Analysis of Microbial Diversity in South Shetland Islands and Antarctic Peninsula Soils Based on Illumina High-Throughput Sequencing and Cultivation-Dependent Techniques
Abstract
1. Introduction
2. Materials and Methods
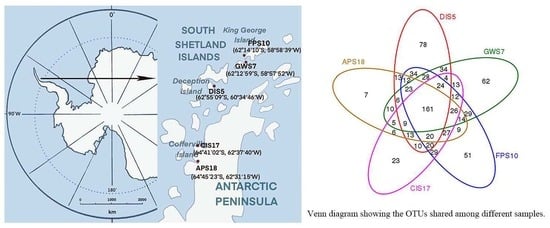
2.1. Sample Collection
2.2. DNA Extraction, PCR Amplification and Sequencing
2.3. Sequence Processing and Analyses
2.4. Statistical Analysis
2.5. Isolation, Purification and Preservation of Strains
2.6. Psychrophilic Bacteria Screening
2.7. 16S rRNA Gene Sequencing and Data Analysis of Isolated Strains
3. Results
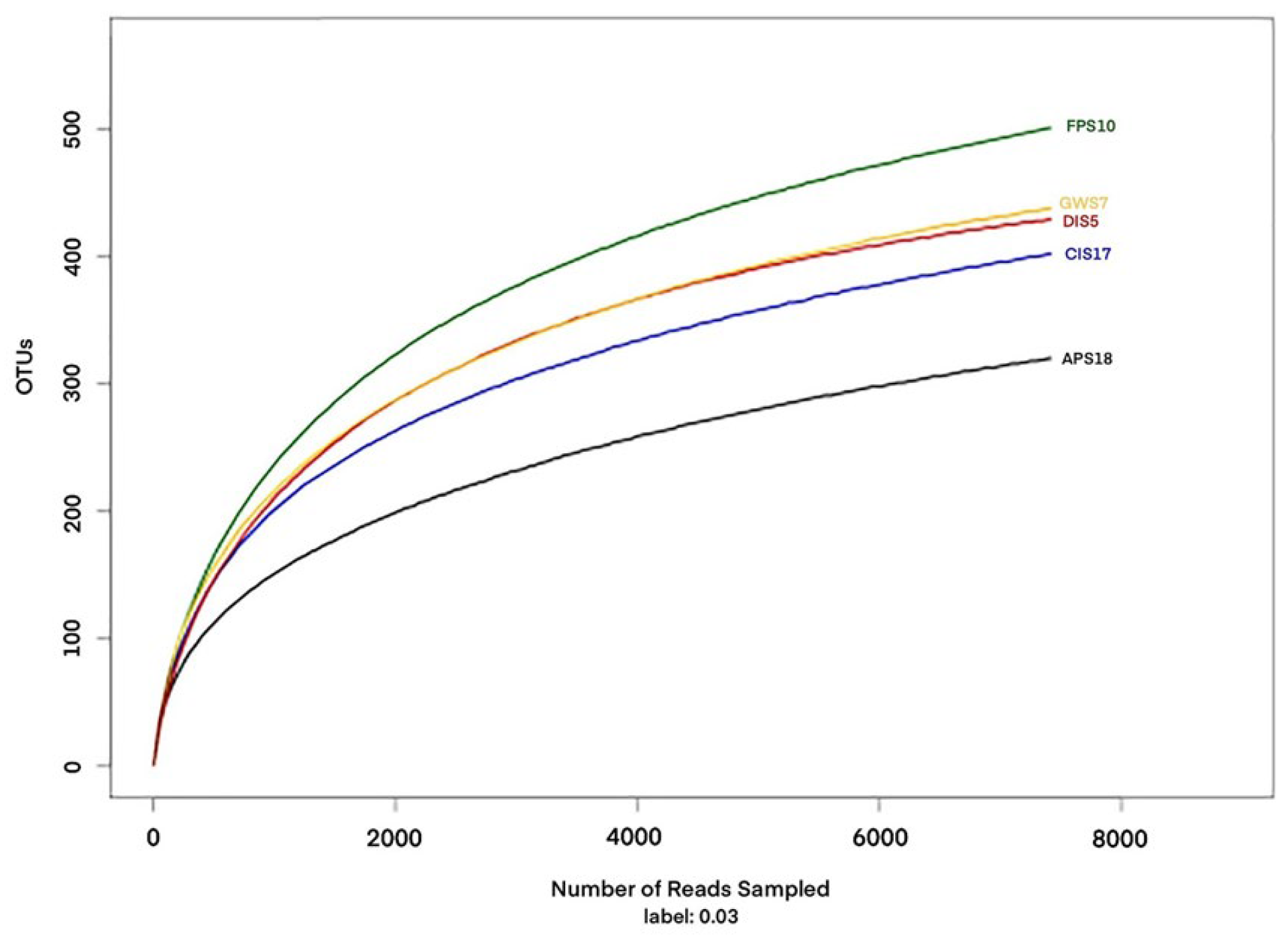
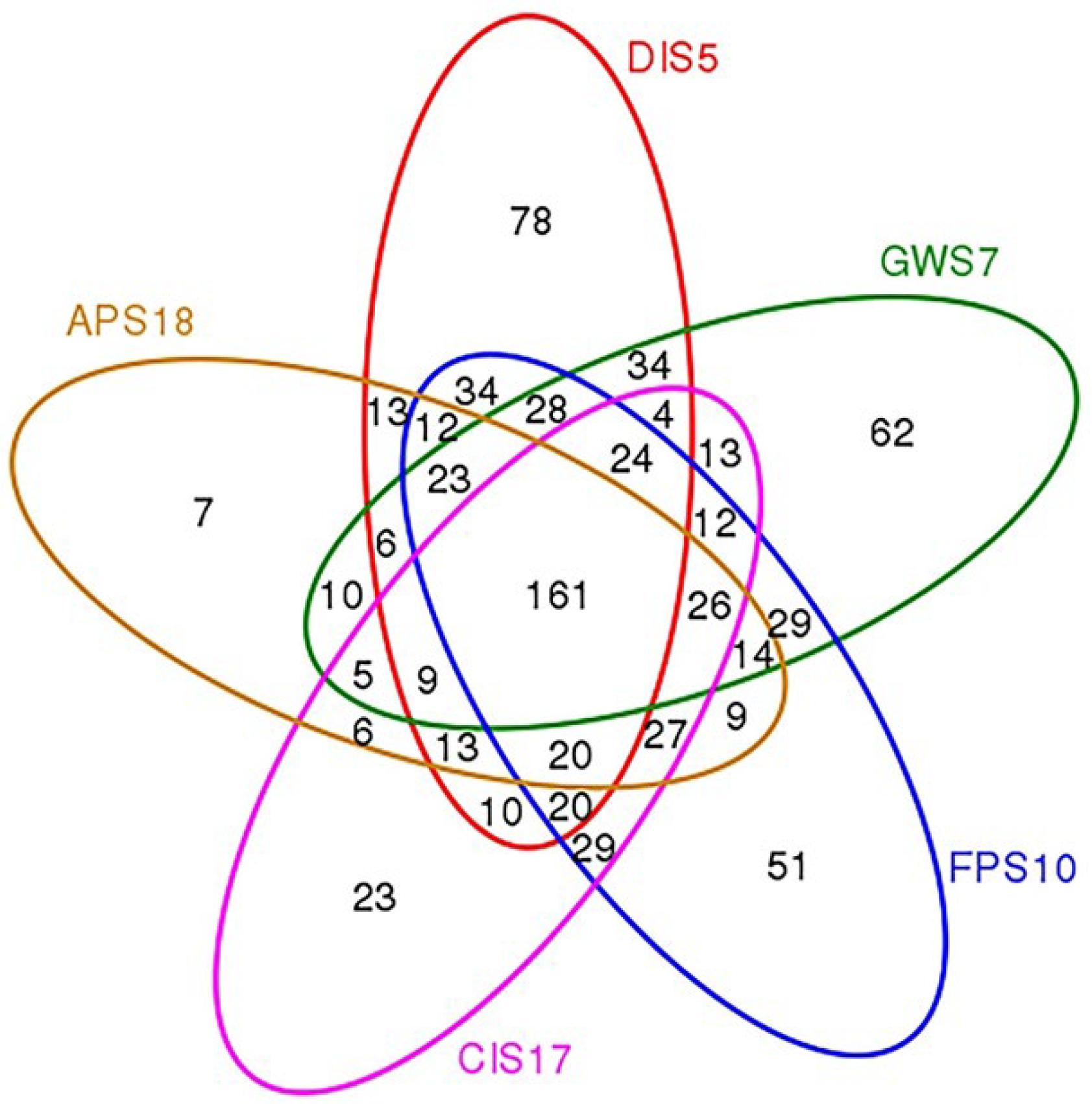
3.1. OTU Clustering and Annotation
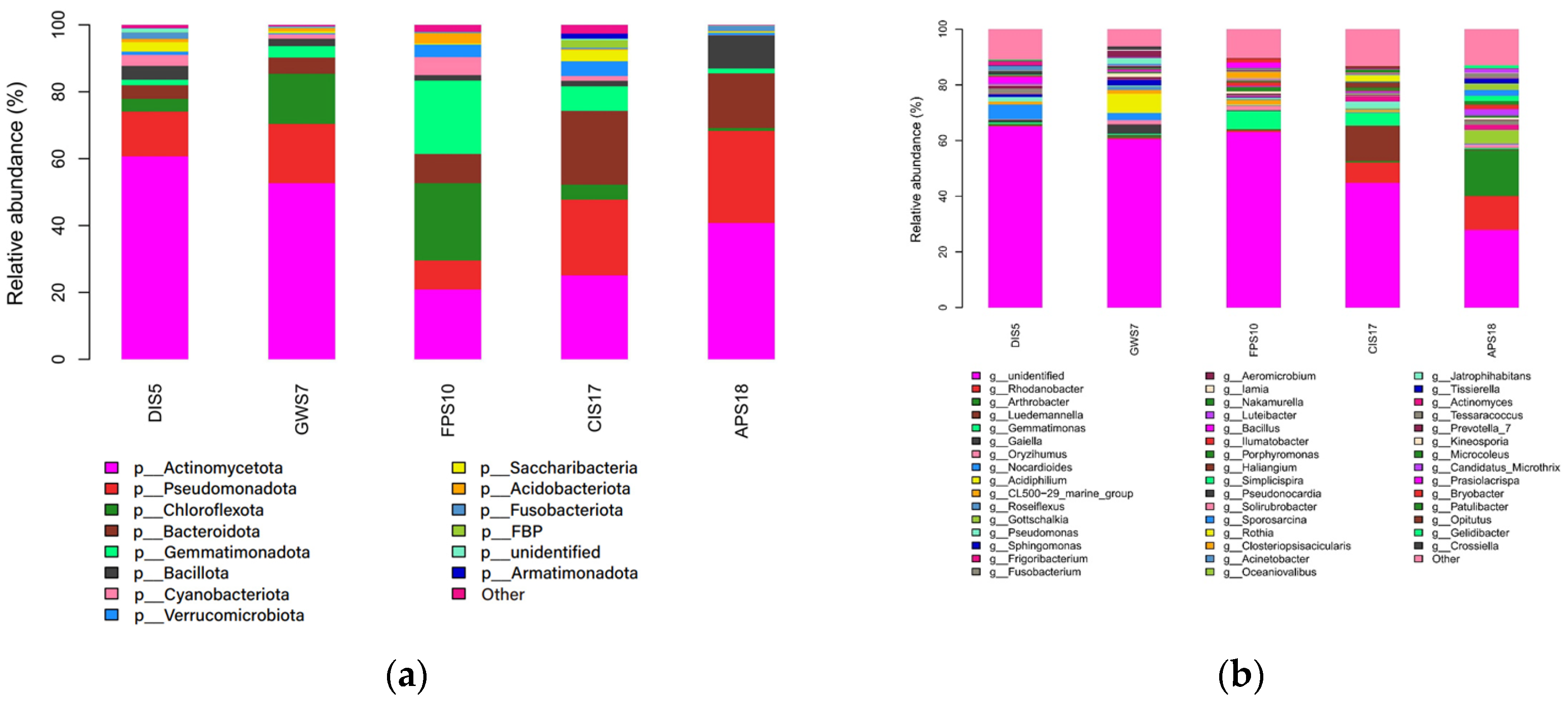
3.2. Composition and Relative Abundance of Microbiota
3.3. Isolation of Cultivable Microorganisms
3.4. Psychrophilic Members in Antarctic Soils
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Cary, S.C.; McDonald, I.R.; Barrett, J.E.; Cowan, D.A. On the rocks: The microbiology of Antarctic Dry Valley soils. Nat. Rev. Microbiol. 2010, 8, 129–138. [Google Scholar] [CrossRef]
- Niederberger, T.D.; McDonald, I.R.; Hacker, A.L.; Soo, R.M.; Barrett, J.E.; Wall, D.H.; Cary, S.C. Microbial community composition in soils of Northern Victoria Land, Antarctica. Environ. Microbiol. 2008, 10, 1713–1724. [Google Scholar] [CrossRef]
- de la Torre, J.R.; Goebel, B.M.; Friedmann, E.I.; Pace, N.R. Microbial diversity of cryptoendolithic communities from the McMurdo Dry Valleys, Antarctica. Appl. Environ. Microbiol. 2003, 69, 3858–3867. [Google Scholar] [CrossRef]
- Ganzert, L.; Lipski, A.; Hubberten, H.W.; Wagner, D. The impact of different soil parameters on the community structure of dominant bacteria from nine different soils located on Livingston Island, South Shetland Archipelago, Antarctica. FEMS Microbiol. Ecol. 2011, 76, 476–491. [Google Scholar] [CrossRef]
- Roesch, L.F.W.; Fulthorpe, R.R.; Pereira, A.B.; Pereira, C.K.; Lemos, L.N.; Barbosa, A.D.; Suleiman, A.K.A.; Gerber, A.L.; Pereira, M.G.; Loss, A.; et al. Soil bacterial community abundance and diversity in ice-free areas of Keller Peninsula, Antarctica. Appl. Soil Ecol. 2012, 61, 7–15. [Google Scholar] [CrossRef]
