Abstract
This study focused on the assessment of the antimicrobial resistance of Staphylococcus aureus (S. aureus) and Escherichia coli (E. coli) isolated from bovine mastitis milk samples and the revealing anti-mastitis potential of phytocompounds of Ziziphus jujube and Acacia nilotica through molecular docking analysis. The mastitis milk samples were collected from various dairy farms for the isolation of the bacteria (S. aureus and E. coli) and their response to antibiotics. Ethanolic extracts of both plants were prepared. Their antibacterial activity was evaluated, and they were processed for phytochemical analysis after which, molecular docking analysis with pathogenic proteins of the bacteria was carried out. Parametric and non-parametric statistical analyses were performed to reach the conclusions of this study. The findings of the study revealed a higher drug resistance (≥40%) of E. coli against ampicillin, amikacin, and vancomycin, while S. aureus exhibited the highest resistance to ampicillin, erythromycin, and ciprofloxacin. The ethanolic extracts of the Ziziphus jujube and Acacia nilotica plants produced a ZOI between 18 and 23 mm against multidrug-resistant S. aureus and E. coli. Gas chromatography–mass spectrophotometry (GC–MS) was used to explore 15 phytocompounds from Ziziphus jujube and 18 phytocompounds from Acacia nilotica. The molecular docking analysis of 2cyclopenten−1-one,3,4,4 trimethyl and Bis (2ethylhexyl) phthalate of Ziziphus jujube showed a binding affinity of −4.8 kcal/mol and −5.3 kcal/mol and −5.9 kcal/mol and −7.1 kcal/mol against the DNA Gyrase and toxic shock syndrome toxin-1 proteins of S. aureus and E. coli, respectively. The suberic acid monomethyl ester of Acacia nilotica showed a binding affinity of −5.9 kcal/mol and −5 kcal/mol against the outer membrane protein A and Topoisomerase IV protein of E. coli and −5.1 kcal/mol and −5.8 kcal/mol against the toxic shock syndrome toxin-1 and Enterotoxin B proteins of S. aureus. Similarly, 2,2,4-trimethyl-1,3-pentanediol di-iso-butyrate showed a binding affinity of −6.5 kcal/mol and −5.3 kcal/mol against the outer membrane protein A and Topoisomerase IV of E. coli and −5.2 kcal/mol and −5.9 kcal/mol against the toxic shock syndrome toxin-1 and Enterotoxin B proteins of S. aureus, respectively. The study concluded that there was an increasing trend for the antimicrobial resistance of S. aureus and E. coli, while the Ziziphus jujube and Acacia nilotica plant extracts expressed significant affinity to tackle this resistance; hence, this calls for the development of novel evidence-based therapeutics.
1. Introduction
Antimicrobial resistance has become the leading cause of the treatment failure of infectious diseases all over the world. Resistance to antimicrobials continues to compromise disease-prevention strategies and to increase the production and economic losses in animal-based food systems. Among the common conditions manifested at the farm level, mastitis is one of the frequent conditions of dairy animals [1]. This disease affects the udders of the animals, leading to a drop in milk quantity and quality, which significantly affects the farm’s profitability [2]. Most infections last for an entire period of lactation or even the cow’s entire life span [3] and are connected to an annual economic loss of around USD 35 billion globally, while losses of USD 2 billion have been noted in the U.S. alone [4]. The data are scarce concerning Pakistan regarding the economic loss caused by mastitis. One of the studies from Pakistan, however, reported an annual loss of USD 3.75 million [5].
Mastitis is an inflammation of the mammary glands caused by physical, chemical, or infectious agents. The latter cause is further consisted of bacterial, viral, fungal, and protozoal agents, but bacterial etiology alone comprises more than 90% of mastitis cases. Among the bacterial etiology, Staphylococcus aureus (S. aureus) and Escherichia coli (E. coli) appear as the major causes of this condition all around the world [6,7]. An increase in the incidence of antimicrobial resistance among pathogens combined with the high costs of treatment have prompted the scientific community to look for substitutes with comparable efficacy to that of antibiotics [8]. In the quest to find alternatives to antibiotics, medicinal plant extracts could be a possible option to treat infections [9]. Unlike antibiotics, plant extracts are widely recognized as useful therapeutics [10] and have proven effective alternative to antibiotics against mastitis pathogens [11]. Mastitis has been traditionally treated with a range of plant species in Pakistan, which includes Allium sativum, Bunium persicum, Oryza sativa, and Triticum aestivum, but further alternatives are required to obtain satisfactory results [12].
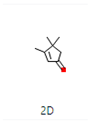
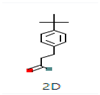
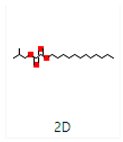
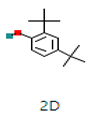
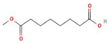
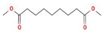
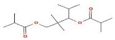
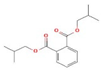
Ziziphus jujube or Chinese dates and Acacia nilotica have been reported to have a broad-spectrum antibacterial activity against a variety of bacterial organisms [13]. The extracts of these plants have been used previously to alleviate or treat a variety of ailments, and the antimicrobial potential of their extracts could be extended to microbes that have developed resistance [9]. A major impediment in using plant-based extracts is that there is little knowledge about their chemical composition. The identification of phytochemicals in plant extracts is the core to understanding the antimicrobial activities of the various components. Moreover, the molecular docking of the active ingredients with the ligands of microbes could further reveal the antibacterial potential of plant-based extracts, which can further bridge the knowledge gap. The phytocompounds from the leaf extract of Ziziphus jujube are 2-Cyclopenten-1-one,3,4,4-trimethyl and Bis(2-ethylhexyl) phthalate, and those from Acacia nilotica include suberic acid monomethyl ester and 2,2,4-trimethyl-1,3-pentanediol di-iso-butyrate. These phytocompounds show greater negative binding energies, they follow Lipinski’s five rules, and have a strong repressive activity against mastitis. Proteins such as toxic shock syndrome toxin-1(2QIL), DNA Gyrase (5L3J), outer membrane protein A (IBXW), Enterotoxin B (3SEB), and Topoisomerase IV (3FV5) represent salient pathogenic characteristics of pathogens, which can serve as representative molecular docking profiles. Similarly, DNA Gyrase has been proven to be a valid target in the design of novel antibacterial drugs, while TSST-1 is a superantigen that stimulates the release of cytokines, causing the leakage of endothelial cells at low concentrations with a cytotoxic effect to the cells at high concentrations. Enterotoxin B helps stimulate cytokine release and inflammation. Outer membrane protein A enables intracellular survival, that is, the evasion from the body’s defenses. Topoisomerase IV helps create pores in the membranes of host cells (cell lysis) and catalyzes DNA double-strand breaks.
Hence, the current study was conducted with the aim to investigate the antibacterial activity of Ziziphus jujube and Acacia nilotica plant extracts against drug-resistant S. aureus and E. coli as well as the characterization of extracts via phytochemical analysis, using the GC–MS and the molecular docking techniques, to provide a prediction of the ligand–receptor complex structure.
