Abstract
A prospective 3-month study carried out in 267 ICUs revealed an S. aureus nosocomial bacteremia in one admitted patient out of 110 in adult and pediatric sectors, and in one out of 230 newborns; 242 S. aureus bacteremias occurred during the study, including 7.9% MRSA-bacteremias. In one ICU out of ten, the molecular characteristics, antimicrobial susceptibility profiles and biofilm production of the strains responsible for S. aureus bacteremia were studied. Of the 53 strains studied, 9.4% were MRSA and 52.8% were resistant to erythromycin. MLST showed the predominance of CC398 (37.7% of the strains) followed by CC8 (17.0%), CC45 (13.2%) and CC30 (9.4%). The lukF/S genes were absent from our isolates and tst-1 was found in 9.4% of the strains. Under static conditions and without exposure to glucose, biofilm production was rare (9.4% of the strains, without any CC398). The percentage increased to 62.3% for strains grown in broth supplemented with 1% glucose (including 7 out of 9 CC8 and 17 out of the 20 CC398). Further study of the CC398, including whole genome sequencing, revealed (1) highly frequent patient death within seven days after CC398 bacteremia diagnosis (47.4%), (2) 95.0% of the strains producing biofilm when exposed to sub-inhibitory concentrations of cloxacillin, (3) a stronger biofilm production following exposure to cloxacillin than that observed in broth supplemented with glucose only (p < 0.001), (4) a high minimum biofilm eradication concentration of cloxacillin (128 mg/L) indicating a low cloxacillin susceptibility of biofilm-growing CC398, (5) 95.0% of the strains carrying a ϕSa-3 like prophage and its particular evasion cluster (i.e., yielding chp and scin genes), and (6) 30.0% of the strains carrying a ϕMR11-like prophage and yielding a higher ability to produce biofilm. Our results provide evidence that active surveillance is required to avoid spreading of this virulent staphylococcal clone.
Keywords:
Staphylococcus aureus; intensive care; CC398; bacteremia; biofilm; cloxacillin; antibiotic tolerance 1. Introduction
S. aureus possesses numerous virulence factors facilitating tissue colonization, immune evasion and tissue destruction. One of its defense mechanisms is the capacity to form biofilms. Bacteria embedded in biofilms are resistant to host immune response and difficult to eradicate with antibiotic [1,2]. Biofilm-forming capacity is a virulence determinant in the development of catheter-related infections, and the effective treatment of staphylococcal infections that share such features has become a challenge [3].
Described initially in livestock, S. aureus of clonal complex 398 (CC398) are increasingly responsible for bacteremia in humans living in animal-free environments [4,5,6,7]. Whole genome sequencing analysis of CC398 strains have demonstrated that livestock-associated and emerging strains differ by their prophage content [8,9]. The emerging strains usually carry ϕSa-3 and ϕMR11-like prophages, which have been shown to be involved in immune escape, adhesion to host cells and extracellular matrix components, along with epithelial cell invasion, i.e., factors contributing to an increased ability of the bacteria to colonize and infect the host [10].
Enhanced biofilm formation ability was recently described in emerging CC398 strains in China [11]. Facing the epidemiological changes observed with this clone in human clinical settings, epidemiological data remain scarce. We conducted a prospective incidence-study of nosocomial bacteremia in 267 intensive care units (ICUs), and analyzed the molecular characteristics and antimicrobial susceptibility profiles of the S. aureus strains responsible for bacteremia in one ICU out of ten. We sequenced the genome of the CC398 strains, analyzed their prophage content, studied their ability to produce biofilm following exposure to sub-inhibitory concentrations of cloxacillin, the first line treatment for patients suffering from MSSA bacteremia, and determined their cloxacillin susceptibility in biofilm, where applicable.
