Evaluation of a Lateral Flow Immunoassay for Rapid Detection of CTX-M Producers from Blood Cultures
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Microbiological Methods
2.2. Working Protocol for Directing Testing of Positive BCs with the NG-Test CTX-M MULTI
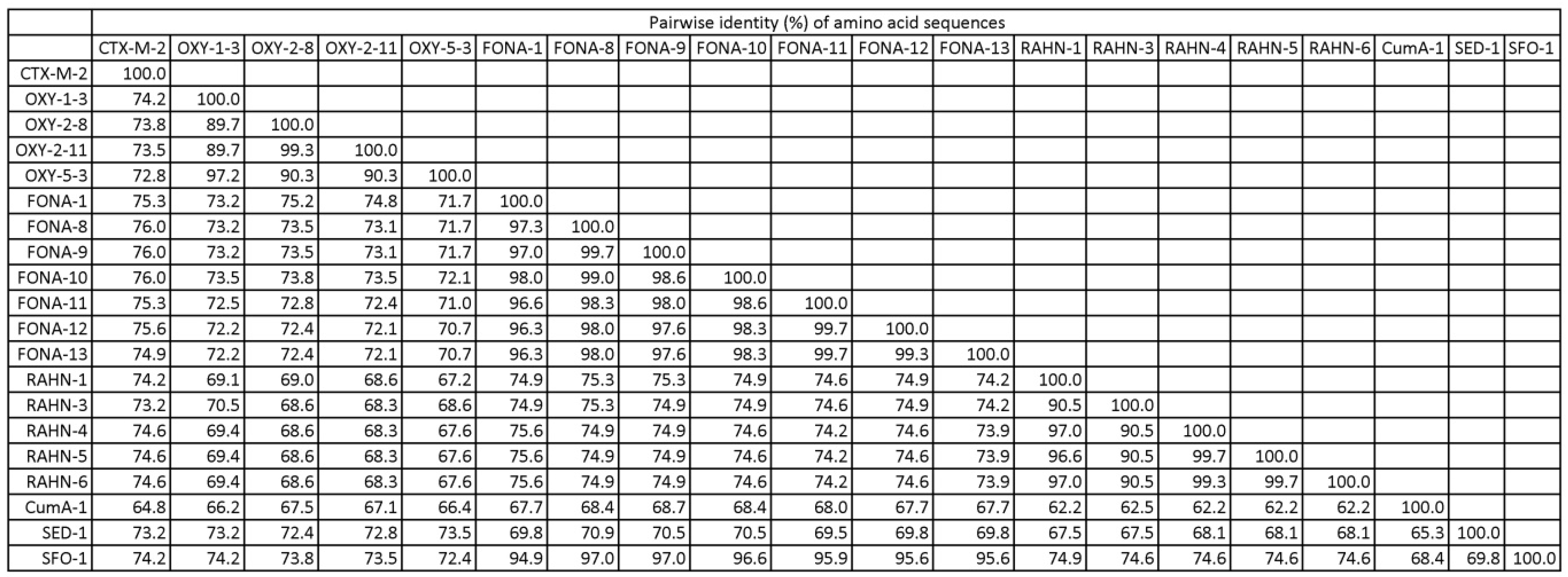
2.3. Whole Genome Sequencing and Bioinformatics
2.4. Data Analysis
3. Results
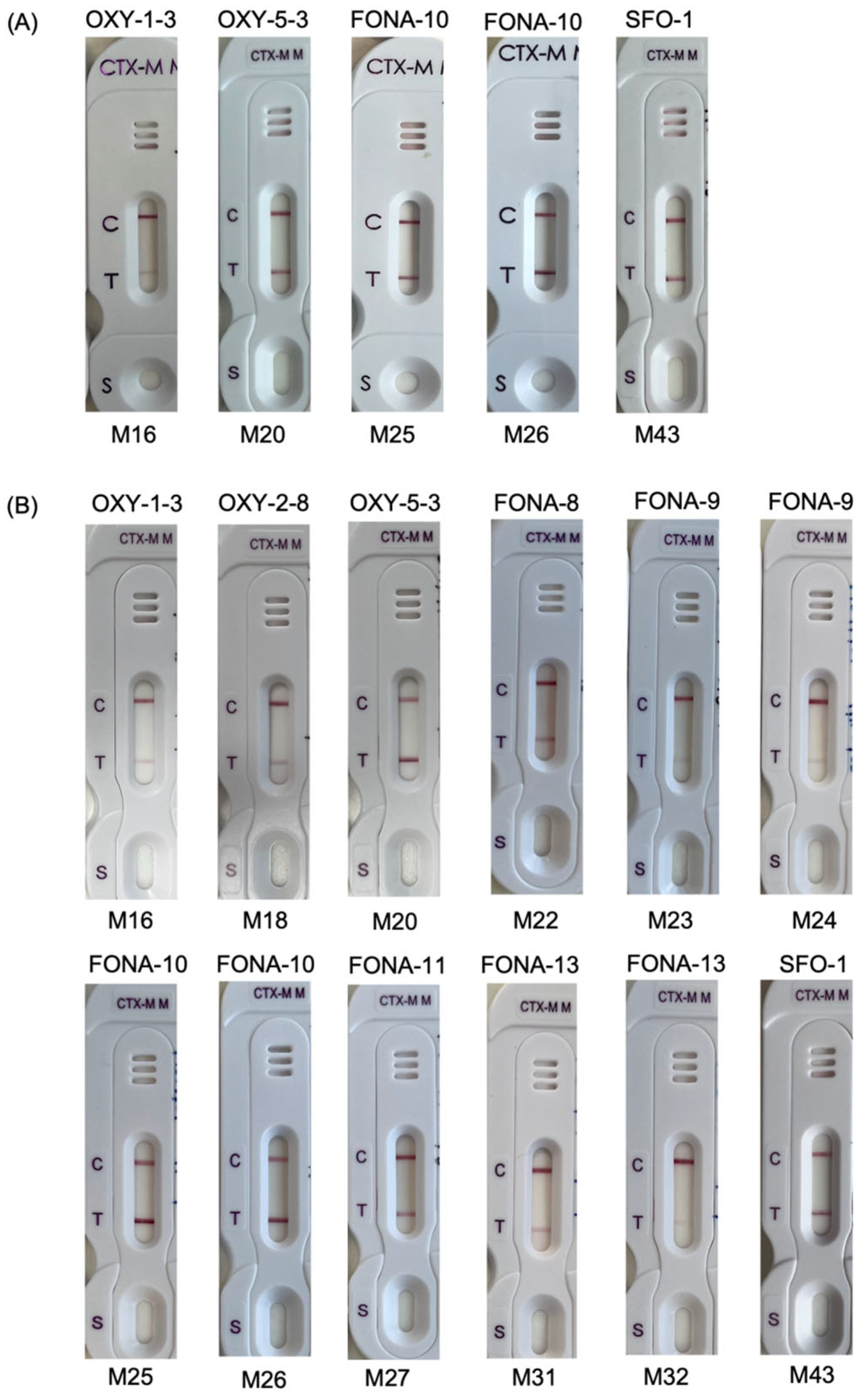
3.1. Spiked Blood Cultures
3.2. Clinical Blood Cultures
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institution Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bevan, E.R.; Jones, A.M.; Hawkey, P.M. Global epidemiology of CTX-M β-lactamases: Temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 2017, 72, 2145–2155. [Google Scholar] [CrossRef] [PubMed]
- Schwaber, M.J.; Carmeli, Y. Mortality and delay in effective therapy associated with extended-spectrum β-lactamase production in Enterobacteriaceae bacteraemia: A systematic review and meta-analysis. J. Antimicrob. Chemother. 2007, 60, 913–920. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, R. Growing group of extended-spectrum β-lactamases: The CTX-M enzymes. Antimicrob. Agents Chemother. 2004, 48, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Ho, P.-L.; Chu, Y.P.-S.; Lo, W.-U.; Chow, K.-H.; Law, P.Y.; Tse, C.W.-S.; Ng, T.-K.; Cheng, V.C.-C.; Que, T.-L. High prevalence of Escherichia coli sequence type 131 among antimicrobial-resistant E. coli isolates from geriatric patients. J. Med. Microbiol. 2015, 64, 243–247. [Google Scholar] [CrossRef][Green Version]
- Ho, P.-L.; Chau, P.-H.; Yan, M.-K.; Chow, K.-H.; Chen, J.H.; Wong, S.C.; Cheng, V.C. High burden of extended-spectrum β-lactamase-positive Escherichia coli in geriatric patients. J. Med. Microbiol. 2014, 63, 878–883. [Google Scholar] [CrossRef]
- To, K.K.; Lo, W.-U.; Chan, J.F.; Tse, H.; Cheng, V.C.; Ho, P.-L. Clinical outcome of extended-spectrum beta-lactamase-producing Escherichia coli bacteremia in an area with high endemicity. Int. J. Infect. Dis. 2013, 17, e120–e124. [Google Scholar] [CrossRef]
- Bernabeu, S.; Ratnam, K.C.; Boutal, H.; Gonzalez, C.; Vogel, A.; Devilliers, K.; Plaisance, M.; Oueslati, S.; Malhotra-Kumar, S.; Dortet, L. A lateral flow immunoassay for the rapid identification of CTX-M-producing Enterobacterales from culture plates and positive blood cultures. Diagnostics 2020, 10, 764. [Google Scholar] [CrossRef]