2. Materials and Methods
2.1. Isolation and Antibiotic Susceptibility Profile of Mastitis Milk Samples
Milk samples (n = 200) of dairy cows suffering from either clinical or subclinical mastitis were aseptically collected as per standard guidelines [10], using convenient sampling techniques. For this purpose, various dairy farms (n = 20) in the district Lahore were included. From each dairy farm, n = 10 cows were selected based on their clinical presentation and complaints of reduced milk production and abnormal milk. The subclinical mastitis was decided based on somatic cell count greater than 200,000 or a positive California Mastitis Test (CMT). The animals that showed clinical signs were also included in this study.
The milk samples were swabbed on a blood agar following incubation at 37 °C for 24 h. The typical colonies of S. aureus (golden yellow) and E. coli (grayish white and moist) were subcultured on mannitol salt agar and eosin methylene blue agar. Biochemical confirmation of the isolates was performed as per the guidelines of Bergey’s’ Manual of Determinative Bacteriology [7]. Further, the isolates were also confirmed via molecular analysis by targeting nuc genes (500 bp with primers as “forward primers 5′AAGGGCAATACGCAAAGAG 3′ and reverse primers 5′AAACATAAGCAACTTTAGCCAAG 3′”) in S. aureus (Figure S1) [14] and 23S rRNA gene (at 231 bp using primers E23S-F: ATCAACCGATTCCCCAGT; E23S-R: TCACTATCGGTCAGTCAGGAG) for E. coli (Figure S2) [15].
The isolates were processed further to assess their responses to a wider range of antibiotics (ampicillin, enrofloxacin, ciprofloxacin, trimethoprim, sulfamethoxazole, amikacin, oxytetracycline, vancomycin, erythromycin, linezolid, and azithromycin) as per Kirby Bauer’s method of disc diffusion. Bacterial cultures adjusted at 1–1.5 × 108 colony forming units (CFU)/mL were swabbed on Mueller Hinton agar following which antibiotic discs were aseptically placed on agar. Incubation at 37 °C for 20–24 h was carried out, followed by a measurement of zones of inhibition (ZOI) using Vernier calipers and compared with standards.
2.2. Selection of Multiple Drug-Resistant Bacteria
The isolates of S. aureus and E. coli showing resistance to more than two classes of antibiotics were considered multiple drug-resistant (MDR) pathogens. To evaluate the antibacterial response of plant extracts, n = 05 MDR S. aureus and E. coli isolates were selected for their further use in the current study.
2.3. Preparation of Plant Extracts and Determination of Antimicrobial Activity
The leaves (2–3 kg) of Ziziphus jujube and flowers of Acacia nilotica were collected from Bahawalpur in 2021, an area where they are commonly found. The leaves were identified by a professional botanist and assigned the voucher number 3859 to aid in identification in the future. The leaves were shade-dried and pulverized into a fine powder. The powders (100 g) were emulsified in 250 mL of absolute ethanol for two weeks and filtered through Whatman paper (0.45 µm pore size). The solvent was evaporated through a rotary evaporator (PG-215-Rotary Water Bath-928) at 55 °C for 1 hour. The extracts were lyophilized in VaCo2 (ZIRBUS).
Ethanolic extracts were applied to MDR S. aureus and E. coli at various concentrations (125 mg/mL, 250 mg/mL, and 500 mg/mL of Ziziphus jujube, and 125 mg/mL and 500 mg/mL of Acacia nilotica). For this purpose, broth cultures of bacteria were adjusted to a concentration of 1-1.5 × 108 CFU/mL. Filter paper discs were impregnated with various concentrations of ethanolic extracts and applied on to Mueller Hinton agar. The fresh growth of E. coli and S. aureus was swabbed before the application of discs. The plates were incubated at 37 °C for 24 h to observe the ZOI. Amoxicillin and erythromycin antibiotics purchased from the market were applied as positive controls. These antibiotics were mostly used on farms with greater efficacy, hence the reason for their selection in this study.
2.4. Phytochemical Analysis of Extracts
The gas chromatography mass spectrophotometry (GC–MS) technique was used for phytochemical analysis to identify the various compounds present in the ethanolic extracts. For this purpose, the GC–MS instrument (Agilent Technologies 7890A GC System, 5301 Stevens Creek Blvd, Santa Clara, CA, USA) fitted with an Elite-5ms fused silica column (30 m × 0.25 mm, film thickness 0.5 m) was operated, and Helium was used as a carrier gas. The leaf extract was mixed with ethanol (1:10 v/v). The injector volume was 1 L, and the injector temperature was set at 250 °C. The oven temperature was kept at 70 °C for 4 min before being raised to 310 °C at a rate of 5 °C/min for 10 min. All samples were evaluated in both full scan and selective ion scan modes (mass range of 40–510 atomic mass units). The temperatures of the injector inlet and transfer lines were 280 °C and 290 °C, respectively. Phytocompounds were obtained along with retention time and peak area percentage. A chromatogram was also achieved that showed the most numerous phytocompounds in the leaf extracts.
2.5. Derivation of Bacterial Pathogenic Protein
The three-dimensional structure of target proteins and their identification numbers were derived from the protein data bank (PDB). The proteins were prepared in the tool BIOVIA/Discovery studio 2021. For in silico protein purification, water and other molecules were removed from the structure. The proteins involved in mastitis were targeted as Topoisomerase IV (PDB ID: 3FV5), outer membrane protein A (PDB ID: 1BXW), and DNA Gyrase (PDB ID: 5L3J) for E. coli, and Enterotoxin B (PDB ID: 3SEB) and toxic shock syndrome toxin-1 (PDB ID: 2QIL) for S. aureus [16].
2.6. Selection of Effective Phytocompounds:
For the identification of the best ligand interaction, minimum binding energy needed to bind with a particular target protein was considered. From PubChem, two-dimensional structures of the active compounds present in extracts were downloaded and then changed into a Pbdqt file using Discovery Studio Vision. The ADMET rule (absorption, distribution, metabolism, excretion, and toxicity) was applied for the designing process. For this purpose, the compound was written in PubChem, and a canonical smile was taken and pasted in SwissADME to obtain ADMET properties. From SwissADME, phytocompounds were screened based on cytochrome inhibitors, whereas CYP inhibitors were favorable and performed activity in favor of drug.
2.7. Molecular Docking of Bacterial Proteins with Ligands of Extract
The aim of molecular docking was to predict the binding energy of small molecular ligand interactions with the binding site of the target protein. Compounds of 2-Cyclopenten-1-one,3,4,4-trimethyl-and-Bis(2-ethylhexyl) phthalate were docked with bacterial proteins (PDB ID: 5L3J for E. coli and 2QIL for S. aureus) that had high negative binding affinity. The molecular docking of phytocompounds was achieved with the help of PyRx software Version 0.9. The phytocompounds were docked by targeting the binding sites of proteins with grid box dimensions that were originated by fixing x, y, and z. In this way, the ligands were set on their binding energy, and bonding interaction strength was yielded between ligands and proteins. Similarly, suberic acid monomethyl ester and 2,2,4-trimethyl-1,3-pentanediol di-iso-butyrate from the extracts were docked with the 1BXW and 3FV5 proteins of E. coli and the 2QIL and 3SEB proteins of S. aureus. Blind docking was applied to these compounds as they show greater negative binding energies [17].