2. Materials and Methods
2.1. Nosocomial Bacteremia Survey, Data Collection and Analysis
In each participating ICU, a 3-month surveillance of nosocomial bacteremia was carried out between 1 January and 15 August 2021. A protocol derived from that of the ECDC HAI light surveillance protocol was used (https://www.ecdc.europa.eu/sites/default/files/documents/HAI-Net-ICU-protocol-v2.2_0.pdf; accessed on 1 January 2021). During the survey period, each positive blood culture was analyzed to determine whether it was associated with a nosocomial bacteremia. The origin of the bacteremia was determined using clinical and biological data. Data were collected (HCF data, patient characteristics (sex, age, birth weight for newborns, the severity index, the immune status, the type of cancer where applicable, and death within 7 days of bacteremia diagnosis), characteristics of the bacteremia (origin (skin (primary cutaneous form or superinfection of a skin wound), surgical site, lungs, urinary tract, intravascular device, intra-abdominal or digestive tract) and microorganisms). Data processing, validation of the database and data analysis were carried out by the national team using R software (version 3.6.1 on Ubuntu; General Public Licence; Vienna, Austria). For the variables studied, percentages were calculated from the numbers, without taking missing data into account. Incidence rates were generated according to patient-days and admitted patients. Here, we report the data regarding S. aureus bacteremias.
2.2. Microbiological Study
Local infection control teams were asked to send the S. aureus strains isolated from bacteremia to the central lab using transport swabs (Copan Italia SPA, Brescia, Italy). The strains were tested for antimicrobial drug susceptibility using the AST-P631 vitek card (oxacillin, cefoxitin, gentamicin, tobramycin, ofloxacin, erythromycin, linezolid, teicoplanin, vancomycin, tetracycline, nitrofurantoin, fusidic acid, rifampicin, trimethoprim-sulfamethoxazol, fosfomycin; bioMérieux, France), Etest® (daptomycin, mupirocine; bioMérieux, Marcy-l’Étoile, France) and Eucast guidelines (http://www.eucast.org/; accessed on 1 January 2021). The presence of mecA/C, mupA/B, qacAB/C, tst-1, and lukF/S genes was determined using PCR. Strain genomic diversity was studied using MLST (https://pubmlst.org/organisms/staphylococcus-aureus; accessed on 1 January 2021). Purified genomic DNA for CC398 strains was sequenced on the Illumina HiSeq (Illumina, San Diego, CA, USA) using 100 base-pair paired-ends read and bar code strategy according to the Nextera XT kit (Illumina, San Diego, CA, USA), following the manufacturer’s recommendations [12]. The ability to produce biofilm was assessed under static conditions using the method of Christensen [13]. Bacteria were grown at 37 °C in Tryptic Soy broth (TSB) or TSB supplemented with 1% D-(+)-glucose (Sigma Aldrich), or TSB supplemented with 1% D-(+)-glucose and cloxacillin at a concentration of MIC/4. After 48 h of growth, the plates were washed three times with Phosphate-buffered saline, prior to staining with a 0.4% crystal violet solution. Each strain was tested three times. A biofilm-positive phenotype was defined as an optical density at 595 nm twice that of the negative control, and the strains were divided into four categories: no biofilm producer, weak, moderate and strong producers [14]. To assess a decrease in susceptibility to cloxacillin, bacteria in biofilm were tested using the Calgary biofilm device (CBD, Innovatech Inc., Edmonton, Alberta, Canada) to determine the minimum biofilm eradication concentration of cloxacillin (MBEC) following the manufacturer’s recommendations [15]. The biofilm inhibitory concentrations were defined as the lowest concentrations of drug resulting in an OD650 nm difference of ≤10% of the mean of two positive control well readings.
2.3. Statistical Analysis and Ethics Approval
For categorical variables, we used Pearson’s chi-squared test to compare groups. All analyses were two-tailed and a p < 0.05 was considered significant. We used Stata version 10.0 software (Stata Corp., College Station, TX, USA) for statistical analysis.
Ethics approval for the survey was obtained from the Réseau national de Prévention des Infections Associées aux soins (REPIAS), Santé Publique France national agency. Written informed consent was exempted, since the study focused on bacteria and patient intervention was not required.
3. Results
3.1. Nosocomial bacteremia 3-Month Survey
The survey was carried out in 267 ICUs at 212 French hospitals. Monitoring focused on 3668 beds (i.e., 52% of French ICU beds) and covered 313,891 patient-days (PDs). A total of 1931 nosocomial bacteremias occurred during the study in the ICUs, and out of these bacteremias, 242 were S. aureus bacteremias (12.5%), including 7.9% MRSA-bacteremias (19/240; 2 nk). An S. aureus bacteremia was detected in one admitted patient out of 110 in adult and pediatric sectors, and in one out of 230 newborns (Table 1).

Table 1.
