Deletions across the SARS-CoV-2 Genome: Molecular Mechanisms and Putative Functional Consequences of Deletions in Accessory Genes
Abstract
1. Introduction
2. Materials and Methods
3. Results
3.1. Description of Dataset
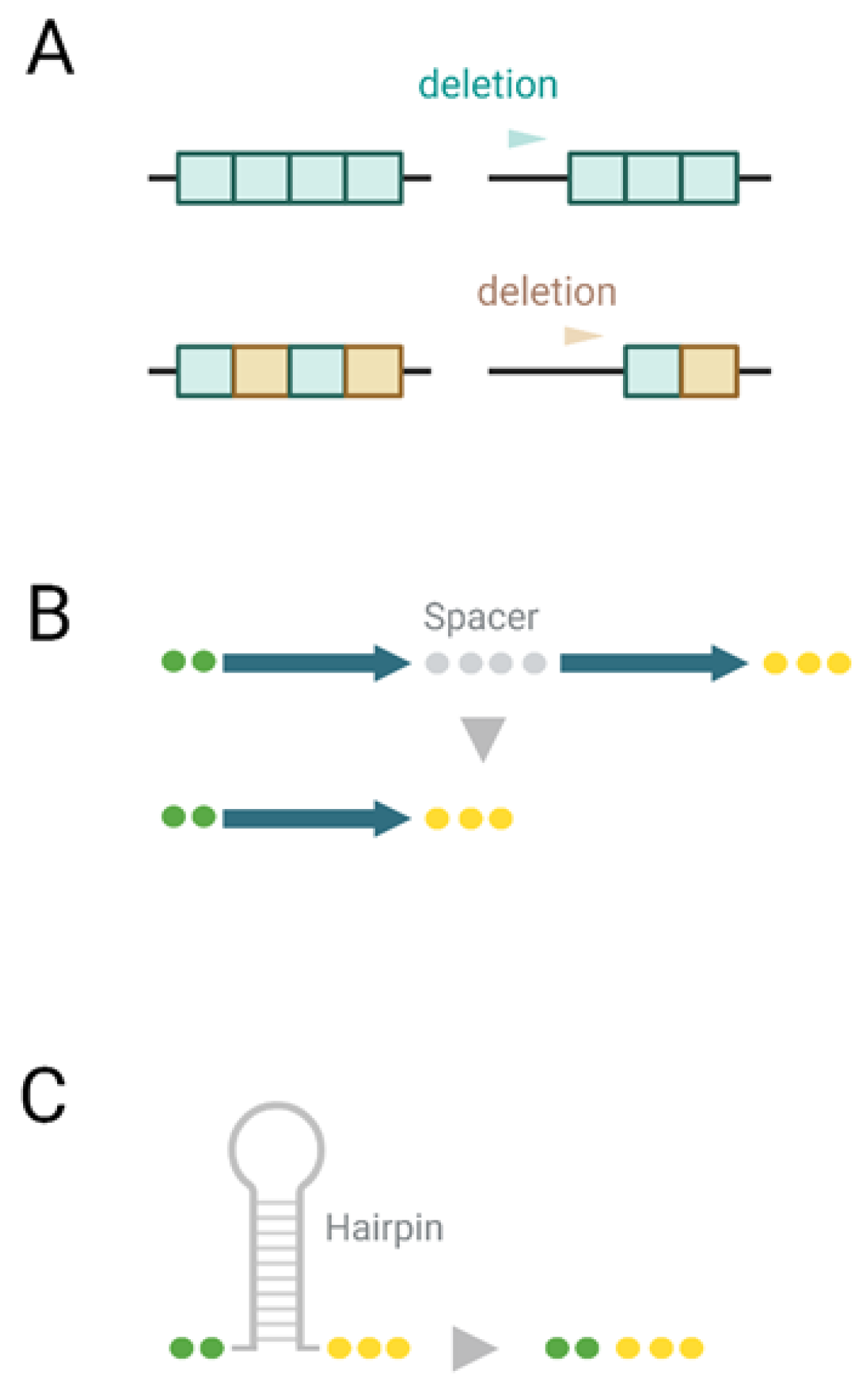
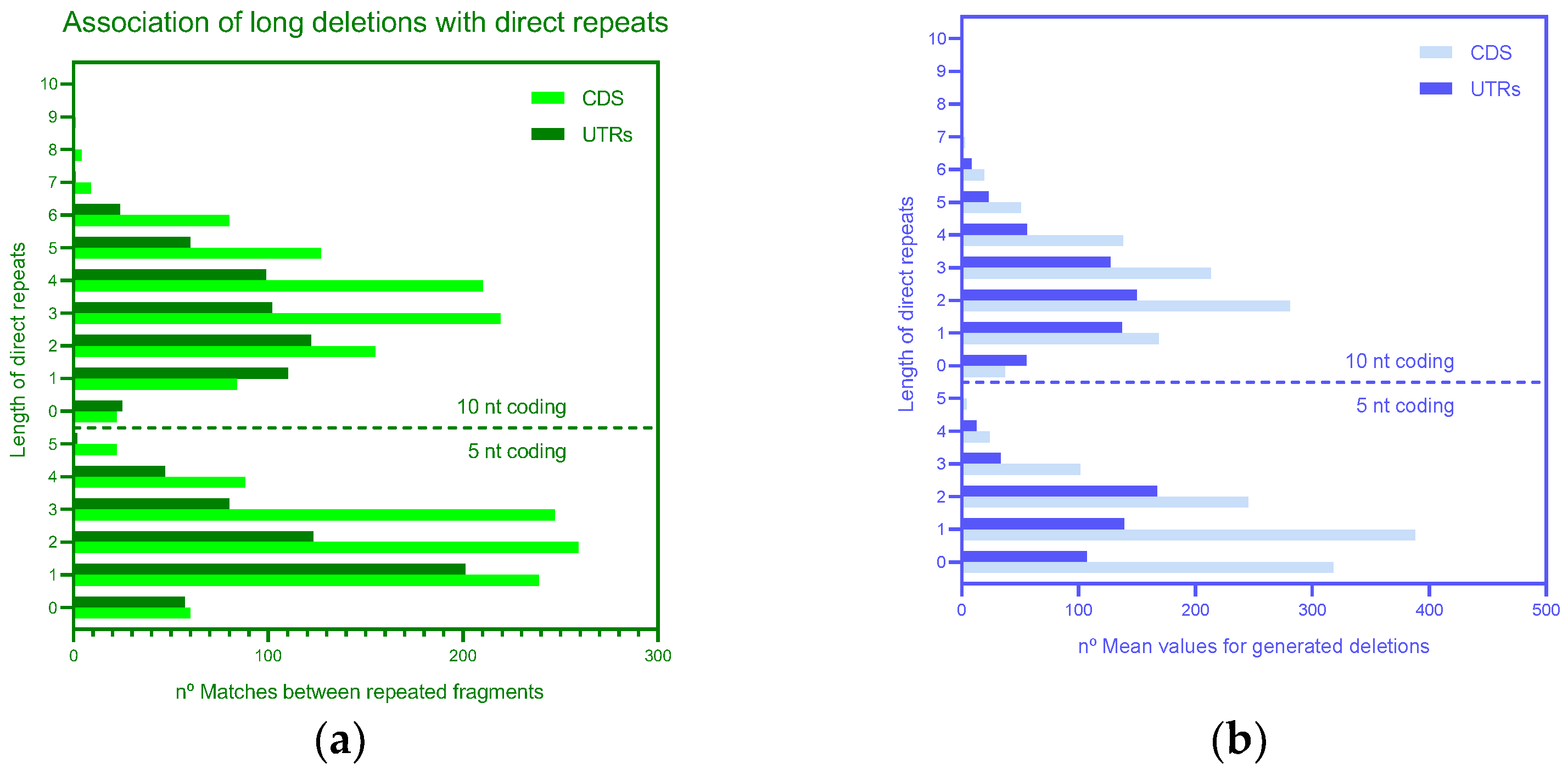
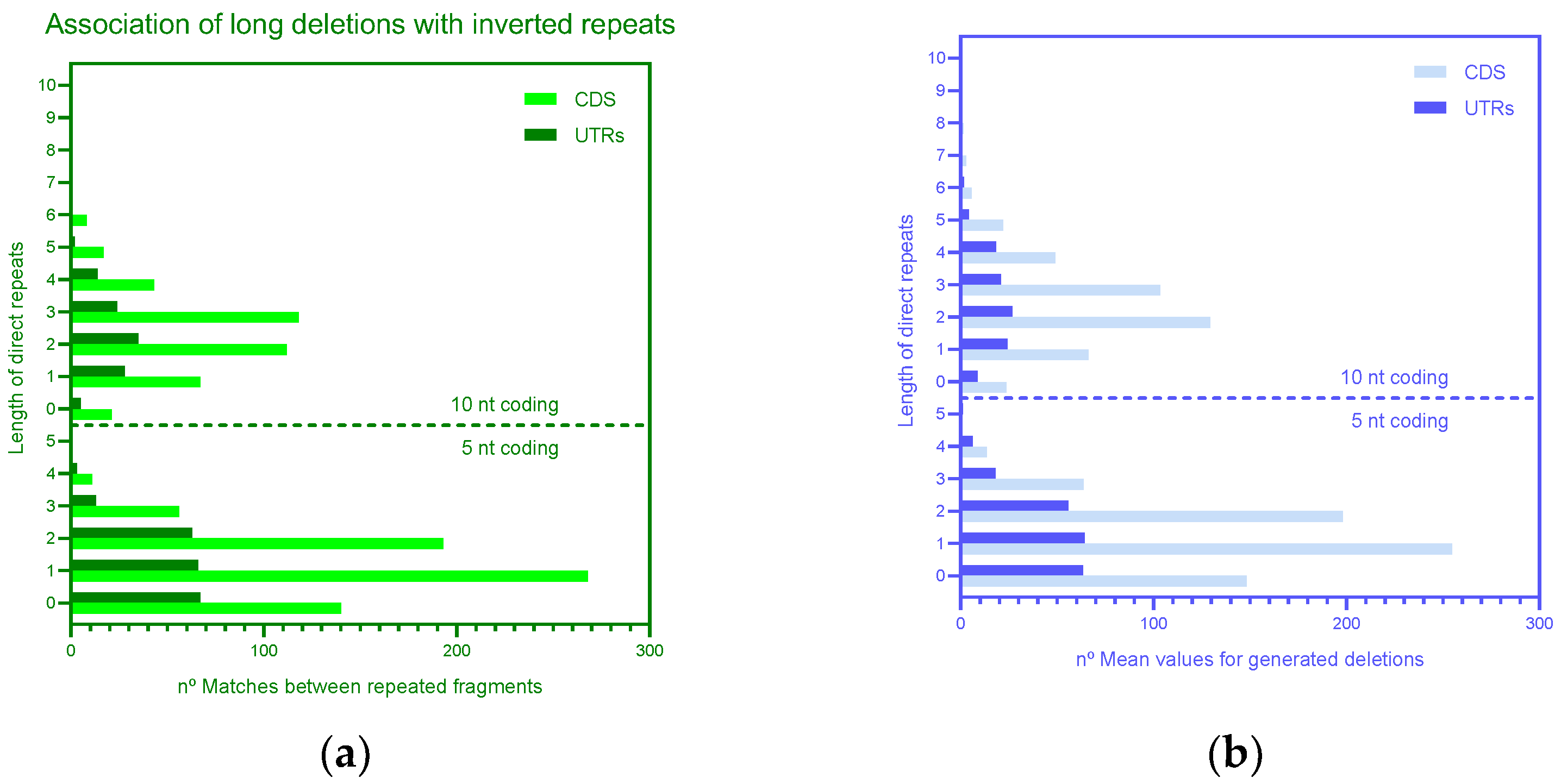
3.2. Mechanisms of Deletions
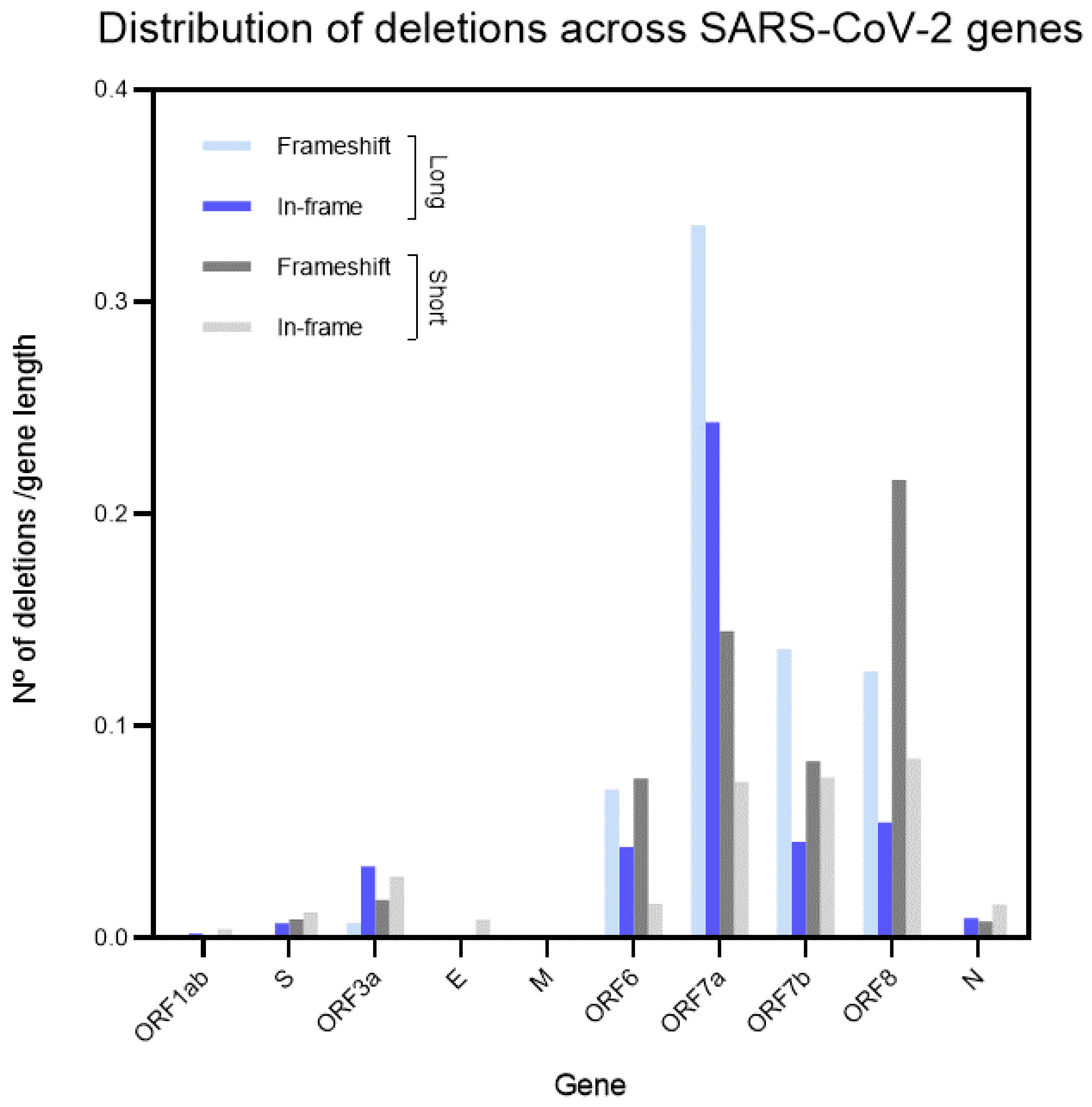
3.3. Distribution of Deletions across Genes
3.4. Deletions in ORF7a and ORF8: Putative Functional Consequences
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lovett, S.T. Encoded errors: Mutations and rearrangements mediated by misalignment at repetitive DNA sequences. Mol. Microbiol. 2004, 52, 1243–1253. [Google Scholar] [CrossRef]
- Bzymek, M.; Lovett, S.T. Instability of repetitive DNA sequences: The role of replication in multiple mechanisms. Proc. Natl. Acad. Sci. USA 2001, 98, 8319–8325. [Google Scholar] [CrossRef]
- Bzymek, M.; Saveson, C.J.; Feschenko, V.V.; Lovett, S.T. Slipped Misalignment Mechanisms of Deletion Formation: In Vivo Susceptibility to Nucleases. J. Bacteriol. 1999, 181, 477–482. [Google Scholar] [CrossRef]
- Bi, X.; Liu, L.F. recA-independent and recA-dependent Intramolecular Plasmid Recombination: Differential Homology Requirement and Distance Effect. J. Mol. Biol. 1994, 235, 414–423. [Google Scholar] [CrossRef]
- Chou, Q. Minimizing deletion mutagenesis artifact during Taq DNA polymerase PCR by E. coli SSB. Nucleic Acids Res. 1992, 20, 4371. [Google Scholar] [CrossRef][Green Version]
- Chédin, F.; Dervyn, E.; Dervyn, R.; Ehrlich, S.D.; Noirot, P. Frequency of deletion formation decreases exponentially with distance between short direct repeats. Mol. Microbiol. 1994, 12, 561–569. [Google Scholar] [CrossRef]