- Bianco, G.; Boattini, M.; Iannaccone, M.; Cavallo, R.; Costa, C. Evaluation of the NG-Test CTX-M MULTI immunochromatographic assay for the rapid detection of CTX-M extended-spectrum-β-lactamase producers from positive blood cultures. J. Hosp. Infect. 2020, 105, 341–343. [Google Scholar] [CrossRef]
- Boattini, M.; Bianco, G.; Comini, S.; Iannaccone, M.; Casale, R.; Cavallo, R.; Nordmann, P.; Costa, C. Direct detection of extended-spectrum-β-lactamase-producers in Enterobacterales from blood cultures: A comparative analysis. Eur. J. Clin. Microbiol. Infect. Dis. 2022, 41, 407–413. [Google Scholar] [CrossRef]
- Cendejas-Bueno, E.; del Pilar Romero-Gómez, M.; Falces-Romero, I.; Aranda-Diaz, A.; García-Ballesteros, D.; Mingorance, J.; García-Rodríguez, J. Evaluation of a lateral flow immunoassay to detect CTX-M extended-spectrum β-lactamases (ESBL) directly from positive blood cultures for its potential use in Antimicrobial Stewardship programs. Rev. Esp. Quimioter. 2022, 35, 284. [Google Scholar] [CrossRef]
- Comini, S.; Bianco, G.; Boattini, M.; Banche, G.; Ricciardelli, G.; Allizond, V.; Cavallo, R.; Costa, C. Evaluation of a diagnostic algorithm for rapid identification of Gram-negative species and detection of extended-spectrum β-lactamase and carbapenemase directly from blood cultures. J. Antimicrob. Chemother. 2022, 77, 2632–2641. [Google Scholar] [CrossRef] [PubMed]
- Keshta, A.S.; Elamin, N.; Hasan, M.R.; Pérez-López, A.; Roscoe, D.; Tang, P.; Suleiman, M. Evaluation of rapid immunochromatographic tests for the direct detection of extended spectrum beta-lactamases and carbapenemases in Enterobacterales isolated from positive blood cultures. Microbiol. Spectr. 2021, 9, e00785-21. [Google Scholar] [CrossRef] [PubMed]
- Walter, A.; Bodendoerfer, E.; Kolesnik-Goldmann, N.; Mancini, S. Rapid identification of CTX-M-type extended-spectrum β-lactamase-producing Enterobacterales from blood cultures using a multiplex lateral flow immunoassay. J. Glob. Antimicrob. Resist. 2021, 26, 130–132. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Siu, G.K.; Yeung, A.S.; Chen, J.H.; Ho, P.L.; Leung, K.; Tsang, J.L.; Cheng, V.C.; Guo, L.; Yang, J. Performance of the VITEK MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system for rapid bacterial identification in two diagnostic centres in China. J. Med. Microbiol. 2015, 64, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboaratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing, 32nd ed.; M100; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2022. [Google Scholar]
- Ho, P.L.; Yeung, M.K.; Lo, W.U.; Tse, H.; Li, Z.; Lai, E.L.; Chow, K.H.; To, K.K.; Yam, W.C. Predominance of pHK01-like incompatibility group FII plasmids encoding CTX-M-14 among extended-spectrum beta-lactamase–producing Escherichia coli in Hong Kong, 1996–2008. Diagn. Microbiol. Infect. Dis. 2012, 73, 182–186. [Google Scholar] [CrossRef] [PubMed]
- Giordano, L.; Fiori, B.; D’Inzeo, T.; Parisi, G.; Liotti, F.M.; Menchinelli, G.; De Angelis, G.; De Maio, F.; Luzzaro, F.; Sanguinetti, M. Simplified testing method for direct detection of carbapenemase-producing organisms from positive blood cultures using the NG-Test Carba 5 assay. Antimicrob. Agents Chemother. 2019, 63, e00550-19. [Google Scholar] [CrossRef]
- Lee, C.-H.; Jiang, S.; Tong, M.-K.; Tse, C.W.-S.; Ho, P.-L. False-positive immunochromatographic CTX-M enzyme test from direct testing of a positive blood culture. Eur. J. Clin. Microbiol. Infect. Dis. 2022, 41, 511–513. [Google Scholar] [CrossRef]
- Cao, H.; Liu, M.C.-J.; Tong, M.-K.; Jiang, S.; Lau, A.; Chow, K.-H.; Tse, C.W.-S.; Ho, P.-L. Diversity of genomic clusters and CfiA/cfiA alleles in Bacteroides fragilis isolates from human and animals. Anaerobe 2022, 75, 102567. [Google Scholar] [CrossRef]
- Cao, H.; Liu, M.C.-J.; Tong, M.-K.; Jiang, S.; Chow, K.-H.; To, K.K.-W.; Tse, C.W.-S.; Ho, P.-L. Comprehensive investigation of antibiotic resistance gene content in cfiA-harboring Bacteroides fragilis isolates of human and animal origins by whole genome sequencing. Int. J. Med. Microbiol. 2022, 312, 151559. [Google Scholar] [CrossRef]