S. aureus bacteremia incidence rates.
The characteristics of the infected patients and their bacteremias are presented in Table 2. The 242 patients suffering S. aureus bacteremia had frequent immunodepression (12.3%), cancer (13.6%), and COVID-19 infection (61.5%). In adults, the IGS II severity score was 40.0. The major sources of the S. aureus bacteremias were the lungs (50.0%) and intravascular-catheters (62; 25.6%). Death within the first week following infection diagnosis was notified in 69 cases (28.8%).

Table 2.
Characteristics of the patients suffering from bacteremia.
3.2. Microbiological Study
Of the 267 ICUs participating in the survey, 30 took part in the microbiological study (11.2%). Despite this modest ICU participation in our study, the number of S. aureus bacteremias detected in these 30 ICUs represented 22.7% of the whole group of 242 bacteremias. In addition, whereas 55 S. aureus bacteremias occurred during the study in these 30 ICUs, 53 S. aureus strains were available (96.4%). As there were no significant differences regarding the source of the 53 bacteremias compared with those of the 242 from the national level, we considered the 53 strains representative of the national set of S. aureus bacteremias (Table 2).
Of the 53 S. aureus strains, 5 were MRSA (9.4%) and 28 were resistant to erythromycin (52.8%) (Table 3). MLST revealed 20 CC398 strains (37.7%), 9 CC8 (17.0%), 7 CC45 (13.2%), 5 CC30 (9.4%), and 12 in 6 other CCs. mupA, qacAB, and qacC were carried by 2 CC8, 1 CC398, and 1 CC8 strain, respectively; tst-1 was found in 5 strains (9.4%); the lukF/S genes encoding PVL were absent from our isolates. An association was found (1) between CC8 and methicillin resistance (p = 0.039), fusidic acid resistance (p < 0.001), and mupirocin resistance (p = 0.026), (2) between CC398 strains and erythromycin resistance (p < 0.001), and (3) between CC30 strains and the tst-1 gene (p < 0.001).

Table 3.
Antimicrobial susceptibility profiles, virulence genes and biofilm production of the 53 S. aureus strains, bacteremia source and patient death within 7 days after bacteremia diagnosis, according to sequence type obtained by MLST.
Under static conditions and without exposure to glucose, a biofilm production was observed in 5 strains (9.4%), including one strong producer. The percentage increased to 60.4% for strains grown in broth supplemented with 1% glucose, including 6 strong producers (18.7%). No association was found between any clone and a particular bacteremia source. Patient death within seven days after bacteremia diagnosis was most frequent with bacteremia associated with a CC398 or a CC45 strain (47.4% and 42.9%, respectively) compared to bacteremia associated with strains from other CCs (11.5%; NS).
The 20 CC398 strains were sequenced and studied with a previously studied collection of 23 CC398 strains (i.e., six livestock-associated strains and 17 emerging strains; Figure 1) [16]. Among the 20 strains recovered during the present study, 19 carried a ϕSa-3 prophage (95.0%) and its particular evasion cluster (8). The ϕSa-3 prophages were inserted in 18 cases into the virulence gene hlb, and in the remaining strain, in ebh, a gene encoding for the giant surface anchored protein Ebh that has been associated with S. aureus complement resistance [17]. Six strains carried a ϕMR11-like prophage (30.0%), all inserted into the smpB virulence gene. Two carried other prophage features (10.0%), and one strain, responsible for a case of endocarditis in a 74 year-old female, did not carry any prophage elements.
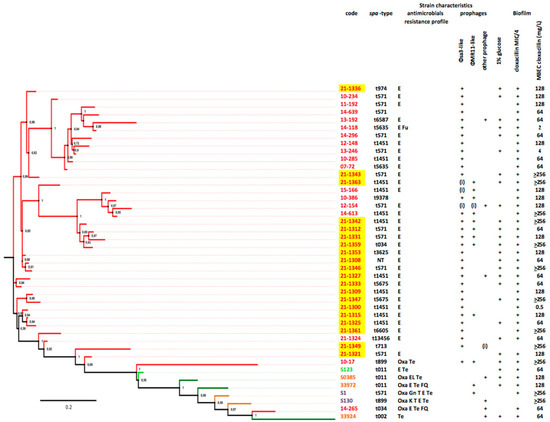
Figure 1.
