CPGminer: An Interactive Dashboard to Explore the Genomic Features and Taxonomy of Complete Prokaryotic Genomes
Abstract
1. Introduction
2. Materials and Methods
2.1. Programming Language and Library
2.2. Customized CPG Metadata
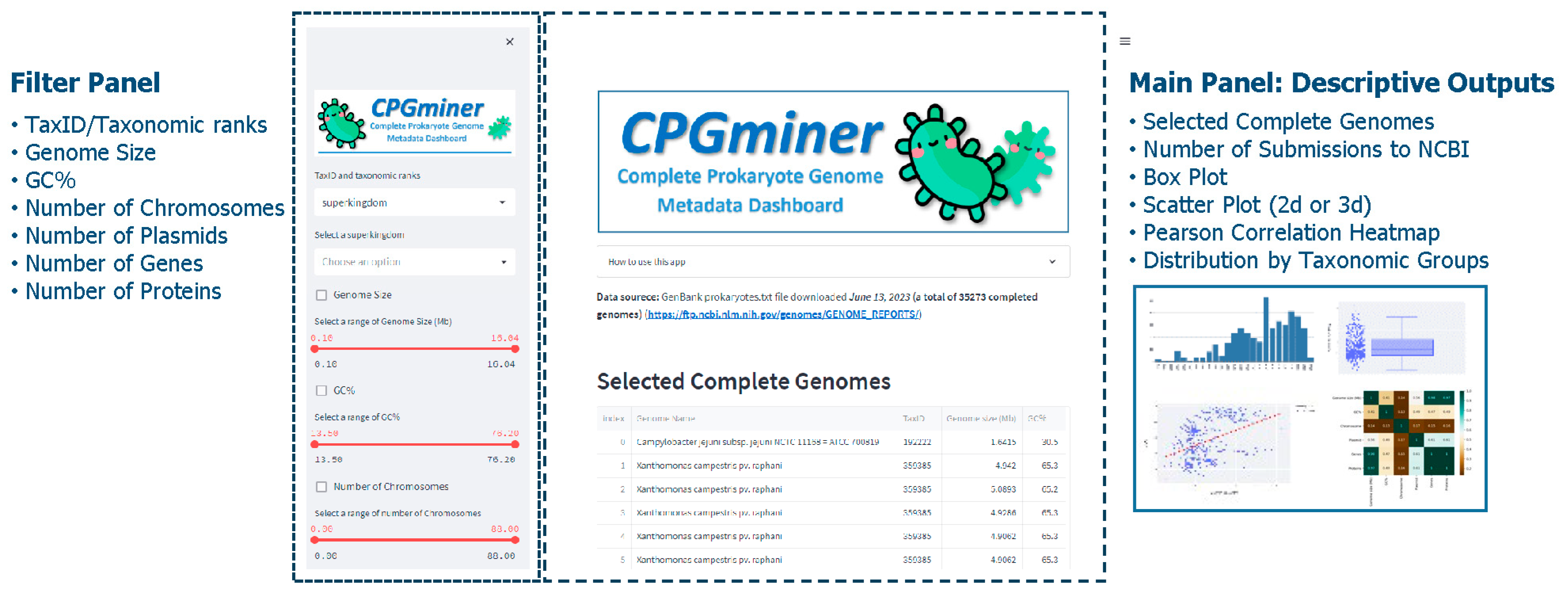
2.3. User Interface
- Selected complete genomes: A downloadable table of selected genomes with 17 metadata fields including genome name, taxID, genome size, GC%, replicons, numbers of chromosomes, plasmids, genes, and proteins, release date, genome download FTP address, and taxonomic information fields from superkingdom to species. This table serves as underlying data for the subsequent plots.
- Box plot for genomic features: genome size, GC%, and numbers of chromosomes, plasmids, genes, and proteins.