- Ruisi, S.; Barreca, D.; Selbmann, L.; Zucconi, L.; Onofri, S. Fungi in Antarctica. Rev. Environ. Sci. Bio/Technol. 2007, 6, 127–141. [Google Scholar] [CrossRef]
- Holdgate, M.W. Terrestrial Ecosystems in the Antarctic. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 1977, 279, 5–25. [Google Scholar]
- Xiao, C.; Dali, L.; Peijin, Z. Preliminary approach on the soil microbial ecological effect in the Great Wall Station area, Antarctic. Biodiv Sci. 1995, 3, 134–138. [Google Scholar]
- Baker, P.E.; McReath, I.; Harvey, M.R.; Roobol, M.J.; Davies, T.G. The Geology of the South. Shetland Islands: V. Volcanic Evolution of Deception Island; British Antarctic Survey Scientific Reports; British Antarctic survey: Cambridge, UK, 1975; No. 78. [Google Scholar]
- Dowd, S.E.; Sun, Y.; Secor, P.R.; Rhoads, D.D.; Wolcott, B.M.; James, G.A.; Wolcott, R.D. Survey of bacterial diversity in chronic wounds using pyrosequencing, DGGE, and full ribosome shotgun sequencing. BMC Microbiol. 2008, 8, 43. [Google Scholar] [CrossRef]
- Poulsen, P.H.B.; Abu Al-Soud, W.; Bergmark, L.; Magid, J.; Hansen, L.H.; Sorensen, S.J. Effects of fertilization with urban and agricultural organic wastes in a field trial—Prokaryotic diversity investigated by pyrosequencing. Soil Biol. Biochem. 2013, 57, 784–793. [Google Scholar] [CrossRef]
- Wu, R.N.; Yu, M.L.; Meng, L.S.; Xu, X.; Yue, X.Q.; Wu, R. PCR-DGGE analysis of the microbial diversity in naturally fermented suan-cai from northeast China. Mod. Food Sci. Technol. 2014, 30, 8–12. [Google Scholar]
- Boucher, D.; Jardillier, L.; Debroas, D. Succession of bacterial community composition over two consecutive years in two aquatic systems: A natural lake and a lake-reservoir. FEMS Microbiol. Ecol. 2006, 55, 79–97. [Google Scholar] [CrossRef] [PubMed]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C.; Haas, B.J.; Clemente, J.C.; Quince, C.; Knight, R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 2011, 27, 2194–2200. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Fouts, D.E.; Szpakowski, S.; Purushe, J.; Torralba, M.; Waterman, R.C.; MacNeil, M.D.; Alexander, L.J.; Nelson, K.E. Next generation sequencing to define prokaryotic and fungal diversity in the bovine rumen. PLoS ONE 2012, 7, e48289. [Google Scholar] [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Schloss, P.D.; Gevers, D.; Westcott, S.L. Reducing the effects of PCR amplification and sequencing artifacts on 16S rRNA-based studies. PLoS ONE 2011, 6, e27310. [Google Scholar] [CrossRef]
- Wang, Y.; Sheng, H.F.; He, Y.; Wu, J.Y.; Jiang, Y.X.; Tam, N.F.; Zhou, H.W. Comparison of the levels of bacterial diversity in freshwater, intertidal wetland, and marine sediments by using millions of illumina tags. Appl. Environ. Microbiol. 2012, 78, 8264–8271. [Google Scholar] [CrossRef]
- Jami, E.; Israel, A.; Kotser, A.; Mizrahi, I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 2013, 7, 1069–1079. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.T.; Peng, X.; Deng, G.H.; Sheng, H.F.; Wang, Y.; Zhou, H.W.; Tam, N.F. Illumina sequencing of 16S rRNA tag revealed spatial variations of bacterial communities in a mangrove wetland. Microb. Ecol. 2013, 66, 96–104. [Google Scholar] [CrossRef]
- Kuczynski, J.; Stombaugh, J.; Walters, W.A.; González, A.; Caporaso, J.G.; Knight, R. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr. Protoc. Bioinform. 2011, 27, 1E-5. [Google Scholar] [CrossRef] [PubMed]
- Köpke, B.; Wilms, R.; Engelen, B.; Cypionka, H.; Sass, H. Microbial Diversity in Coastal Subsurface Sediments: A Cultivation Approach Using Various Electron Acceptors and Substrate Gradients. Appl. Environ. Microbiol. 2005, 71, 7819–7830. [Google Scholar] [CrossRef] [PubMed]
- Alain, K.; Querellou, J. Cultivating the uncultured: Limits, advances and future challenges. Extremophiles 2009, 13, 583–594. [Google Scholar] [CrossRef]
- Kim, S.B.; Yoon, J.H.; Kim, H.; Lee, S.T.; Park, Y.H.; Goodfellow, M. A phylogenetic analysis of the genus Saccharomonospora conducted with 16S rRNA gene sequences. Int. J. Syst. Bacteriol. 1995, 45, 351–356. [Google Scholar] [CrossRef]