The docked complex was applied to find the ligand–protein interactions. Discovery Studio was used to visualize the protein–ligand complex obtained by docking through PyRx. In the Discovery Studio, binding energies, receptor–ligand interaction, H-bond, and the distance-like properties were visualized, which helped to screen phytocompounds that have the highest antibacterial activity and could be a candidate for drug design.
3. Results
3.1. Antimicrobial Resistance Profile of S. aureus and E. coli
The responses of E. coli and S. aureus against antibiotics showed considerable resistance (Table 1). The study found that more than 40% of E. coli isolates were resistant to ampicillin, trimethoprim, sulfamethoxazole, amikacin, and vancomycin. Around 40% of the S. aureus isolates were found resistant to ampicillin, ciprofloxacin, and erythromycin. The study showed a higher percentage of intermediate-susceptible isolates of S. aureus as well as E. coli. It was noteworthy that E. coli isolates were resistant to the majority of the antibiotics (equal to or more than 30%), except for erythromycin, oxytetracycline, and ciprofloxacin. In the case of S. aureus, amikacin and linezolid were found to be the most effective candidates. The percentage-sensitive isolates remained more than 40% against all antibiotics except ampicillin (Table 1).

Table 1.
Response of S. aureus and E. coli against antibiotics.
3.2. Response of Multiple Drug-Resistant E. coli and S. aureus against Ethanolic Plants’ Extracts
The ZOI shown by the leaf extract of Ziziphus jujube revealed that plants pose antibacterial properties against Gram-positive (S. aureus) and Gram-negative (E. coli) bacteria (Table 2, Figure 1). The ethanolic leaf extract of Ziziphus jujube showed ZOI values of 21 mm, 19 mm, and 18 mm, and 23 mm, 21 mm, and 18 mm at concentrations of 500 mg, 225 mg, and 125 mg for S. aureus and E. coli, respectively. Ethanolic extracts of A. nilotica showed 20 mm and 18 mm ZOI at concentrations of 500 mg and 125 mg, respectively, against S. aureus, while amoxicillin showed a 17 mm ZOI at 125 mg concentration. Against E. coli, the same extract concentrations of 500 mg and 125 mg were used, which showed ZOI values of 23 mm and 21 mm, respectively (Table 2, Figure 2).

Table 2.
Zone of inhibition (mm) for E. coli and S. aureus against extracts of plants.
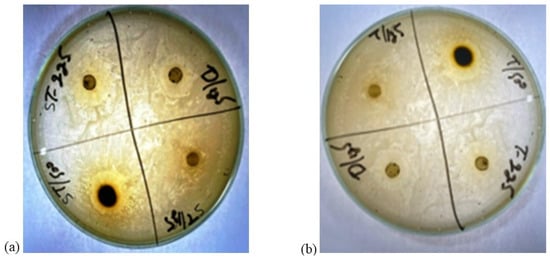
Figure 1.
Zones of inhibition of leaf extract of Ziziphus jujube plant. (a) zone of inhibition(mm) of Ziziphus jujube for S. aureus (b) zone of inhibition (mm) of Ziziphus jujube for E. coli.
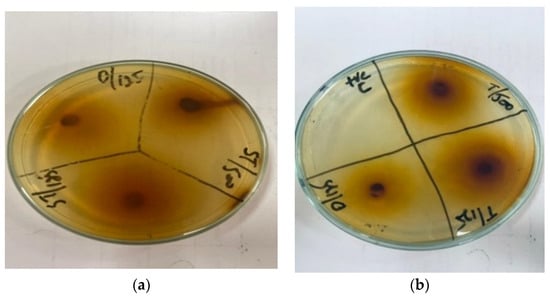
Figure 2.
Antibacterial activity of Acacia nilotica flower’s extract at 500 and 125 mg/mL of plant’s concentration and Amoxicillin concentration 125 mg. (a) Zone of inhibition (mm) of Acacia nilotica for S. aureus; (b) zone of inhibition (mm) of Acacia nilotica for E. coli.
3.3. Phytochemical Analysis of Plant Extract
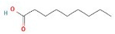
The GC–MS results of Ziziphus jujube revealed that phytol was the most abundant compound (with a 17.73% intensity) detected at 35 min (Table 3) on the GC–MS column, and n-hexadecanoic acid with a 15.16% intensity was detected at 30 minutes (with the attribute of being antibacterial). Fifteen peaks were detected in GC–MS chromatogram for ethanolic extracts of the leaf extract of Ziziphus jujube (Figure 3). Eighteen peaks were detected in GC–MS chromatogram for ethanolic extracts of the flowers of Acacia nilotica (Figure 4, Table 4).

Table 3.
GC–MS results showing phytocompounds in ethanolic extract of Ziziphus jujube.
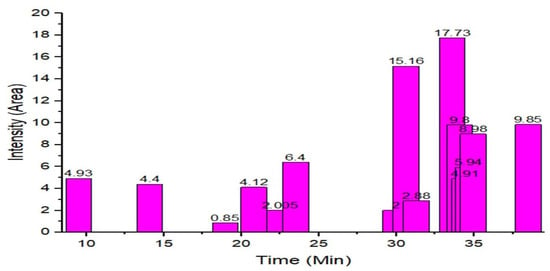
Figure 3.
GC–MS chromatogram of ethanolic phytocompounds of Ziziphus jujube leaf extract.
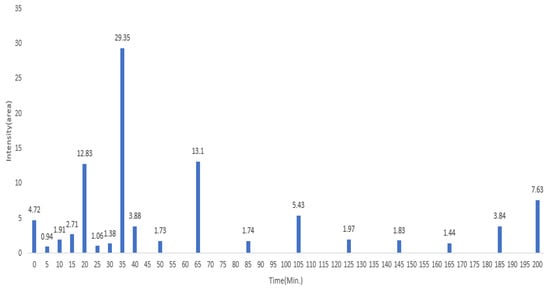
Figure 4.
GC–MS chromatogram of ethanolic phytocompounds of Acacia nilotica flowers extract.

Table 4.
GC–MS results showing phytocompounds in ethanol extract of Acacia nilotica.
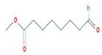
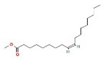
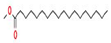
2-Cyclopenten-1-one,3,4,4-trimethyl-, and Bis(2-ethylhexyl)phthalate were two screened compounds fulfilling ADMET properties (Table 5). Two compounds (2,2,4-trimethyl-1,3-pentanediol diisobutyrate and suberic acid monomethyl ester) showed the highest antibacterial activity and followed Lipinski’s five rules (Table 6). The former molecule has a molecular weight of 124.18 g/mol and has high GI absorption along with antibacterial activity. The latter molecule was attributed to having a molecular weight of 376.53 g/mol and a high GI absorption with antibacterial activity. The Lipinski’s rule of five was employed for all 13 ligands, which was different for each ligand. Out of the 13 phytocompounds, 2 compound (2-Cyclopenten-1-one,3,4,4-trimethyl- and Bis(2-ethylhexyl)) phthalates were screened out for molecular docking (Table 7).