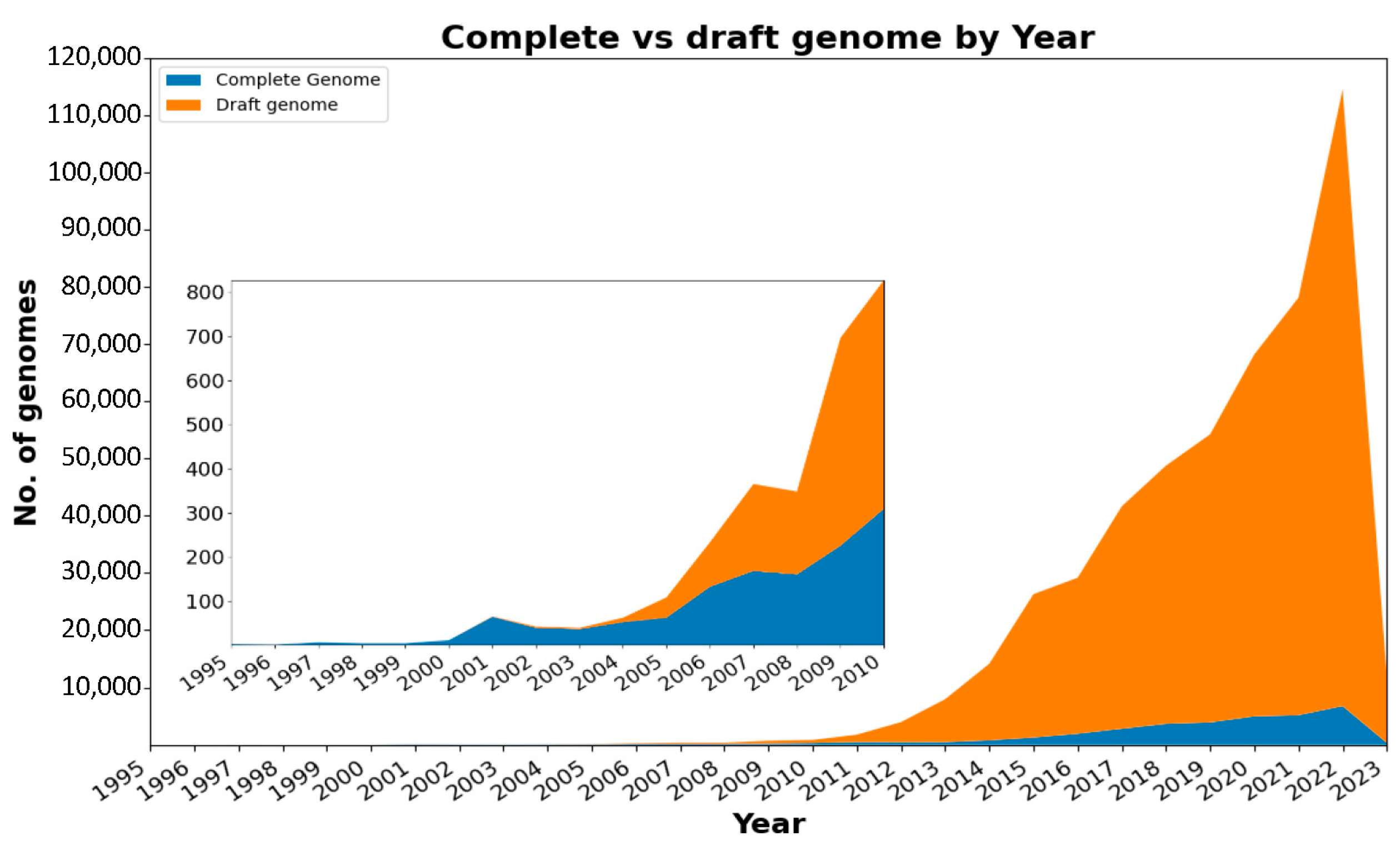
- Number of submissions to NCBI by year.
- Scatter plot for genomic features: Users can select two or three genomic features and it will display a 2D or 3D scatter plot depending on the number of genomic features users select.
- Pearson correlation heatmap of genomic features.
- Distribution by taxonomic groups: A downloadable table of selected genomes that shows their count in each taxonomical grouping. Users can select one level from superkingdom to species and the resulting table displays all available taxonomical categories to the selected level and its count.