- Yadav, A.N.; Verma, P.; Kumar, M.; Pal, K.K.; Dey, R.; Gupta, A.; Padaria, J.C.; Gujar, G.T.; Kumar, S.; Suman, A.; et al. Diversity and phylogenetic profiling of niche-specific Bacilli from extreme environments of India. Ann. Microbiol. 2015, 65, 611–629. [Google Scholar] [CrossRef]
- Rainey, F.A.; Ward-Rainey, N.; Kroppenstedt, R.M.; Stackebrandt, E. The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: Proposal of Nocardiopsaceae fam. nov. Int. J. Syst. Bacteriol. 1996, 46, 1088–1092. [Google Scholar] [CrossRef]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Cowan, D.A.; Russell, N.J.; Mamais, A.; Sheppard, D.M. Antarctic Dry Valley mineral soils contain unexpectedly high levels of microbial biomass. Extremophiles 2002, 6, 431–436. [Google Scholar] [CrossRef]
- Brinkmeyer, R.; Knittel, K.; Jürgens, J.; Weyland, H.D.; Amann, R.I.; Helmke, E. Diversity and Structure of Bacterial Communities in Arctic versus Antarctic Pack Ice. Appl. Environ. Microbiol. 2003, 69, 6610–6619. [Google Scholar] [CrossRef] [PubMed]
- Tindall, B.J. Prokaryotic Diversity in the Antarctic: The Tip of the Iceberg. Microb. Ecol. 2004, 47, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Aislabie, J.; Chhour, K.-L.; Saul, D.J.; Miyauchi, S.; Ayton, J.; Paetzold, R.; Balks, M.R. Dominant bacteria in soils of Marble Point and Wright Valley, Victoria Land, Antarctica. Soil Biol. Biochem. 2006, 38, 3041–3056. [Google Scholar] [CrossRef]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64(Pt. 2), 346–351. [Google Scholar] [CrossRef]
- Ellis-Evans, J.C.; Laybourn-Parry, J.E.M.; Bayliss, P.; Perriss, S.J. Physical, chemical and microbial community characteristics of lakes of the Larsemann Hills, Continental Antarctica. Arch. Fur Hydrobiol. 1998, 141, 209–230. [Google Scholar] [CrossRef]
- Tiao, G.; Lee, C.K.; McDonald, I.R.; Cowan, D.A.; Cary, S.C. Rapid microbial response to the presence of an ancient relic in the Antarctic Dry Valleys. Nat. Commun. 2012, 3, 660. [Google Scholar] [CrossRef][Green Version]
- Dennis, P.G.; Newsham, K.K.; Rushton, S.P.; Ord, V.J.; O’donnell, A.G.; Hopkins, D.W. Warming constrains bacterial community responses to nutrient inputs in a southern, but not northern, maritime Antarctic soil. Soil Biol. Biochem. 2013, 57, 248–255. [Google Scholar] [CrossRef]