Table 5.
Phytocompounds of Ziziphus jujube showing ADMET properties for molecular docking.

Table 6.
Phytocompounds of Acacia nilotica showing ADMET properties for molecular docking.

Table 7.
Screened phytocompounds of Ziziphus jujube.
Same is the case with phytochemical analysis of Acacia nilotica based on ADMET parameters as indicated in Table 8.

Table 8.
Phytocompounds of Acacia nilotica screened on the basis of ADMET parameters for molecular docking.
3.4. Derivation of Bacterial Proteins
The study derived DNA Gyrase (5L3J) protein of E. coli because of its proven action to serve as target for drug designs (Table 9). Similarly, toxic shock syndrome toxin-1(TSST-1 2QIL) of S. aureus proteins were derived with the fact that it being superantigen could provoke cytokines storm and cytotoxic effects to cells when used. Similarly, 1BXW outer membrane protein A and topoisomerase IV from E. coli; and Toxic shock syndrome toxin-1, and 3SEB Enterotoxin B from S. aureus protein were found (Table 10).

Table 9.
Protein screening based on mode of action for molecular docking of Ziziphus jujube.

Table 10.
Mode of action of S. aureus and E. coli proteins for molecular docking of Acacia nilotica.
3.5. Molecular Docking of Bacterial Protein with Ligands of Phytocompounds
All the compounds showed different binding energies when docked with the ligands of phytocompounds. The binding energies of phytocompounds of Ziziphus jujube leaf extracts with S. aureus and E. coli proteins showed higher minus-binding energies, indicating a greater stability of the compounds (Table 11). 2-Cyclopenten-1-one,3,4,4-trimethyl showed −4.8 kcal/mol and −5.3 kcal/mol binding energies with proteins from E. coli and S. aureus, respectively, while Bis(2-ethylhexyl) phthalate showed −5.9 kcal/mol and −7.1 kcal/mol binding energies against E. coli and S. aureus proteins, respectively (Table 11). These energies indicate that the screened phytocompounds have sufficient suppressive ability against mastitis and hold a greater stability for their sustained efficacy.

Table 11.
Binding energies of ligands of plant extracts with bacterial proteins of E. coli and S. aureus.
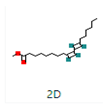
The docking interaction of toxic shock syndrome toxin (TSST-1) with the 2-Cyclopenten-1-one,3,4,4-trimethyl-ligand PDB structure (Figure 5) and Bis(2-ethylhexyl) phthalate ligand PDB structure of TSST-1 protein (Figure 6) revealed strong interactions with each other and with residue molecules. Similarly, the docking interaction of DNA Gyrase target protein PDB structure with 2-Cyclopenten-1-one,3,4,4-trimethyl-ligand PDB structure (Figure 7) and Bis(2-ethylhexyl) phthalate ligand PDB structure (Figure 8) showed unique interactions with each other and other residue molecules.
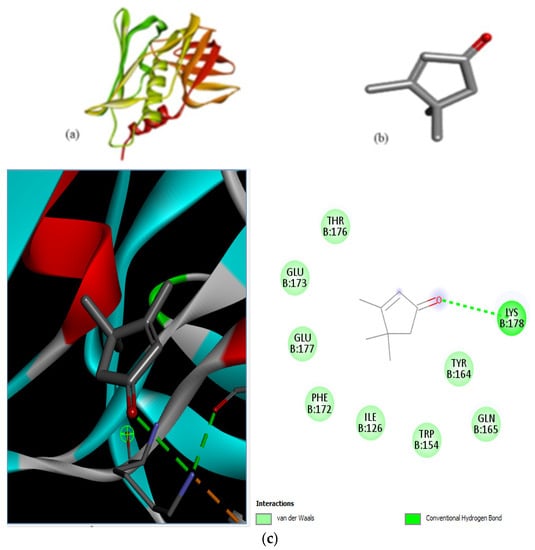
Figure 5.
Molecular docking of Staphylococcus aureus protein with cyclopentene ligands of Ziziphus jujube plant. (a) Toxic shock syndrome toxin-1 target protein PDB structure. (b) 2-Cyclopenten-1-one,3,4,4-trimethyl-ligand PDB structure. (c) 2-Cyclopenten-1-one,3,4,4-trimethyl showing interaction with target protein and 2D structure showing interaction with residues.
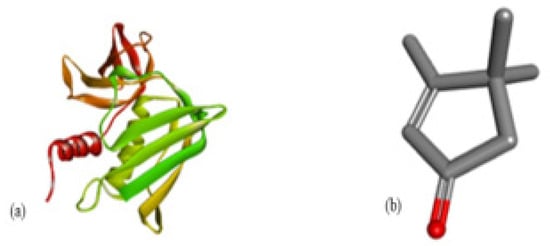
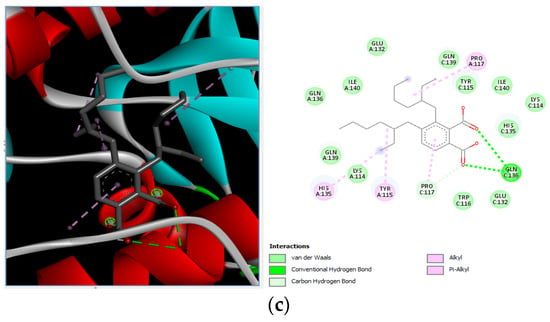
Figure 6.
Molecular docking of Staphylococcus aureus protein with phthalate ligands of Ziziphus jujube plant. (a) Toxic shock syndrome toxin-1 target protein PDB structure. (b) Bis(2-ethylhexyl) phthalate ligand PDB structure. (c) Bis(2-ethylhexyl) phthalate—showing interaction with target protein and 2D structure showing interaction with residues.
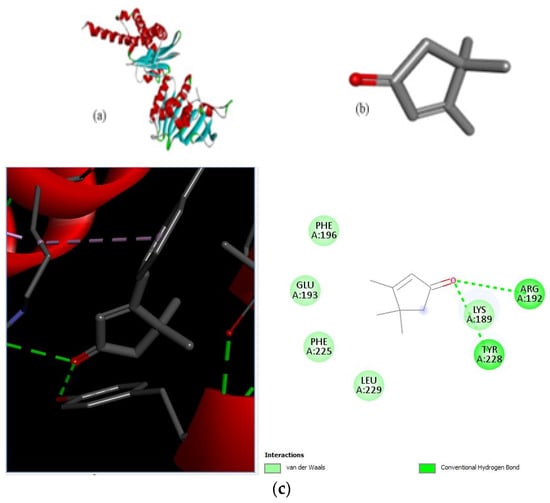
Figure 7.
Molecular docking of E. coli protein with phthalate ligands of Ziziphus jujube plant. (a) DNA Gyrase target protein PDB structure. (b) 2-Cyclopenten-1-one,3,4,4-trimethyl-ligand PDB structure. (c) 2-Cyclopenten-1-one,3,4,4-trimethyl showing interaction with target protein and 2D structure showing interaction with residues.