3. Results and Discussion
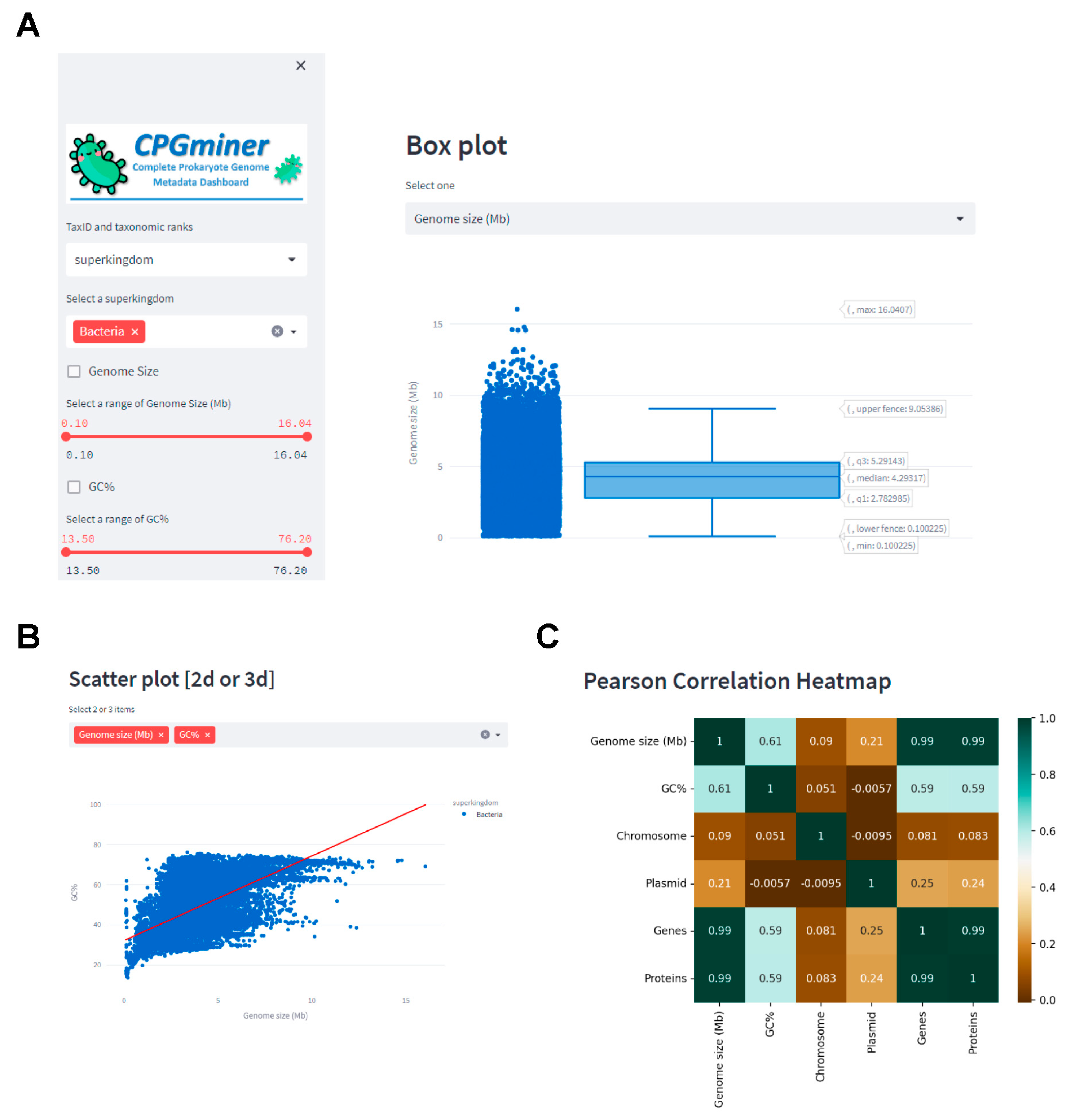
3.1. Question 1: What Are the Median Values of Genome Size and Guanine + Cytosine (GC) Content of Bacterial Genomes and the Correlation between Them?