Phylogeny of CC398 strains, prophage content, biofilm production, and MBEC of cloxacillin. Emerging strains (i.e., those from human bacteremia contracted in an animal-free environment) are shown in red, whereas the remaining strains are livestock-associated (animal colonization in light green, animal infection in dark green, and human colonization in yellow). Prophages are indicated by “+” when complete and by “(i)” when incomplete. Note that the emerging strains isolated in 2021 are highlighted in yellow.
Biofilm production of CC398 strains was further studied in the presence of sub-inhibitory concentrations of cloxacillin. Whereas glucose-induced biofilm production was demonstrated in 85.0% of the CC398 strains, all but one strains produced biofilm in broth supplemented with 1% glucose and cloxacillin at a concentration of MIC/4 (95.0%; NS). Strong producers were more frequent following exposure to sub-inhibitory concentrations of cloxacillin (18/19; 94.7%) than in the case of strains grown with glucose only (3/17; 17.6%; p < 0.001), suggesting that cloxacillin at sub-inhibitory concentration may have increased biofilm production in CC398 strains.
Biofilm eradication assay of CC398 strains. The minimum biofilm eradication concentration (MBEC) of cloxacillin, determined for the 20 CC398 strains, ranged from 0.5 to >256 mg/L according to the strain (Figure 1), with a median value of 128. Strains carrying a ϕMR11-like prophage more frequently presented a MBEC > 64 mg/L rather than strains lacking this prophage (1/5 vs. 6/14; NS), suggesting an impact of lysogeny by ϕMR11-like on the susceptibility to cloxacillin of sessile CC398 bacteria.
4. Discussion
To our knowledge, this is the first multicenter study depicting the incidence of CC398 S. aureus bacteremia in ICUs. The survey revealed one nosocomial bacteremia for 230 newborns, and twice more in adult and pediatric patients, a situation close to the one described in a French ICU in 2011, where a 5-month study revealed 1 case of CC398 nosocomial bacteremia in 89 patients [7]. We observed one third of bacteremias associated with a CC398 strain. This prevalence, obtained in the ICU setting, was higher than that shown in a recent study carried out in all the departments of 17 Spanish hospitals (i.e., medical, surgical, and ICU) [18], in which CC398 strains represented 4.3% of the S. aureus recovered from bacteremias during a 6–12-month period in 2018–2019. Due to the similarity of the characteristics of both infected patients and bacteremia in the 30 ICUs and in the 267 ICUs that participated in the French survey, we believe our results may reflect the general situation. Further studies should be carried out, both inside and outside the ICU, to investigate the current incidence of the CC398 clone in hospitals worldwide.
As usual in Europe [18,19], the CC398 strains responsible for bacteremia were mostly of spa-types t1451 and t571, methicillin and tetracycline-susceptible and resistant to erythromycin. The lack of MRSA is reassuring, as is the rarity of strains carrying a qac gene, as recent studies mostly carried out in Asia have described an increasing incidence of severe infections caused by livestock-independent MRSA [11,20,21].
A high mortality rate was associated with CC398 bacteremias (47.4%), a concordant result with previous studies [22]. However, in contrast to the strains recently described in China [23] and Australia [24], none of the 20 CC398 studied carried the lukF/S or tst-1 genes in their genome. Patient risk factors such as extreme age, immunosuppression (16.7%), and cancer (15.0%), or other unknown compounds present on mobile genetic elements and that should be carefully studied, might play a major role in infection outcome. Note that the number of events is moderate and would need analysis of a larger number of strains.
WGS confirmed typical prophage content of the CC398 emerging strains, with ϕSa-3 in all but one isolate, and ϕMR11 in one third of these [8,9]. Consequently, the strains carried the putative virulence genes chp and scn in ϕSa-3 encoding the chemotaxis inhibitor protein (CHIPS) and the staphylococcal complement inhibitor (SCIN), respectively, and seb encoding a putative superantigen similar to enterotoxin B in ϕMR11. As previously demonstrated, CC398 lysogens carrying these prophage genes may benefit from the production of the three immune-modulating proteins, CHIPS, SCIN, and SEB, when exposed to conditions favoring prophage induction [25,26]. Analysis of the insertion sites of the prophages identified three different loci in the bacterial genomes studied, located in all cases in bacterial virulence genes (i.e., hlb, smpB, or ebh). As shown with a prophage favoring S. aureus colonization of diabetic feet [27], lysogeny of the strains with ß-converting prophages may limit S. aureus virulence and favor colonization, and thus bacteremia [28]. In the ICU, where risk factors of infections are numerous, whether patient-related or healthcare-related, it would seem likely that in S. aureus, the acquisition of such prophages could contribute to a further increase of the ability to colonize human flora and devices and infect humans.