- Lei, X.; Sheng-guo, Z.; Nan, Z.; Song-li, L.; Jia-qi, W. Advance in isolation and culture techniques of uncultured microbes: A review. Microbiol. China 2017, 44, 3053–3066. [Google Scholar]
- Prasad, S.; Manasa, P.; Buddhi, S.; Tirunagari, P.; Begum, Z.; Rajan, S.; Shivaji, S. Diversity and bioprospective potential (cold-active enzymes) of cultivable marine bacteria from the subarctic glacial Fjord, Kongsfjorden. Curr. Microbiol. 2014, 68, 233–238. [Google Scholar] [CrossRef]
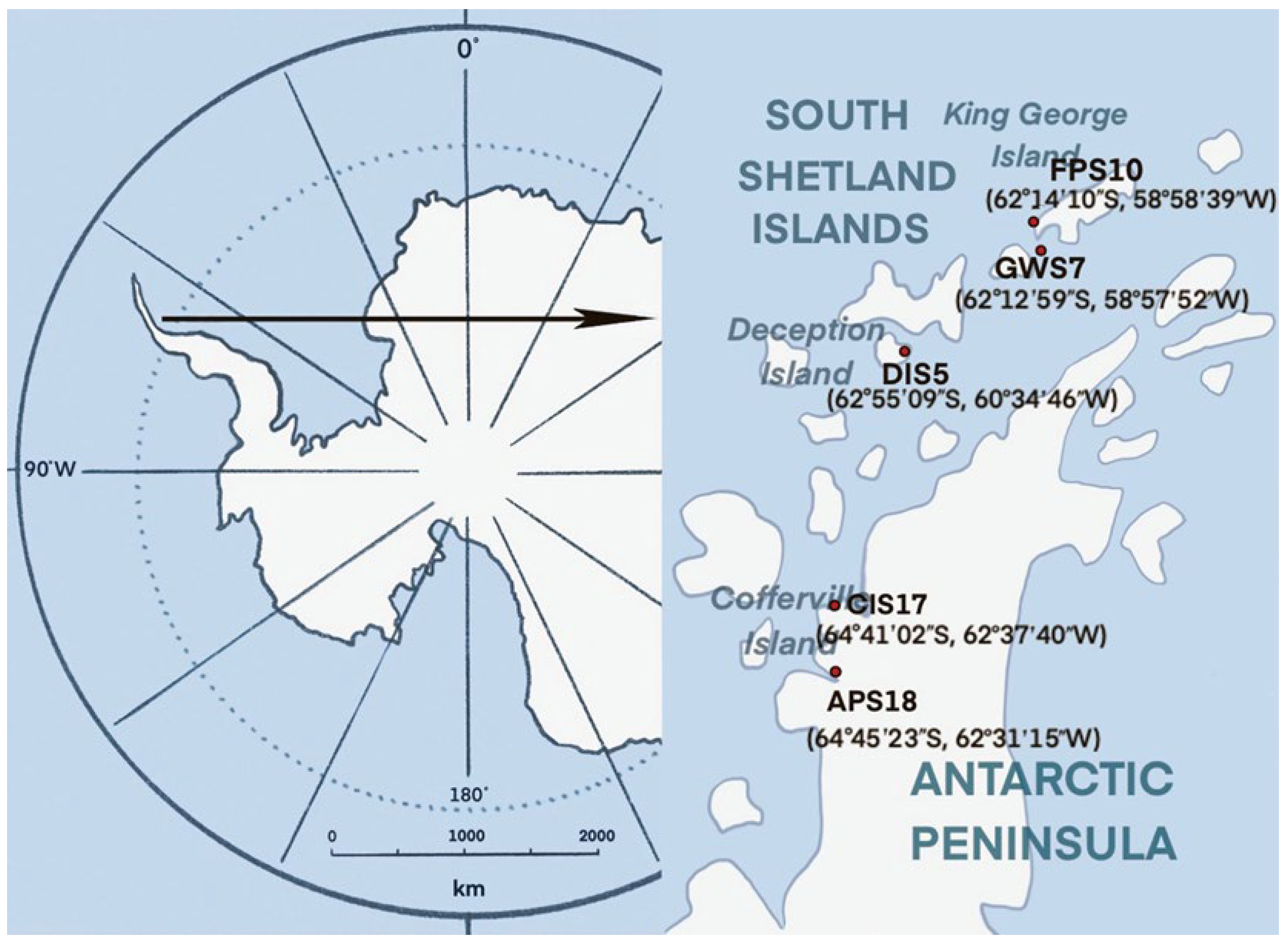


| Sample Name | Sample Color | Soil Sampling Location | Geographical Coordinates S/W |
|---|---|---|---|
| DIS5 | black | Deception Island (pendulum bay) | 62°55′09″/60°34′46″ |
| GWS7 | black brown | King George Island (Great Wall Station) | 62°12′59″/58°57′52″ |
| FPS10 | yellowish-brown | King George Island (the southernmost tip of the Fildes Peninsula) | 62°14′10″/58°58′39″ |
| CIS17 | black brown | Antarctic Peninsula (Cofferville island) | 64°41′02″/62°37′40″ |
| APS18 | brown | Antarctic Peninsula | 64°45′23″/62°31′15″ |
| Sample Name | Carbon Contents | Nitrogen Contents | pH | Main Chemical Elements |
|---|---|---|---|---|
| DIS5 | 1.19% | 0.16% | 6.8 | Fe, Mn |
| GWS7 | 0.73% | 0.09% | 6.3 | Al, Ca, Mg, Fe |
| FPS10 | 0.70% | 0.12% | 6.1 | Al, Cu, Fe |
| CIS17 | 10.96% | 1.32% | 5.8 | Al, Fe, Cu, Zn |
| APS18 | 9.50% | 1.33% | 6.0 | Al, Cu, Zn |
| Sample | Lean Tags | OTUs | Shannon | Chao1 | Coverage (%) |
|---|---|---|---|---|---|
| DIS5 | 27,577 | 489 | 6.10 | 498 | 98.5 |
| GWS7 | 23,542 | 460 | 6.97 | 545 | 98.1 |
| FPS10 | 26,032 | 519 | 6.77 | 606 | 98.0 |
| CIS17 | 24,571 | 361 | 5.96 | 427 | 98.5 |
| APS18 | 16,539 | 402 | 6.52 | 506 | 98.3 |
| Phylum | Genus | Strain Number | Site | Closest Match | GenBank ID | Similarity (%) |
|---|---|---|---|---|---|---|
| Actinobacteria | Arthrobacter | LB8 | DIS5 | Arthrobacter alpinus DSM 22274T | GQ227413 | 98.98 |
| T10 | DIS5 | Arthrobacter oryzae NRRL B-24478T | CLG_48533 | 99.60 | ||