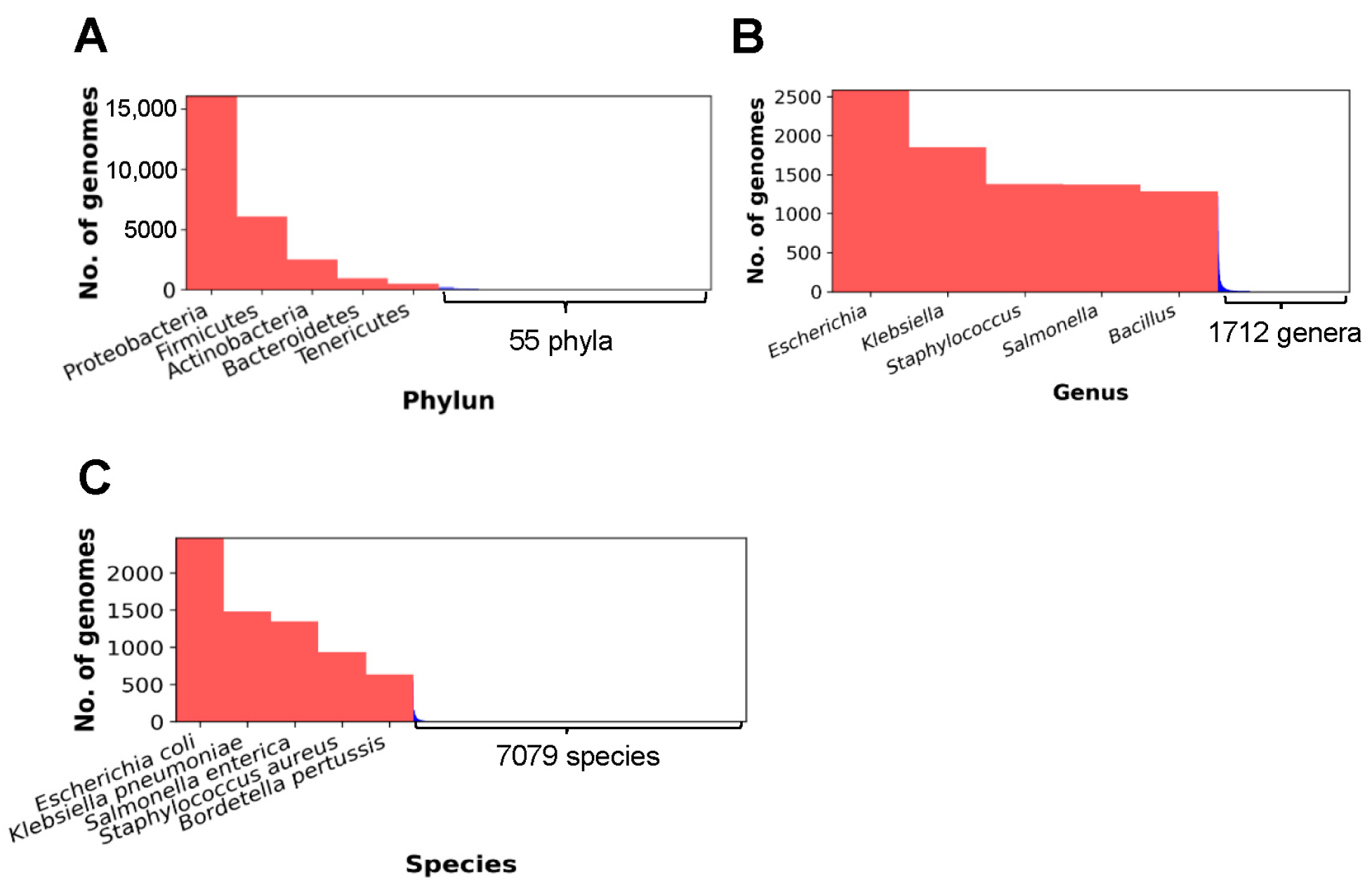
3.2. Question 2: Which Phyla, Genera, and Species of Bacteria Represent the Top Five Complete Genome Sequences?
3.3. Question 3: What Percentage of Prokaryotes Have Multiple Chromosomes? What Percentage of Prokaryotes Have Plasmid(s)?