- Lovett, S.; Gluckman, T.J.; Simon, P.J.; Sutera, V.A.; Drapkin, P.T. Recombination between repeats in Escherichia coli by a recA-independent, proximity-sensitive mechanism. Mol. Gen. Genet. 1994, 245, 294–300. [Google Scholar] [CrossRef]
- Dianov, G.L.; Kuzminov, A.V.; Mazin, A.V.; Salganik, R.I. Molecular mechanisms of deletion formation in Escherichia coli plasmids. I. Deletion formation mediated by long direct repeats. Mol. Gen. Genet. 1991, 228, 153–159. [Google Scholar] [CrossRef]
- Mazin, A.V.; Kuzminov, A.V.; Dianov, G.L.; Salganik, R.I. Mechanisms of deletion formation in Escherichin coli plasmids. II. Deletions mediated by short direct repeats. Mol. Gen. Genet. 1991, 228, 209–214. [Google Scholar] [CrossRef]
- Albertini, A.M.; Hofer, M.; Calos, M.P.; Miller, J.H. On the formation of spontaneous deletions: The importance of short sequence homologies in the generation of large deletions. Cell 1982, 29, 319–328. [Google Scholar] [CrossRef]
- Efstratiadis, A.; Posakony, J.W.; Maniatis, T.; Lawn, R.M.; O’Connell, C.; Spritz, R.A.; Deriel, J.K.; Forget, B.G.; Weissman, S.M.; Slightom, J.L.; et al. The structure and evolution of the human β-globin gene family. Cell 1980, 21, 653–668. [Google Scholar] [CrossRef] [PubMed]
- Streisinger, G.; Okada, Y.; Emrich, J.; Newton, J.; Tsugita, A.; Terzaghi, E.; Inouye, M. Frameshift Mutations and the Genetic Code. Cold Spring Harb. Symp. Quant. Biol. 1966, 31, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Lovett, S.T.; Feschenko, V.V. Stabilization of diverged tandem repeats by mismatch repair: Evidence for deletion formation via a misaligned replication intermediate. Proc. Natl. Acad. Sci. USA 1996, 93, 7120–7124. [Google Scholar] [CrossRef] [PubMed]
- Trinh, T.Q.; Sinden, R.R. Preferential DNA secondary structure mutagenesis in the lagging strand of replication in E. coli. Nature 1991, 352, 544–547. [Google Scholar] [CrossRef]