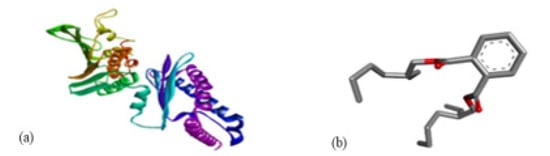
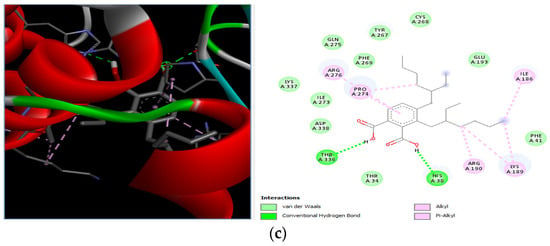
Figure 8.
Molecular docking of E. coli protein with phthalate ligands of Ziziphus jujube plant. (a) DNA Gyrase target protein PDB structure. (b) Bis(2-ethylhexyl) phthalate ligand PDB structure. (c) Bis(2-ethylhexyl) phthalate—showing interaction with target protein and 2D structure showing interaction with residues.
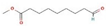
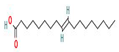
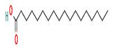
Different binding energies when the proteins of E. coli and S. aureus were docked with the binding sites of ligands are shown in Figure 9 and Figure 10. The binding energies of suberic acid monomethyl ester were higher when docked against outer membrane protein A (−5.9 kcal/mol) of E. coli and Enterotoxin B (−5.8 kcal/mol) of S. aureus and when following Lipinski’s five rules, while Topoisomerase IV and TSST-1 showed lower binding energies at −5 kcal/mol and −5.1 kcal/mol, respectively. 2,2,4-trimethyl-1,3-pentanediol di-isobutyrate showed higher negative binding energies (−6.5 kcal/mol) for E. coli protein (PDB ID: IBXW) and (−5.9 kcal/mol) against Enterotoxin B, while lower binding energies were evident for Topoisomerase IV and TSST-1 (−5.3 kcal/mol and −5.2 kcal/mol, respectively). These two phytocompounds showed higher negative binding energies and followed Lipinski’s five rules, and had strong repressive action against mastitis, while other compounds possessed less than −5 kcal/mol binding energies and were found to be less stable (Table 11).
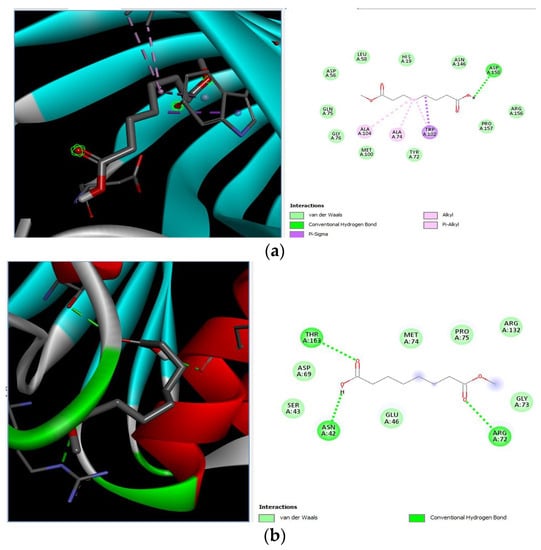
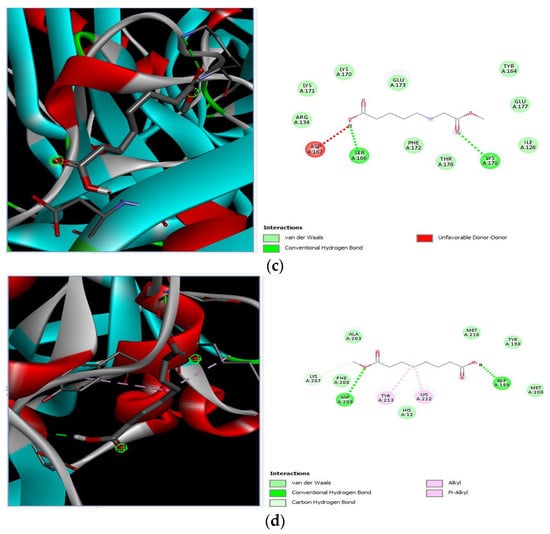
Figure 9.
Molecular docking of suberic acid monomethyl ester with S. aureus and E. coli proteins: (a) 2D and 3D representations of greater negative binding energy between suberic acid monomethyl ester and outer membrane protein A (PDB ID: IBXW). (b) Docking interaction between suberic acid monomethyl ester and Topoisomerase IV (PDB ID: 3FV5). (c) Docking interaction between suberic acid monomethyl ester and toxic shock syndrome toxin-1 (PDB ID: 2QIL). (d) 2D and 3D representations of greater negative binding energy between suberic acid monomethyl ester and Enterotoxin B (3SEB).
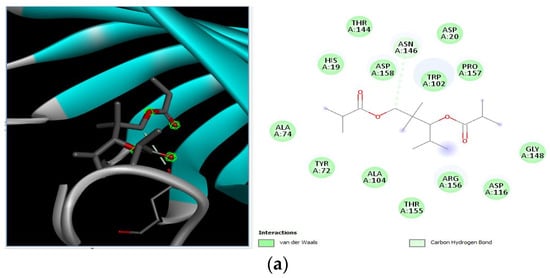
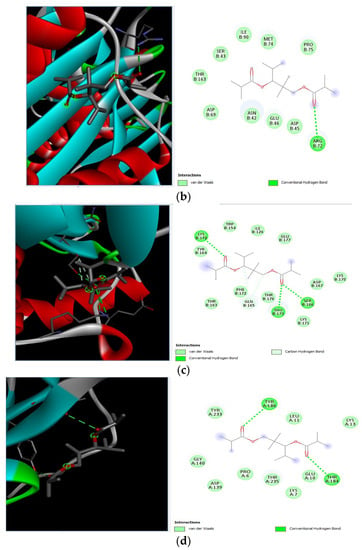
Figure 10.
Molecular docking of 2,2,4-trimethyl-1,3-pentanediol di-isobutyrate with S. aureus and E. coli proteins. (a) 2D and 3D representations of greater binding energy between 2,2,4-trimethyl-1,3-pentanediol di-isobutyrate and outer membrane protein A. (b) Docking interaction between 2,2,4-trimethyl-1,3-pentanediol di-isobutyrate and Topoisomerase IV. (c) Docking interaction between 2,2,4-trimethyl-1,3-pentanediol di-isobutyrate and toxic shock syndrome toxin-1. (d) 2D and 3D representations of greater binding energy between 2,2,4-trimethyl-1,3-pentanediol di-isobutyrate and Enterotoxin B.