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Torsvik, V.; Ovreas, L.; Thingstad, T.F. Prokaryotic diversity—Magnitude, dynamics, and controlling factors. Science 2002, 296, 1064–1066. [Google Scholar] [CrossRef] [PubMed]
- Fleischmann, R.D.; Adams, M.D.; White, O.; Clayton, R.A.; Kirkness, E.F.; Kerlavage, A.R.; Bult, C.J.; Tomb, J.F.; Dougherty, B.A.; Merrick, J.M.; et al. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science 1995, 269, 496–512. [Google Scholar] [CrossRef] [PubMed]
- Bult, C.J.; White, O.; Olsen, G.J.; Zhou, L.; Fleischmann, R.D.; Sutton, G.G.; Blake, J.A.; FitzGerald, L.M.; Clayton, R.A.; Gocayne, J.D.; et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 1996, 273, 1058–1073. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, J.; Wang, J.; Wang, J.; Li, Y. Estimate of the sequenced proportion of the global prokaryotic genome. Microbiome 2020, 8, 134. [Google Scholar] [CrossRef]
- Land, M.; Hauser, L.; Jun, S.R.; Nookaew, I.; Leuze, M.R.; Ahn, T.H.; Karpinets, T.; Lund, O.; Kora, G.; Wassenaar, T.; et al. Insights from 20 years of bacterial genome sequencing. Funct. Integr. Genom. 2015, 15, 141–161. [Google Scholar] [CrossRef] [PubMed]
- Holmes, C.; Carlson, S.M.; McDonald, F.; Jones, M.; Graham, J. Exploring the post-genomic world: Differing explanatory and manipulatory functions of post-genomic sciences. New Genet. Soc. 2016, 35, 49–68. [Google Scholar] [CrossRef] [PubMed]
- Ko, K.K.K.; Chng, K.R.; Nagarajan, N. Metagenomics-enabled microbial surveillance. Nat. Microbiol. 2022, 7, 486–496. [Google Scholar] [CrossRef]
- Ochman, H.; Davalos, L.M. The nature and dynamics of bacterial genomes. Science 2006, 311, 1730–1733. [Google Scholar] [CrossRef]
- Mardis, E. What is Finished, and Why Does it Matter. Genome Res. 2002, 12, 669–671. [Google Scholar] [CrossRef]
- Prentice, M.B. Bacterial comparative genomics. Genome Biol. 2004, 5, 338. [Google Scholar] [CrossRef][Green Version]
- Karp, P.D.; Ivanova, N.; Krummenacker, M.; Kyrpides, N.; Latendresse, M.; Midford, P.; Ong, W.K.; Paley, S.; Seshadri, R. A Comparison of Microbial Genome Web Portals. Front. Microbiol. 2019, 10, 208. [Google Scholar] [CrossRef] [PubMed]
- Napoles-Duarte, J.M.; Biswas, A.; Parker, M.I.; Palomares-Baez, J.P.; Chavez-Rojo, M.A.; Rodriguez-Valdez, L.M. Stmol: A component for building interactive molecular visualizations within streamlit web-applications. Front. Mol. Biosci. 2022, 9, 990846. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Lin, J.; Prokop, A.; Gopalakrishnan, V.; Hanna, R.N.; Papa, E.; Freeman, A.; Patel, S.; Yu, W.; Huhn, M.; et al. StarGazer: A Hybrid Intelligence Platform for Drug Target Prioritization and Digital Drug Repositioning Using Streamlit. Front. Genet. 2022, 13, 868015. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, Analysis, and Visualization of Phylogenomic Data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef]
- Lassalle, F.; Perian, S.; Bataillon, T.; Nesme, X.; Duret, L.; Daubin, V. GC-Content evolution in bacterial genomes: The biased gene conversion hypothesis expands. PLoS Genet. 2015, 11, e1004941. [Google Scholar] [CrossRef] [PubMed]
