shedders/Low shedders | 1 | 0.785 |
| 21 | 0.539 |
| Shedding Level | 1 DPI | 21 DPI | p-Value | |
|---|---|---|---|---|
| Observed OTUs | High shedders | 433 | 606 | 3 × 10−5 |
| Low shedders | 381 | 635 | 3 × 10−6 | |
| Noninfected | 459 | 665 | 0.008 | |
| Inverted Simpson’s | High shedders | 13.58 | 32.91 | 0.001 |
| Low shedders | 10.64 | 30.95 | 6 × 10−6 | |
| Noninfected | 12.07 | 28.34 | 0.008 | |
| Shannon evenness | High shedders | 0.645 | 0.728 | 6 × 10−4 |
| Low shedders | 0.623 | 0.736 | 6 × 10−6 | |
| Noninfected | 0.641 | 0.737 | 0.008 |
| Animal Infection Status | PERMANOVA (p-Value) |
|---|---|
| Noninfected | 0.007 |
| High shedders | 0.000 |
| Low shedders | 0.000 |
| Days Postinfection | Seronegative | Seropositive | ||
|---|---|---|---|---|
| Observed OTUs | Infected pigs | 1 DPI | 417 | 403 |
| 21 DPI | 630 | 613 | ||
| High shedder | 1 DPI | 417 | 446 | |
| 21 DPI | 618 | 599 | ||
| Low shedder | 1 DPI | 416 | 352 | |
| 21 DPI | 639 | 632 | ||
| Inverted Simpson’s | Infected pigs | 1 DPI | 10.52 | 13.68 |
| 21 DPI | 32.68 | 31.52 | ||
| High shedder | 1 DPI | 9.6 | 16.98 | |
| 21 DPI | 32.49 | 33.13 | ||
| Low shedder | 1 DPI | 11.63 | 9.82 | |
| 21 DPI | 32.83 | 29.39 | ||
| Shannon evenness | Infected pigs | 1 DPI | 0.625 | 0.643 |
| 21 DPI | 0.737 | 0.728 | ||
| High shedder | 1 DPI | 0.612 | 0.673 | |
| 21 DPI | 0.730 | 0.727 | ||
| Low shedder | 1 DPI | 0.639 | 0.609 | |
| 21 DPI | 0.743 | 0.730 |
| Animal Shedding Status | PERMANOVA (p-Value) | |
|---|---|---|
| 1 DPI | 21 DPI | |
| All shedders | 0.619 | 0.198 |
| High shedders | 0.231 | 0.927 |
| Low shedders | 0.783 | 0.123 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Larivière-Gauthier, G.; Kerouanton, A.; Mompelat, S.; Bougeard, S.; Denis, M.; Fravalo, P. Monophasic Variant of Salmonella Typhimurium Infection Affects the Serum Metabolome in Swine. Microorganisms 2023, 11, 2565. https://doi.org/10.3390/microorganisms11102565
Larivière-Gauthier G, Kerouanton A, Mompelat S, Bougeard S, Denis M, Fravalo P. Monophasic Variant of Salmonella Typhimurium Infection Affects the Serum Metabolome in Swine. Microorganisms. 2023; 11(10):2565. https://doi.org/10.3390/microorganisms11102565
Chicago/Turabian StyleLarivière-Gauthier, Guillaume, Annaëlle Kerouanton, Sophie Mompelat, Stéphanie Bougeard, Martine Denis, and Philippe Fravalo. 2023. "Monophasic Variant of Salmonella Typhimurium Infection Affects the Serum Metabolome in Swine" Microorganisms 11, no. 10: 2565. https://doi.org/10.3390/microorganisms11102565
APA StyleLarivière-Gauthier, G., Kerouanton, A., Mompelat, S., Bougeard, S., Denis, M., & Fravalo, P. (2023). Monophasic Variant of Salmonella Typhimurium Infection Affects the Serum Metabolome in Swine. Microorganisms, 11(10), 2565. https://doi.org/10.3390/microorganisms11102565