- Bierne, H.; Vilette, D.; Ehrlich, S.D.; Michel, B. Isolation of a dnaE mutation which enhances RecA-independent homologous recombination in the Escherichia coli chromosome. Mol. Microbiol. 1997, 24, 1225–1234. [Google Scholar] [CrossRef] [PubMed]
- Saveson, C.J.; Lovett, S.T. Enhanced Deletion Formation by Aberrant DNA Replication in Escherichia coli. Genetics 1997, 146, 457–470. [Google Scholar] [CrossRef]
- Hu, X.; Worton, R.G. Partial gene duplication as a cause of human disease. Hum. Mutat. 1992, 1, 3–12. [Google Scholar] [CrossRef]
- Krawczak, M.; Cooper, D.N. Gene deletions causing human genetic disease: Mechanisms of mutagenesis and the role of the local DNA sequence environment. Hum. Genet. 1991, 86, 425–441. [Google Scholar] [CrossRef]
- Kondrashov, A.S.; Rogozin, I.B. Context of deletions and insertions in human coding sequences. Hum. Mutat. 2004, 23, 177–185. [Google Scholar] [CrossRef]
- Warren, S.T. The Expanding World of Trinucleotide Repeats. Science 1996, 271, 1374–1375. [Google Scholar] [CrossRef]
- Wu, A.; Wang, L.; Zhou, H.-Y.; Ji, C.-Y.; Xia, S.Z.; Cao, Y.; Meng, J.; Ding, X.; Gold, S.; Jiang, T.; et al. One year of SARS-CoV-2 evolution. Cell Host Microbe 2021, 29, 503–507. [Google Scholar] [CrossRef] [PubMed]
- Garushyants, S.K.; Rogozin, I.B.; Koonin, E.V. Template switching and duplications in SARS-CoV-2 genomes give rise to insertion variants that merit monitoring. Commun. Biol. 2021, 4, 1343. [Google Scholar] [CrossRef]
- Zinzula, L. Lost in deletion: The enigmatic ORF8 protein of SARS-CoV-2. Biochem. Biophys. Res. Commun. 2021, 538, 116–124. [Google Scholar] [CrossRef] [PubMed]
- Peacock, T.P.; Penrice-Randal, R.; Hiscox, J.A.; Barclay, W.S. SARS-CoV-2 one year on: Evidence for ongoing viral adaptation. J. Gen. Virol. 2021, 102, 001584. [Google Scholar] [CrossRef]