- Hu, E.Z.; Lan, X.R.; Liu, Z.L.; Gao, J.; Niu, D.K. A positive correlation between GC content and growth temperature in prokaryotes. BMC Genom. 2022, 23, 110. [Google Scholar] [CrossRef]
- Meziti, A.; Rodriguez, R.L.; Hatt, J.K.; Pena-Gonzalez, A.; Levy, K.; Konstantinidis, K.T. The Reliability of Metagenome-Assembled Genomes (MAGs) in Representing Natural Populations: Insights from Comparing MAGs against Isolate Genomes Derived from the Same Fecal Sample. Appl. Environ. Microbiol. 2021, 87, e02593-20. [Google Scholar] [CrossRef]
- Mackenzie, C.; Simmons, A.E.; Kaplan, S. Multiple chromosomes in bacteria. The yin and yang of trp gene localization in Rhodobacter sphaeroides 2.4.1. Genetics 1999, 153, 525–538. [Google Scholar] [CrossRef]
- Jha, J.K.; Baek, J.H.; Venkova-Canova, T.; Chattoraj, D.K. Chromosome dynamics in multichromosome bacteria. Biochim. Biophys. Acta 2012, 1819, 826–829. [Google Scholar] [CrossRef]
- diCenzo, G.C.; Finan, T.M. The Divided Bacterial Genome: Structure, Function, and Evolution. Microbiol. Mol. Biol. Rev. 2017, 81, e00019-17. [Google Scholar] [CrossRef]
- Fournes, F.; Val, M.E.; Skovgaard, O.; Mazel, D. Replicate Once Per Cell Cycle: Replication Control of Secondary Chromosomes. Front. Microbiol. 2018, 9, 1833. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Liao, Q.; Karaboja, X.; Barton, I.S.; Schantz, E.G.; Mejia-Santana, A.; Fuqua, C.; Wang, X. Conformation and dynamic interactions of the multipartite genome in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 2022, 119, e2115854119. [Google Scholar] [CrossRef] [PubMed]
- Adam, Y.; Brezellec, P.; Espinosa, E.; Besombes, A.; Naquin, D.; Paly, E.; Possoz, C.; van Dijk, E.; Barre, F.X.; Ferat, J.L. Plesiomonas shigelloides, an Atypical Enterobacterales with a Vibrio-Related Secondary Chromosome. Genome Biol. Evol. 2022, 14, evac011. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Yoon, S.; Kondakala, S.; Foley, S.L.; Hart, M.; Baek, D.-H.; Wang, W.; Kim, S.-K.; Sutherland, J.B.; Kim, S.-J.; et al. CPGminer: An Interactive Dashboard to Explore the Genomic Features and Taxonomy of Complete Prokaryotic Genomes. Microorganisms 2023, 11, 2556. https://doi.org/10.3390/microorganisms11102556
Kim J, Yoon S, Kondakala S, Foley SL, Hart M, Baek D-H, Wang W, Kim S-K, Sutherland JB, Kim S-J, et al. CPGminer: An Interactive Dashboard to Explore the Genomic Features and Taxonomy of Complete Prokaryotic Genomes. Microorganisms. 2023; 11(10):2556. https://doi.org/10.3390/microorganisms11102556
Chicago/Turabian StyleKim, Jaehyun, Sunghyun Yoon, Sandeep Kondakala, Steven L. Foley, Mark Hart, Dong-Heon Baek, Wenjun Wang, Sung-Kwan Kim, John B. Sutherland, Seong-Jae Kim, and et al. 2023. "CPGminer: An Interactive Dashboard to Explore the Genomic Features and Taxonomy of Complete Prokaryotic Genomes" Microorganisms 11, no. 10: 2556. https://doi.org/10.3390/microorganisms11102556
APA StyleKim, J., Yoon, S., Kondakala, S., Foley, S. L., Hart, M., Baek, D.-H., Wang, W., Kim, S.-K., Sutherland, J. B., Kim, S.-J., & Kweon, O. (2023). CPGminer: An Interactive Dashboard to Explore the Genomic Features and Taxonomy of Complete Prokaryotic Genomes. Microorganisms, 11(10), 2556. https://doi.org/10.3390/microorganisms11102556