4. Discussion
The studies have proven that the regular use of antibiotics results in the development of antimicrobial resistance (AMR) among core mastitis pathogens like E. coli and S. aureus. Moreover, the use of plant extracts has been found to be effective in achieving optimum control over AMR mastitis pathogens. One of the studies conducted in France has concluded antimicrobial resistance against three primary isolated bacteria (Streptococcus uberis, E. coli, and Coagulase-positive staphylococci) isolated from dairy cattle and found high levels of resistance against commonly used antimicrobials [26]. These findings are in accordance with our results, where a higher resistance against antibiotics was found. The bacterium S. aureus is one of the reasons for chronic mastitis that could be prevented by on-time diagnosis, ensuring good farm management, and improving udder hygiene. The production of metabolizing enzymes and toxins by S. aureus that damage the milking tissue resulted in a deeper penetration of bacteria into the tissue. Hence, it became difficult for antibiotics to reach the affected tissue at the minimum inhibitory concentration (MIC), leading to the development of AMR [13]. Therefore, alternatives to antibiotics that could suppress bacterial capability to colonize are recommended. Moreover, the overuse of antibiotics to manage AMR contributes to the development of multidrug resistance, which has severe animal and public health implications.
In a previous study, Ziziphus jujube showed effective antibacterial activity against Gram-positive and Gram-negative bacteria such as S. aureus and E. coli [6], which is in line with the current study. Similar results were reported by another study that concluded the antibacterial activity of Ziziphus leaves extract at 10mg/mL against S. aureus, Listeria. monocytogenes, Salmonella typhimurium, and E. coli with ZOI ranging from 10 mm to 14.2 mm for various plant extracts. Likewise, another study reported a zone of inhibition of Ziziphus leaf extract, ranging between 7 mm and 15 mm against S. aureus, E. coli, and S. typhimurium. These antibacterial properties of the plant extracts are due to the presence of enriched constituents like polyphenols, flavonoids, and tannins [27]. The higher contents of phenols in the plant extracts were detected in the current study. These findings are consistent with a previously reported study, where high concentrations of phenolic contents in the leaves of Ziziphus jujube plants were found with potential antibacterial activity [28]. Similarly, higher concentrations of total flavonoids were detected as found by Song et al. [29]. A higher concentration of phenols in the leaves might be due to the presence of phenolic compounds in vacuoles of the colored tissues of plants [30]. The presence of phenolic compounds with antibacterial activity as identified in our study has also been determined in coffee and other plant species. The isolated compounds from the phenolic class were quinic acid, gallic acid, catechin, chlorogenic acid, 4-0-caffeoylquinic acid, caffeic acid, syringic acid, rutin, 1,3-di-O-caffeoyulquinic acid, epicatechin, p-coumaric acid, trans-ferulic acid, hyperoside, and others [31]. Phenolic compounds like 3,4-dihydroxyphenyl ethanol glucoside have been proven to express the highest bacteriostatic activity against Bacillus cereus, S. typhimurium, S. aureus, and E. coli.
Our results indicated that Ziziphus jujube has effective antibacterial activity against Gram-positive and Gram-negative bacteria such as S. aureus and E. coli, as was also found by Kher et al. [32]. Around 15 phytocompounds from Ziziphus jujube leaf extract and 18 phytocompounds from the flower extract of Acacia nilotica were obtained from GC–MS and screened by Swiss ADME on the basis of CYP inhibitors. The following phytocompounds were selected for molecular docking that have an area sum% 2-cyclopenten-1-one,3,4,4-trimethyl (4.93%), Bis(2-ethylhexyl) phthalate (5.94%), suberic acid monomethyl ester (2.71%), and 2,2,4-trimethyl-1-3-pentanediol di-isobutyrate (1.06%). The phytocompounds selected have CYP inhibitors favorable for proteins and perform activity in favor of drugs, while the approach to select drugs on the basis of CYP inhibitors were in agreement with other studies [16]. The compound 3,4-dihydroxyphenyl ethanol glucoside expressed the highest bacteriostatic activity against Bacillus cereus, Salmonella typhimurium, S. aureus, and E. coli [16]. Among other compounds, flavonoids are believed to be the most active compounds that act by intercalation into DNA, leading to frameshift changes [33], whereas another study [34] has reported the promotion of Topoisomerase IV-dependent DNA cleavage. Moreover, geographical area could be an important factor causing variation in the phenolic composition of these plants’ extracts, but this factor could not create any bias in our study as all plants were sourced from a region with a uniform climate. Given the strong antibacterial potential of extracts of Acacia nilotica flowers and Ziziphus jujube leaves, it could be stated that the extracts of these plants could be used as an alternative to antibiotics if used in optimum concentrations. The use of Acacia nilotica and Ziziphus jujube extracts to prevent, control, or treat many infectious diseases of bacterial origin has been well recognized in homeopathic medicine [7]. Acacia nilotica flowers and Ziziphus jujube leaf extracts have shown greater antibacterial activity comparable to commonly used antibacterials on dairy farms, providing an auspicious tool for mastitis control.
Extracts and phytocompounds from Humulus lupulus showed significant activity against S. aureus compared to their response against Lactobacillus acidophilus [35]. Similarly, the antimicrobial activity of the tannins of the chestnut extract was noted, while this response was pronounced in the case of E. coli, and hence reflected the availability of potential alternative to antimicrobials [36]. The In silico inverse molecular docking procedures have been used previously to measure antimicrobial activity of rosemary extracts [37]. Xanthohumol and its metabolites were effectively docked with most human pathogenic proteins to aid in anticancer and antimicrobial activities [38]. In extension to the previous study [37], protein–ligand complexes were observed for K-RAS, HIV-1, and factor X was found to explore the simulation of molecular dynamics and free energy calculations. Carnosic acid and rosmanol were found to be significant binders with the HIV-1 protease, while carnasol showed prominent binding with the protein K-RAS [39].
5. Conclusions
The study found an increasing resistance in S. aureus and E. coli against a wider range of antibiotics from various classes depicting the narrowing window of the efficacy of antibiotics available at present. The plant extracts of Ziziphus jujube and Acacia nilotica have shown antibacterial activity against multiple drug-resistant S. aureus and E. coli equivalent to the positive control (erythromycin and amoxicillin). The extract of Ziziphus jujube and Acacia nilotica plants have a variety of phytocompounds that agreed with ADMET and Lipinski rules and have proven their antibacterial activity. The molecular docking of the two most appropriately screened phytocompounds 2-cyclopenten-1-one,3,4,4-trimethyl- and Bis(2-ethylhexyl) phthalate showed a greater binding affinity with pathogenic proteins of S. aureus (toxic shock syndrome toxin-1) and E. coli (DNA Gyrase). Similarly, suberic acid monomethyl ester and 2,2,4-trimethyl-1,3-pentanediol di-iso-butyrate of Acacia nilotica proved to be effective molecular ligands with pathogenic proteins. The study thus reached to the conclusion that there was an increase in resistance against antibiotics, while a greater antibacterial scope was noted in using phytocompounds of plant extracts that can pave the way to develop alternative therapeutics from natural resources.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms11102450/s1, Figure S1. Molecular identification (amplicons of PCR on agarose gel) of S. aureus by targeting nuc gene. M indicates a marker of 1 kb, 1–5 are samples, P means positive control. Figure S2 Molecular identification of E. coli targeting 23S RNA gene. M indicates marker of 1000 bp, 1–3 are samples, P indicates positive control, N indicate negative control
Author Contributions
W.Z. performed the methodology, prepared initial draft, and analyzed the data; I.H. and B.A. performed the methodology, prepared initial draft, reviewed the final draft, and analyzed the data; H.S. conceived the idea, performed the resourcing and methodology, and prepared the initial draft; A.A. prepared the final draft, analyzed the data, performed the resourcing; A.I.A. conceived the idea, performed the methodology, analyzed the data, prepared the final draft; H.B.A. performed the formal analysis and reviewed the manuscript; M.H.A. performed the methodology, analyzed the data, and revised the manuscript; K.L. conceived the idea, methodology, resourcing, and prepared the final draft. All authors have read and agreed to the published version of the manuscript.