- Ceraolo, C.; Giorgi, F.M. Genomic variance of the 2019-nCoV coronavirus. J. Med. Virol. 2020, 92, 522–528. [Google Scholar] [CrossRef]
- Michel, C.J.; Mayer, C.; Poch, O.; Thompson, J.D. Characterization of accessory genes in coronavirus genomes. Virol. J. 2020, 17, 131. [Google Scholar] [CrossRef]
- Panzera, Y.; Calleros, L.; Goñi, N.; Marandino, A.; Techera, C.; Grecco, S.; Ramos, N.; Frabasile, S.; Tomás, G.; Condon, E.; et al. Consecutive deletions in a unique Uruguayan SARS-CoV-2 lineage evidence the genetic variability potential of accessory genes. PLoS ONE 2022, 17, e0263563. [Google Scholar] [CrossRef]
- Grubaugh, N.D.; Petrone, M.E.; Holmes, E.C. We shouldn’t worry when a virus mutates during disease outbreaks. Nat. Microbiol. 2020, 5, 529–530. [Google Scholar] [CrossRef]
- McCarthy, K.R.; Rennick, L.J.; Nambulli, S.; Robinson-McCarthy, L.R.; Bain, W.G.; Haidar, G.; Duprex, W.P. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science 2021, 371, 1139–1142. [Google Scholar] [CrossRef]
- Narayanan, K.; Huang, C.; Makino, S. SARS coronavirus accessory proteins. Virus Res. 2008, 133, 113–121. [Google Scholar] [CrossRef]
- Muth, D.; Corman, V.M.; Roth, H.; Binger, T.; Dijkman, R.; Gottula, L.T.; Gloza-Rausch, F.; Balboni, A.; Battilani, M.; Rihtarič, D.; et al. Attenuation of replication by a 29 nucleotide deletion in SARS-coronavirus acquired during the early stages of human-to-human transmission. Sci. Rep. 2018, 8, 15177. [Google Scholar] [CrossRef] [PubMed]
- Addetia, A.; Xie, H.; Roychoudhury, P.; Shrestha, L.; Loprieno, M.; Huang, M.-L.; Jerome, K.R.; Greninger, A.L. Identification of multiple large deletions in ORF7a resulting in in-frame gene fusions in clinical SARS-CoV-2 isolates. J. Clin. Virol. 2020, 129, 104523. [Google Scholar] [CrossRef] [PubMed]