| D3 | GWS7 | Arthrobacter pascens DSM 20545T | X80740 | 99.58 | ||
| LB13, LB14, K11 | DIS5 | Arthrobacter psychrochitiniphilus GP3T | AJ810896 | 98.86 | ||
| Actinobacteria | Kocuria | LB4 | DIS5 | Kocuria palustris DSM 11925T | Y16263 | 100.00 |
| C21 | FPS10 | Kocuria rosea DSM 20447T | X87756 | 99.73 | ||
| Leifsonia | T9 | DIS5 | Leifsonia kafniensis KFC-22T | AM889135 | 99.73 | |
| Microbacterium | H3-2 | FPS10 | Microbacterium aurum KACC 15219T | CP018762 | 99.86 | |
| O3 | GWS7 | Microbacterium hydrocarbonoxydans BNP48T | AJ698726 | 99.74 | ||
| C7, C8 | FPS10, CIS17 | Microbacterium paraoxydans NBRC 103076T | AJ491806 | 99.86 | ||
| B11 | FPS10 | Microbacterium rhizomatis DCY102T | KP161851 | 98.88 | ||
| Micrococcus | H3-1 | FPS10 | Micrococcus aloeverae AE-6T | KF524364 | 100.00 | |
| Microterricola | LBN | DIS5 | Microterricola gilv SSWW-21T | AM286414 | 99.04 | |
| Paeniglutamicibacter | ON | DIS5 | Paeniglutamicibacter cryotolerans LI3T | GQ406812 | 99.59 | |
| T2, LB5 | DIS5 | Paeniglutamicibacter sulfureus DSM 20167T | X83409 | 99.58 | ||
| T3, T8, LB6 | DIS5 | Paeniglutamicibacter sulfureus DSM 20167T | X83409 | 99.45 | ||
| LB10 | DIS5 | Paeniglutamicibacter sulfureus DSM 20167T | X83409 | 98.62 | ||
| Pseudarthrobacter | Z5 | APS18 | Pseudarthrobacter sulfonivorans ALLT | AF235091 | 98.07 | |
| H2 | FPS10 | Pseudarthrobacter sulfonivorans ALLT | AF235091 | 99.59 | ||
| Rhodococcus | L1 | GWS7 | Rhodococcus fascians LMG 3623T | X79186 | 100.00 | |
| Actinobacteria | Rhodococcus | H4 | FPS10 | Rhodococcus kyotonensis JCM 23211T | AB269261 | 99.18 |
| P1, B18, B21, E1 | GWS7, CIS17, APS18 | Rhodococcus qingshengii JCM 15477T | DQ090961 | 100.00 | ||
| Bacteroidetes | Flavobacterium | NJA2 | GWS7 | Flavobacterium circumlabens CCM 8828T | AM177392 | 100.00 |
| Hymenobacter | 3F2 | DIS5 | Hymenobacter sedentarius DG5BT | CP013909 | 97.03 | |
| Dyadobacter | 3J3 | DIS5 | Dyadobacter koreensis DSM19938T | jgi.1055180 | 98.08 | |
| Firmicutes | Bacillus | H1 | FPS10 | Bacillus cereus ATCC 14579T | AE016877 | 100.00 |
| T4, LB3, K18, O8 | DIS5 | Bacillus oceanisediminis H2T | GQ292772 | 99.30 | ||