Funding
This research received no external funding.
Data Availability Statement
All the data is available in article.
Acknowledgments
The authors extend their appreciation to the Researchers Supporting Project number (RSP2023R191), King Saud University, Riyadh, Saudi Arabia.
Conflicts of Interest
The authors declare no conflict of interest.
References
- AbdelKhalek, A.; Alanzi, J. Prevalence of Subclinical Mastitis in some Dairy Cattle Farms in Kuwait. Mansoura Veter. Med. J. 2019, 20, 46–51. [Google Scholar] [CrossRef]
- De Vries, A.; Marcondes, M.I. Review: Overview of factors affecting productive lifespan of dairy cows. Animal 2020, 14, s155–s164. [Google Scholar] [CrossRef] [PubMed]
- Dahl, G.E. Physiology of lactation in dairy cattle—Challenges to sustainable production. In Animal Agriculture; Academic Press: Cambridge, MA, USA, 2020; pp. 121–129. [Google Scholar]
- Khan, T.; Sankhe, K.; Suvarna, V.; Sherje, A.; Patel, K.; Dravyakar, B. DNA gyrase inhibitors: Progress and synthesis of potent compounds as antibacterial agents. Biomed. Pharmacother. 2018, 103, 923–938. [Google Scholar] [CrossRef] [PubMed]
- Bilal, M.Q.; Iqbal, M.U.; Muhammad, G.; Avais, M.; Sajid, M.S. Factors affecting the prevalence of clinical mastitis in buffaloes around Faisalabad district (Pakistan). Int. J. Agric. Biol. 2004, 6, 185–187. [Google Scholar]
- Mubarack, H.M.; Doss, A.; Vijayasanthi, M.; Venkataswamy, R. Antimicrobial drug susceptibility of Staphylococcus aureus from subclinical bovine mastitis in Coimbatore, Tamilnadu, South India. Vet. World 2012, 5, 352. [Google Scholar] [CrossRef]
- Lansky, E.P.; Paavilainen, H.M.; Lansky, S. Acacias: The Genus Acacia (Sensu Lato); CRC Press: Boca Raton, FL, USA, 2023; Volume 1. [Google Scholar]
- Salam, M.A.; Al-Amin, M.Y.; Salam, M.T.; Pawar, J.S.; Akhter, N.; Rabaan, A.A.; Alqumber, M.A. Antimicrobial Resistance: A Growing Serious Threat for Global Public Health. Healthcare 2023, 11, 1946. [Google Scholar] [CrossRef] [PubMed]
- Dattaray, D. Traditional Uses and Pharmacology of Plant Tridax procumbens: A Review. Syst. Rev. Pharm. 2022, 13, 476–482. [Google Scholar]
- Firyal, S.; Awan, A.R.; Sattar, H.; Ullah, H.K.R.; Tayyab, M.; Wasim, M.; Saeed, S.; Naseer, R.; Umer, M.O.; Rashid, I. Identification of polymorphism in bovine tumor necrosis factor alpha and toll-like receptor 4 genes and its association with mastitis in Sahiwal cows. Int. J. Agric. Biol. 2018, 20, 750–752. [Google Scholar]
- Kumari, S.; Naraian, R. Enhanced growth and yield of oyster mushroom by growth-promoting bacteria Glutamicibacter arilaitensis MRC119. J. Basic Microbiol. 2021, 61, 45–54. [Google Scholar] [CrossRef]
- Amber, R.; Adnan, M.; Tariq, A.; Khan, S.N.; Mussarat, S.; Hashem, A.; Al-Huqail, A.A.; Al-Arjani, A.F.; Abd Allah, E.F. Antibacterial activity of selected medicinal plants of northwest Pakistan traditionally used against mastitis in livestock. Saudi J. Biol. Sci. 2018, 25, 154–161. [Google Scholar] [CrossRef]
- Mitra, S.D.; Bandopadhyay, S.; Jadhao, S.; Shome, R.; Shome, B.R. Genetic characterization and comparative genomics of a multi drug resistant (MDR) Escherichia coli SCM-21 isolated from a subclinical case of bovine mastitis. Comp. Immunol. Microbiol. Infect. Dis. 2022, 85, 101799. [Google Scholar]
- Murtaza, M.; Aqib, A.I.; Khan, S.R.; Muneer, A.; Ali, M.M.; Waseem, A.; Zaheer, T.; Al-Keridis, L.A.; Alshammari, N.; Saeed, M. Sodium Alginate-Based MgO Nanoparticles Coupled Antibiotics as Safe and Effective Antimicrobial Candidates against Staphylococcus aureus of Houbara Bustard Birds. Biomedicines 2023, 11, 1959. [Google Scholar] [CrossRef] [PubMed]
- Muneer, A.; Kumar, S.; Aqib, A.I.; Khan, S.R.; Shah, S.Q.A.; Zaheer, I.; Rehman, T.U.; Abbas, A.; Hussain, K.; Rehman, A.; et al. Evaluation of Sodium Alginate Stabilized Nanoparticles and Antibiotics against Drug Resistant Escherichia coli Isolated from Gut of Houbara Bustard Bird. Oxidative Med. Cell. Longev. 2022, 2022, 7627759. [Google Scholar] [CrossRef]
- Roy, A.; Anand, A.; Garg, S.; Khan, M.S.; Bhasin, S.; Asghar, M.N.; Emran, T.B. Structure-based in silico investigation of agonists for proteins involved in breast cancer. Evid. Based Complement. Altern. Med. 2022, 2022, 7278731. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Yang, X.; Gan, J.; Chen, S.; Xiao, Z.X.; Cao, Y. CB-Dock2: Improved protein–ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res. 2022, 50, W159–W164. [Google Scholar] [CrossRef] [PubMed]
- Anonymous. NIST Chemistry WebBook; NIST Standard Reference Database Number 69; Linstrom, P.J., Mallard, W.G., Eds.; National Institute of Standards and Technology: Gaithersburg, MD, USA, 2023. Available online: https://webbook.nist.gov/chemistry/ (accessed on 12 September 2023).