- Panzera, Y.; Ramos, N.; Frabasile, S.; Calleros, L.; Marandino, A.; Tomás, G.; Techera, C.; Grecco, S.; Fuques, E.; Goñi, N.; et al. A deletion in SARS-CoV-2 ORF7 identified in COVID-19 outbreak in Uruguay. Transbound. Emerg. Dis. 2021, 68, 3075–3082. [Google Scholar] [CrossRef] [PubMed]
- Alisoltani, A.; Jaroszewski, L.; Iyer, M.; Iranzadeh, A.; Godzik, A. Increased Frequency of Indels in Hypervariable Regions of SARS-CoV-2 Proteins—A Possible Signature of Adaptive Selection. Front. Genet. 2022, 13, 875406. [Google Scholar] [CrossRef] [PubMed]
- Pancer, K.; Milewska, A.; Owczarek, K.; Dabrowska, A.; Kowalski, M.; Łabaj, P.P.; Branicki, W.; Sanak, M.; Pyrc, K. The SARS-CoV-2 ORF10 is not essential in vitro or in vivo in humans. PLOS Pathog. 2020, 16, e1008959. [Google Scholar] [CrossRef] [PubMed]
- Poliakov, E.; Koonin, E.V.; Rogozin, I.B. Impairment of translation of in neurons as a putative causative factor for autism. Biol. Direct 2014, 10, 16. [Google Scholar] [CrossRef]
- Khromov-Borisov, N.N.; Rogozin, I.; Henriques, J.A.P.; de Serres, F.J. Similarity pattern analysis in mutational distributions. Mutat. Res. 1999, 430, 55–74. [Google Scholar] [CrossRef]
- Rogozin, I.B.; Babenko, V.N.; Milanesi, L.; Pavlov, Y.I. Computational analysis of mutation spectra. Briefings Bioinform. 2003, 4, 210–227. [Google Scholar] [CrossRef][Green Version]
- Li, J.-Y.; Liao, C.-H.; Wang, Q.; Tan, Y.-J.; Luo, R.; Qiu, Y.; Ge, X.-Y. The ORF6, ORF8 and nucleocapsid proteins of SARS-CoV-2 inhibit type I interferon signaling pathway. Virus Res. 2020, 286, 198074. [Google Scholar] [CrossRef]
- Stadler, K.; Masignani, V.; Eickmann, M.; Becker, S.; Abrignani, S.; Klenk, H.-D.; Rappuoli, R. SARS—Beginning to understand a new virus. Nat. Rev. Microbiol. 2003, 1, 209–218. [Google Scholar] [CrossRef]