- Li, Y.; Xu, Z.; Han, W.; Cao, H.; Umarov, R.; Yan, A.; Fan, M.; Chen, H.; Duarte, C.M.; Li, L. HMD-ARG: Hierarchical multi-task deep learning for annotating antibiotic resistance genes. Microbiome 2021, 9, 1–12. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.-X.; Yu, T.; Wang, Y.; Zhang, R. Detection of CTX-M-64 in Escherichia coli isolates from human patients in China. Antimicrob. Agents Chemother. 2015, 59, 1371–1372. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ho, P.-L.; Liu, M.C.-J.; Lo, W.-U.; Lai, E.L.-Y.; Lau, T.C.-K.; Law, O.-K.; Chow, K.-H. Prevalence and characterization of hybrid blaCTX-M among Escherichia coli isolates from livestock and other animals. Diagn. Microbiol. Infect. Dis. 2015, 82, 148–153. [Google Scholar] [CrossRef] [PubMed]
- Ortiz de la Rosa, J.M.; Demord, A.; Poirel, L.; Greub, G.; Blanc, D.; Nordmann, P. False immunological detection of CTX-M enzymes in Klebsiella oxytoca. J. Clin. Microbiol. 2021, 59, e00609-21. [Google Scholar] [CrossRef] [PubMed]
- Fuentes-Castillo, D.; Power, P.; Cerdeira, L.; Cardenas-Arias, A.; Moura, Q.; Oliveira, F.A.; Levy, C.E.; Gutkind, G.; Catão-Dias, J.L.; Lincopan, N. FONA-7, a novel extended-spectrum β-lactamase variant of the FONA family identified in Serratia fonticola. Microb. Drug Resist. 2021, 27, 585–589. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Wang, P.; Ma, Y.; Yang, Y.; Ye, X.; Wang, M. Co-production of SFO-1 and DHA-1 β-lactamases and 16S rRNA methylase ArmA in clinical isolates of Klebsiella pneumoniae. J. Antimicrob. Chemother. 2012, 67, 2361–2366. [Google Scholar] [CrossRef]
- Morgenthaler, N.G.; Kostrzewa, M. Rapid identification of pathogens in positive blood culture of patients with sepsis: Review and meta-analysis of the performance of the sepsityper kit. Int. J. Microbiol. 2015, 2015, 827416. [Google Scholar] [CrossRef]
- Chen, J.H.; Ho, P.-L.; Kwan, G.S.; She, K.K.; Siu, G.K.; Cheng, V.C.; Yuen, K.-Y.; Yam, W.-C. Direct bacterial identification in positive blood cultures by use of two commercial matrix-assisted laser desorption ionization–time of flight mass spectrometry systems. J. Clin. Microbiol. 2013, 51, 1733–1739. [Google Scholar] [CrossRef]
| Group | Organism | ESBL | n | Spiked Blood Culture (No.) | Culture Growth (No.) | ||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | ||||
| CTX-M-1G | E. coli | CTX-M-1 | 2 | 2 | |||
| E. coli | CTX-M-15 | 2 | 2 | ||||
| E. coli | CTX-M-55 | 1 | 1 | ||||
| CTX-M-9G | E. coli | CTX-M-13 | 1 | 1 | |||
| E. coli | CTX-M-14 | 1 | 1 | ||||
| E. coli | CTX-M-27 | 1 | 1 | ||||
| E. coli | CTX-M-65 | 1 | 1 | ||||