- Javed, M.R.; Salman, M.; Tariq, A.; Tawab, A.; Zahoor, M.K.; Naheed, S.; Shahid, M.; Ijaz, A.; Ali, H. The Antibacterial and Larvicidal Potential of Bis-(2-Ethylhexyl) Phthalate from Lactiplantibacillus plantarum. Molecules 2022, 27, 7220. [Google Scholar] [CrossRef]
- Cholewiński, G.; Iwaszkiewicz-Grześ, D.; Prejs, M.; Głowacka, A.; Dzierzbicka, K. Synthesis of the inosine 5′-monophosphate dehydrogenase (IMPDH) inhibitors. J. Enzym. Inhib. Med. Chem. 2015, 30, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Reece, R.J.; Maxwell, A. DNA gyrase: Structure and function. Crit. Rev. Biochem. Mol. Biol. 1991, 26, 335–375. [Google Scholar] [CrossRef]
- Simon-Baram, H.; Roth, S.; Niedermayer, C.; Huber, P.; Speck, M.; Diener, J.; Richter, M.; Bershtein, S. A High-Throughput Continuous Spectroscopic Assay to Measure the Activity of Natural Product Methyltransferases. ChemBioChem 2022, 23, e202200162. [Google Scholar] [CrossRef] [PubMed]
- Pautsch, A.; Schulz, G.E. Structure of the outer membrane protein A transmembrane domain. Nat. Struct. Biol. 1998, 5, 1013–1017. [Google Scholar] [CrossRef]
- Kaushik, V.; Tiwari, M.; Tiwari, V. Interaction of RecA mediated SOS response with bacterial persistence, biofilm formation, and host response. Int. J. Biol. Macromol. 2022, 217, 931–943. [Google Scholar] [CrossRef] [PubMed]
- Varshney, A.K.; Wang, X.; Scharff, M.D.; MacIntyre, J.; Zollner, R.S.; Kovalenko, O.V.; Martinez, L.R.; Byrne, F.R.; Fries, B.C. Staphylococcal enterotoxin B–specific monoclonal antibody 20b1 successfully treats diverse Staphylococcus aureus Infections. J. Infect. Dis. 2013, 208, 2058–2066. [Google Scholar] [CrossRef] [PubMed]
- Audah, K.A.; Ettin, J.; Darmadi, J.; Azizah, N.N.; Anisa, A.S.; Hermawan, T.D.F.; Tjampakasari, C.R.; Heryanto, R.; Ismail, I.S.; Batubara, I. Indonesian Mangrove Sonneratia caseolaris Leaves Ethanol Extract Is a Potential Super Antioxidant and Anti Methicillin-Resistant Staphylococcus aureus Drug. Molecules 2022, 27, 8369. [Google Scholar] [CrossRef]
- Vanlalveni, C.; Lallianrawna, S.; Biswas, A.; Selvaraj, M.; Changmai, B.; Rokhum, S.L. Green synthesis of silver nanoparticles using plant extracts and their antimicrobial activities: A review of recent literature. RSC Adv. 2021, 11, 2804–2837. [Google Scholar] [CrossRef]
- Zemouri-Alioui, S.; Bachirbey, M.; Kurt, B.Z.; Sonmez, F.; Louaileche, H. Optimization of ultrasound-assisted extraction of total phenolic contents and antioxidant activity using response surface methodology from jujube leaves (Ziziphus jujuba) and evaluation of anticholinesterase inhibitory activity. J. Food Meas. Charact. 2019, 13, 321–329. [Google Scholar] [CrossRef]
- Song, L.; Liu, P.; Yan, Y.; Huang, Y.; Bai, B.; Hou, X.; Zhang, L. Supercritical CO2 fluid extraction of flavonoid compounds from Xinjiang jujube (Ziziphus jujuba Mill.) leaves and associated biological activities and flavonoid compositions. Ind. Crops Prod. 2019, 139, 111508. [Google Scholar] [CrossRef]
- Marques Casanova, J.; dos Santos Nascimento, L.B.; Marques Casanova, L.; Daflón Castricini, S.; Elis de Souza, J.; Kao Yien, R.M.; Sônia, S.C.; Schwartz Tavares, E. Kalanchoe fedtschenkoi R. Hamet & H. Perrier, a non-conventional food plant in Brazil: HPLC-DAD-ESI-MS/MS profile and leaf histochemical location of flavonoids. J. Appl. Bot. Food Qual. 2022, 95, 154. [Google Scholar]
- Charalambous, D.; Christoforou, M.; Kitiri, E.N.; Andreou, M.; Partassides, D.; Papachrysostomou, C.; Frantzi, M.; Karikas, G.A.; Pantelidou, M. Antimicrobial Activities of Saponaria cypria Boiss. Root Extracts, and the Identification of Nine Saponins and Six Phenolic Compounds. Molecules 2022, 27, 5812. [Google Scholar] [CrossRef]
- Kher, M.N.; Sheth, N.R.; Bhatt, V.D. In vitro antibacterial evaluation of Terminalia chebula as an alternative of antibiotics against bovine subclinical mastitis. Anim. Biotechnol. 2019, 30, 151–158. [Google Scholar] [CrossRef]
- Wink, M.; Botschen, G.C.; Schafer, H.; Waterman, P.G. Chemotaxonomy seen from a phylogenetic perspective and evolution of secondary metabolism. In Annual Plant Reviews; Biochemistry of Plant Secondary Metabolism; Chapter 7; Wink, M., Ed.; Blackwell: Oxford, UK, 2010; Volume 40. [Google Scholar]
- Bernard, F.X.; Sablé, S.; Cameron, B.; Provost, J.; Desnottes, J.F.; Crouzet, J.; Blanche, F. Glycosylated flavones as selective inhibitors of topoisomerase IV. Antimicrob. Agents Chemother. 1997, 41, 992–998. [Google Scholar] [CrossRef]
- Kolenc, Z.; Langerholc, T.; Hostnik, G.; Ocvirk, M.; Štumpf, S.; Pintarič, M.; Košir, I.J.; Čerenak, A.; Garmut, A.; Bren, U. Antimicrobial Properties of Different Hop (Humulus lupulus) Genotypes. Plants 2023, 12, 120. [Google Scholar] [CrossRef] [PubMed]
- Štumpf, S.; Hostnik, G.; Langerholc, T.; Pintarič, M.; Kolenc, Z.; Bren, U. The Influence of Chestnut Extract and Its Components on Antibacterial Activity against Staphylococcus aureus. Plants 2023, 12, 2043. [Google Scholar] [CrossRef] [PubMed]
- Lešnik, S.; Bren, U. Mechanistic Insights into Biological Activities of Polyphenolic Compounds from Rosemary Obtained by Inverse Molecular Docking. Foods 2022, 11, 67. [Google Scholar] [CrossRef] [PubMed]
- Kores, K.; Kolenc, Z.; Furlan, V.; Bren, U. Inverse Molecular Docking Elucidating the Anticarcinogenic Potential of the Hop Natural Product Xanthohumol and Its Metabolites. Foods 2022, 11, 1253. [Google Scholar] [CrossRef] [PubMed]
- Lešnik, S.; Jukič, M.; Bren, U. Mechanistic Insights of Polyphenolic Compounds from Rosemary Bound to Their Protein Targets Obtained by Molecular Dynamics Simulations and Free-Energy Calculations. Foods 2023, 12, 408. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).