| B16 | CIS17 | Bacillus paralicheniformis KJ-16T | LBMN01 | 100.00 | ||
| B14, C12, B22 | FPS10, CIS17 | Bacillus paramycoides NH24A2T | KJ812444 | 100.00 | ||
| C8 | CIS17 | Bacillus siamensis KCTC 13613T | GQ281299 | 99.46 | ||
| C4, B24 | FPS10, CIS17 | Bacillus siamensis KCTC 13613T | GQ281299 | 100.00 | ||
| C19 | CIS17 | Bacillus siamensis KCTC 13613T | GQ281299 | 99.86 | ||
| C22 | FPS10 | Bacillus subtilis subsp. subtilis NCIB 3610T | AJ276351 | 99.89 | ||
| B23, C14 | CIS17, FPS10 | Bacillus wiedmannii FSL W8-0169T | KU198626 | 100.00 | ||
| K20, C10, E2 | DIS5, CIS17, APS18 | Bacillus zhangzhouensis DW5-4T | JX680133 | 100.00 | ||
| C20 | FPS10 | Bacillus zhangzhouensis DW5-4T | JX680133 | 99.86 | ||
| Firmicutes | Filibacter | W6 | APS18 | Filibacter limicola ATCC 43646T | AJ292316 | 98.77 |
| Paenisporosarcina | L10 | DIS5 | Paenisporosarcina indica PN2TT | FN397659 | 99.52 | |
| Planococcus | NJC23 | FPS10 | Planococcus salinarum DSM 23820T | FJ765415 | 98.47 | |
| Sporosarcina | K12, | DIS5 | Sporosarcina globispora DSM 4T | X68415 | 100.00 | |
| B30, B31, B32 | GWS7 | Sporosarcina globispora DSM 4T | X68415 | 99.86 | ||
| B7 | APS18 | Sporosarcina globispora DSM 4T | X68415 | 99.15 | ||
| 6E9 | DIS5 | Sporosarcina pasteurii NCIMB 8841T | X60631 | 97.61 | ||
| Proteobacteria | Acinetobacter | C24 | FPS10 | Acinetobacter lwoffii NCTC 5866T | X81665 | 99.86 |
| Aurantimonas | ON4, ON5 | DIS5 | Aurantimonas endophytica EGI 6500337T | KM114215 | 100.00 | |
| Duganella | L8 | GWS7 | Duganella zoogloeoides IAM 12670T | D14256 | 98.65 | |
| Ensifer | LB2 | DIS5 | Ensifer meliloti LMG 6133T | X67222 | 99.88 | |
| Janthinobacterium | B28 | CIS17 | Janthinobacterium lividum DSM 1522T | Y08846 | 99.73 | |
| Jiella | R10 | DIS5 | Jiella aquimaris LZB041T | KJ620984 | 96.35 | |
| Massilia | C9 | CIS17 | Massilia varians CCUG 35299T | AM774587 | 99.34 | |
| Methylobacterium | R12 | DIS5 | Methylobacterium hispanicum GP34T | AJ635304 | 99.86 | |
| Microvirga | 3D7 | DIS5 | Microvirga subterranean DSM 14364T | FR733708 | 96.75 | |