| E. coli | CTX-M-191 | 1 | 1 | ||||
| CTX-M hybrid | E. coli | CTX-M-64 | 1 | 1 | |||
| E. coli | CTX-M-123 | 2 | 2 | ||||
| E. coli | CTX-M-132 | 2 | 2 | ||||
| OXY | K. michiganensis | OXY-1-3 | 1 | 1 | 1 | ||
| K. oxytoca | OXY-2-8 | 2 | 2 | 1 | 1 | ||
| K. oxytoca | OXY-2-11 | 1 | 1 | 1 | |||
| K. oxytoca | OXY-5-3 | 1 | 1 | 1 | |||
| FONA | S. fonticola | FONA-1 | 1 | 1 | 1 | ||
| S. fonticola | FONA-8 | 1 | 1 | 1 | |||
| S. fonticola | FONA-9 | 2 | 2 | 2 | |||
| S. fonticola | FONA-10 | 2 | 2 | 2 | |||
| S. fonticola | FONA-11 | 2 | 2 | 1 | 1 | ||
| S. fonticola | FONA-12 | 2 | 2 | 2 | |||
| S. fonticola | FONA-13 | 2 | 2 | 2 | |||
| RAHN | R. aquatilis | RAHN-1 | 2 | 2 | 2 | ||
| R. aquatilis | RAHN-3 | 1 | 1 | 1 | |||
| R. aquatilis | RAHN-4 | 1 | 1 | 1 | |||
| R. aquatilis | RAHN-5 | 1 | 1 | 1 | |||
| R. aquatilis | RAHN-6 | 2 | 2 | 2 | |||
| Others | P. vulgarus | CumA-1 | 1 | 1 | 1 | ||
| C. sedlakii | SED-1 | 1 | 1 | 1 | |||
| K. aerogenes | SFO-1 | 1 | 1 | 1 | |||
| No. (%) of Clinical Blood Cultures | No. of Species in the Blood Cultures | |
|---|---|---|
| Hospital source | ||
| KWH-lab | 180 (51.4) | 26 |
| QMH-lab | 170 (48.6) | 29 |
| Sample origin | ||
| Aerobic bottle | 178 (50.9) | 27 |
| Anaerobic bottle | 172 (49.1) | 28 |
| Detected organisms | ||
| Escherichia coli | 197 (56.3) | 1 |
| Klebsiella pneumoniae | 40 (11.4) | 1 |
| Proteus mirabilis | 19 (5.4) | 1 |
| Other Enterobacterales a | 28 (8.0) | 16 |
| Pseudomonas | 29 ((8.2) | 6 |
| Anaerobes | 13 (3.7) | 8 |
| Acinetobacter | 9 (2.6) | 3 |
| Other bacteria a | 15 (4.3) | 10 |
| Number of organisms | ||
| Monomicrobial | 342 (97.7) | 42 |
| Polymicrobial | 8 (2.3) | 11 |
| CTX-M identified | ||
| CTX-M M1 group | 40 (11.4) | 4 |
| CTX-M M9 group | 50 (14.3) | 3 |
| Negative for CTX-M | 260 (74.3) | 45 |
| Total | 350 (100) | 46 |
| Sample Description | Result (No.) | Test Performance a | ||||
|---|---|---|---|---|---|---|
| TP | FP | FN | TN | Sensitivity | Specificity | |
| Testing performed the next day on 130 clinical BCs following bacterial identification | 38 | 0 | 0 | 92 | 100% (91.0–100%) | 100% (96.1–100%) |
| Testing performed on 220 clinical BCs immediately after Gram stain showing Gram negative bacilli | 52 | 1 | 0 | 167 | 100% (93.1–100%) | 99.4% (96.7–100%) |