- Mohammed, M.E.A. The percentages of SARS-CoV-2 protein similarity and identity with SARS-CoV and BatCoV RaTG13 proteins can be used as indicators of virus origin. J. Proteins Proteom. 2021, 12, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Nelson, C.A.; Pekosz, A.; Lee, C.A.; Diamond, M.S.; Fremont, D.H. Structure and Intracellular Targeting of the SARS-Coronavirus Orf7a Accessory Protein. Structure 2005, 13, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.; Schneider, T.; Leong, M.; Aravind, L.; Zhang, D. Novel Immunoglobulin Domain Proteins Provide Insights into Evolution and Pathogenesis of SARS-CoV-2-Related Viruses. mBio 2020, 11, e00760-20. [Google Scholar] [CrossRef] [PubMed]
- Lau, S.K.P.; Feng, Y.; Chen, H.; Luk, H.K.H.; Yang, W.-H.; Li, K.S.M.; Zhang, Y.-Z.; Huang, Y.; Song, Z.-Z.; Chow, F.W.-N.; et al. Severe Acute Respiratory Syndrome (SARS) Coronavirus ORF8 Protein Is Acquired from SARS-Related Coronavirus from Greater Horseshoe Bats through Recombination. J. Virol. 2015, 89, 10532–10547. [Google Scholar] [CrossRef] [PubMed]
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452. [Google Scholar] [CrossRef]
- Rasmussen, A.L. On the origins of SARS-CoV-2. Nat. Med. 2021, 27, 9. [Google Scholar] [CrossRef]
- Postnikova, O.A.; Uppal, S.; Huang, W.; Kane, M.A.; Villasmil, R.; Rogozin, I.B.; Poliakov, E.; Redmond, T.M. The Functional Consequences of the Novel Ribosomal Pausing Site in SARS-CoV-2 Spike Glycoprotein RNA. Int. J. Mol. Sci. 2021, 22, 6490. [Google Scholar] [CrossRef]
- Seyran, M.; Pizzol, D.; Adadi, P.; El-Aziz, T.M.A.; Hassan, S.S.; Soares, A.; Kandimalla, R.; Lundstrom, K.; Tambuwala, M.; Aljabali, A.A.A.; et al. Questions concerning the proximal origin of SARS-CoV-2. J. Med. Virol. 2020, 93, 1204–1206. [Google Scholar] [CrossRef]