| Proteobacteria | Paracoccus | P13, R15, R17, LB12, LB1 | DIS5 | Paracoccus aerius 011410T | KX664462 | 100.00 |
| K15 | DIS5 | Paracoccus marinus KKL-A5T | AB185957 | 98.72 | ||
| Pseudomonas | P6, K3, C30 | GWS7 | Pseudomonas caspiana FBF102T | NR_152639 | 99.76 | |
| P2 | GWS7 | Pseudomonas frederiksbergensis JAJ28T | AJ249382 | 99.17 | ||
| C15, B17 | CIS17 | Pseudomonas gessardii DSM 17152T | AF074384 | 100.00 | ||
| B13 | CIS17 | Pseudomonas gessardii DSM 17152T | AF074384 | 99.88 | ||
| R5 | DIS5 | Pseudomonas mandelii CIP 105273T | AF058286 | 99.17 | ||
| C29 | GWS7 | Pseudomonas prosekii LMG 26867T | LT629762 | 98.86 | ||
| C5 | FPS10 | Pseudomonas prosekii LMG 26867T | LT629762 | 99.43 | ||
| Rhodoferax | 3D4 | DIS5 | Rhodoferax koreense DCY110T | CP019236 | 99.09 | |
| Sphingomonas | G1 | DIS5 | Sphingomonas panni C52T | AJ575818 | 98.86 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, S.; Du, J.; Zhu, L.; Xin, D.; Xin, Y.; Zhang, J. Analysis of Microbial Diversity in South Shetland Islands and Antarctic Peninsula Soils Based on Illumina High-Throughput Sequencing and Cultivation-Dependent Techniques. Microorganisms 2023, 11, 2517. https://doi.org/10.3390/microorganisms11102517
Cui S, Du J, Zhu L, Xin D, Xin Y, Zhang J. Analysis of Microbial Diversity in South Shetland Islands and Antarctic Peninsula Soils Based on Illumina High-Throughput Sequencing and Cultivation-Dependent Techniques. Microorganisms. 2023; 11(10):2517. https://doi.org/10.3390/microorganisms11102517
Chicago/Turabian StyleCui, Siqi, Jie Du, Lin Zhu, Di Xin, Yuhua Xin, and Jianli Zhang. 2023. "Analysis of Microbial Diversity in South Shetland Islands and Antarctic Peninsula Soils Based on Illumina High-Throughput Sequencing and Cultivation-Dependent Techniques" Microorganisms 11, no. 10: 2517. https://doi.org/10.3390/microorganisms11102517
APA StyleCui, S., Du, J., Zhu, L., Xin, D., Xin, Y., & Zhang, J. (2023). Analysis of Microbial Diversity in South Shetland Islands and Antarctic Peninsula Soils Based on Illumina High-Throughput Sequencing and Cultivation-Dependent Techniques. Microorganisms, 11(10), 2517. https://doi.org/10.3390/microorganisms11102517