| Total | 90 | 1 | 0 | 259 | 100 (96.0–100%) | 99.6% (97.9–100%) |
| Source | Year | No. of Clinical (Spike) BCs Tested | Organisms in BCs (No. of Species) | Reference Method g | No. | Sensitivity, % (95% CI) | Specificity, % (95% CI) | |||
|---|---|---|---|---|---|---|---|---|---|---|
| TP | TN | FP | FN | |||||||
| [8] | 2020 | 166 | Enterobacterales (11) a | G | 43 | 123 | 0 | 0 | 100 (91.8–100) | 100 (97.1–100) |
| [7] | 2020 | 100 | Gram negative bacilli b | G | 10 | 90 | 0 | 0 | 100 (69.2–100) | 100 (96.1–100) |
| [12] | 2021 | 49 (32) | Enterobacterales (6) c | P | 26 | 55 | 0 | 0 | 100 (86.8–100) | 100 (93.5–100) |
| [13] | 2021 | 0 (167) | Enterobacterales (9) d | G | 124 | 38 | 2 | 3 h | 97.6 (93.3–99.5) | 95.0 (83.1–99.4) |
| [10] | 2022 | 61 (19) | Enterobacterales (3) e | P | 26 | 53 | 0 | 1 h | 96.3 (81.0–99.9) | 100 (93.3–100) |
| [9] | 2022 | 142 | Enterobacterales (2) f | P | 34 | 105 | 0 | 3 i | 91.9 (78.1–98.3) | 100 (96.6–100) |
| [11] | 2022 | 1055 | Enterobacterales (2) f | P | 273 | 757 | 0 | 25 | 91.6 (87.9–94.5) | 100 (99.5–100) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, H.; Lee, C.-H.; Cao, H.; Jiang, S.; So, S.Y.-C.; Tse, C.W.-S.; Cheng, V.C.-C.; Ho, P.-L. Evaluation of a Lateral Flow Immunoassay for Rapid Detection of CTX-M Producers from Blood Cultures. Microorganisms 2023, 11, 128. https://doi.org/10.3390/microorganisms11010128
Fang H, Lee C-H, Cao H, Jiang S, So SY-C, Tse CW-S, Cheng VC-C, Ho P-L. Evaluation of a Lateral Flow Immunoassay for Rapid Detection of CTX-M Producers from Blood Cultures. Microorganisms. 2023; 11(1):128. https://doi.org/10.3390/microorganisms11010128
Chicago/Turabian StyleFang, Hanshu, Chung-Ho Lee, Huiluo Cao, Shuo Jiang, Simon Yung-Chun So, Cindy Wing-Sze Tse, Vincent Chi-Chung Cheng, and Pak-Leung Ho. 2023. "Evaluation of a Lateral Flow Immunoassay for Rapid Detection of CTX-M Producers from Blood Cultures" Microorganisms 11, no. 1: 128. https://doi.org/10.3390/microorganisms11010128
APA StyleFang, H., Lee, C.-H., Cao, H., Jiang, S., So, S. Y.-C., Tse, C. W.-S., Cheng, V. C.-C., & Ho, P.-L. (2023). Evaluation of a Lateral Flow Immunoassay for Rapid Detection of CTX-M Producers from Blood Cultures. Microorganisms, 11(1), 128. https://doi.org/10.3390/microorganisms11010128