- Oostra, M.; de Haan, C.A.M.; Rottier, P.J.M. The 29-Nucleotide Deletion Present in Human but Not in Animal Severe Acute Respiratory Syndrome Coronaviruses Disrupts the Functional Expression of Open Reading Frame 8. J. Virol. 2007, 81, 13876–13888. [Google Scholar] [CrossRef]
- Liu, D.X.; Fung, T.S.; Chong, K.K.-L.; Shukla, A.; Hilgenfeld, R. Accessory proteins of SARS-CoV and other coronaviruses. Antivir. Res. 2014, 109, 97–109. [Google Scholar] [CrossRef]
- Guan, Y.; Zheng, B.J.; He, Y.Q.; Liu, X.L.; Zhuang, Z.X.; Cheung, C.L.; Luo, S.W.; Li, P.H.; Zhang, L.J.; Guan, Y.J.; et al. Isolation and Characterization of Viruses Related to the SARS Coronavirus from Animals in Southern China. Science 2003, 302, 276–278. [Google Scholar] [CrossRef] [PubMed]
- The Chinese SARS Molecular Epidemiology Consortium. Molecular Evolution of the SARS Coronavirus During the Course of the SARS Epidemic in China. Science 2004, 303, 1666–1669. [Google Scholar] [CrossRef] [PubMed]
- Mohammad, S.; Bouchama, A.; Mohammad Alharbi, B.; Rashid, M.; Saleem Khatlani, T.; Gaber, N.S.; Malik, S.S. SARS-CoV-2 ORF8 and SARS-CoV ORF8ab: Genomic Divergence and Functional Convergence. Pathogens 2020, 9, 677. [Google Scholar] [CrossRef] [PubMed]


| Statistics of Short Deletions (1–6 nt) | |||
|---|---|---|---|
| Coding | UTRs | ||
| Length | # deletions | Length | # deletions |
| 1 | 103 | 1 | 53 |
| 2 | 74 | 2 | 29 |
| 3 | 178 | 3 | 24 |
| 4 | 36 | 4 | 10 |
| 5 | 23 | 5 | 17 |
| 6 | 80 | 6 | 12 |
| Total | 494 | Total | 145 |
| Statistics of long deletions (>6 nt) | |||
| Coding | |||
| # deletions | Mean length | In-frame | Out-of-frame |
| 453 | 33.1 | 237 | 216 |
| UTRs | |||
| # deletions | Mean length | In-frame | Out-of-frame |
| 137 | 35.2 | 48 | 89 |
| Short Deletions (1 nt) | |||||
|---|---|---|---|---|---|
| Coding | UTRs | ||||
| Length | # deletions | # stretches 1 | Length | # deletions | # stretches 1 |
| 1 2 | 59 | 15,663 | 1 2 | 37 | 265 |
| 2 | 20 | 4269 | 2 | 8 | 62 |
| 3 | 12 | 1092 | 3 | 4 | 16 |
| 4 | 8 | 341 | 4 | 3 | 6 |
| 5 | 4 | 106 | |||
| Total | 103 | Total | 52 | ||
| Short deletions (2 nt) | |||||
| Coding | UTRs | ||||
| Dinucleotide stretches | # deletions | Dinucleotide stretches | # deletions | ||
| Yes | 10 | Yes | 5 | ||
| No | 64 | No | 24 | ||
| Short deletions (3–6 nt) | |||||
| Coding | UTRs | ||||
| Polynucleotide stretches | # deletions | Polynucleotide stretches | # deletions | ||
| Yes | 46 | Yes | 10 | ||
| No | 271 | No | 53 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rogozin, I.B.; Saura, A.; Bykova, A.; Brover, V.; Yurchenko, V. Deletions across the SARS-CoV-2 Genome: Molecular Mechanisms and Putative Functional Consequences of Deletions in Accessory Genes. Microorganisms 2023, 11, 229. https://doi.org/10.3390/microorganisms11010229
Rogozin IB, Saura A, Bykova A, Brover V, Yurchenko V. Deletions across the SARS-CoV-2 Genome: Molecular Mechanisms and Putative Functional Consequences of Deletions in Accessory Genes. Microorganisms. 2023; 11(1):229. https://doi.org/10.3390/microorganisms11010229
Chicago/Turabian StyleRogozin, Igor B., Andreu Saura, Anastassia Bykova, Vyacheslav Brover, and Vyacheslav Yurchenko. 2023. "Deletions across the SARS-CoV-2 Genome: Molecular Mechanisms and Putative Functional Consequences of Deletions in Accessory Genes" Microorganisms 11, no. 1: 229. https://doi.org/10.3390/microorganisms11010229
APA StyleRogozin, I. B., Saura, A., Bykova, A., Brover, V., & Yurchenko, V. (2023). Deletions across the SARS-CoV-2 Genome: Molecular Mechanisms and Putative Functional Consequences of Deletions in Accessory Genes. Microorganisms, 11(1), 229. https://doi.org/10.3390/microorganisms11010229