The ability of S. aureus to form biofilm is a significant factor that enhances the pathogenicity of this species [1,3,29]. Concordant with previous studies [3], a minority of our strains was capable of biofilm development under standard conditions in TSB, but enhanced biofilm formation was obtained when strains were grown in broth supplemented with 1% glucose [30]. We confirmed a strong biofilm formation associated with CC8 [31], one of the major lineages associated with catheter-related bacteremia [32]. To our knowledge, our study is the first showing that the CC398 lineage also has a high ability to produce biofilm.
Due to slow diffusion into the biofilm, biofilm bacteria are exposed to sub-inhibitory concentrations of antibiotics [33]. Previous studies have demonstrated that biofilm formation is increased when S. aureus are cultured in the presence of sub-inhibitory concentrations of antibiotics [34], and especially oxacillin, i.e., the antibiotic of choice in the course of severe MSSA infections [35]. Our study is the first to show a strong production of biofilm by CC398 strains following exposure to sub-inhibitory concentrations of cloxacillin. Biofilm helps bacteria to escape immune response, contributes to bacterial persistence in the environment [36,37,38,39], and increases phenotypic resistance to antimicrobials [40]. Biofilm-grown microorganisms have an inherent lack of susceptibility to antibiotics, whereas planktonic cultures of the same organism do not [2,33,41,42,43]. We demonstrated extremely high minimal biofilm eradication cloxacillin concentrations of the CC398 MSSA studied. The resulting tolerance of microbial biofilms to in principle adequate antibiotic therapy may lead to problems in their eradication and in the management of infected patients. Our preliminary data, showing cloxacillin-induced strong production of biofilm by CC398 strains, and a tolerance of S. aureus to cloxacillin should be explored further.
The determinants carried by the ϕSa-3 and ϕMR-11 prophages have previously been associated with high levels of transmissibility [10], and high potential to persist and disperse in the hospital environment [7]. In this study, the strains recovered from a same ICU were genetically distant, allowing us a priori to exclude intra-ICU transmission. The design of our study did not allow us to investigate the mechanisms of acquisition of the S. aureus strains. The genetic diversity observed in our study argues for (but does not prove) dissemination of the CC398 human-adapted subpopulation in human, and nosocomial bacteremia likely caused by multiple strains colonizing the patients before the hospital stay or acquired during hospital stay. Our results suggest the spread of a virulent clone in the hospital settings, confirming the need to investigate the mechanisms involved with the epidemiological success of CC398 in humans.
Author Contributions
Conceptualization, N.v.d.M.-M.; methodology, N.v.d.M.-M., S.D.S. and A.-S.V.; software, S.M.D.; validation, N.v.d.M.-M., L.M. and P.F.; formal analysis, S.D.S., I.D. (biofilm assays), N.v.d.M.-M. and A.-S.V.; investigation, A.-S.V.; writing—original draft preparation, N.v.d.M.-M.; writing—review and editing, N.v.d.M.-M. and P.F.; supervision of the WGS approach: S.M.D. and P.F. The members of the collaborative groups (doctors and nurses in the ICUs, members of the local infection control teams) carried out the survey in their respective ICU and send the S. aureus strains to the national centre. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by subside from the Swiss National Science Foundation (to P.F.) Project 310030_197734.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki, and approved by the Réseau national de Prévention des Infections Associées aux soins (REPIAS), Santé Publique France national agency (decret n° 2017-129).
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Resuscitor: Hervé HYVERNAT, Michael NOVAIN, Rémy PLATTIER (CHU Nice); François BARBIER (CHR Orléans); Pierre MOINE (HU Paris Site R. Poincaré); Marc-Antoine BADAIRE, Julien CALLES, Julie CHANTREUIL, Denis GAROT (CHRU Tours); Benoit THIPHAGNE (CHIC Alencon-Mamers); Gael PRADEL (CH Aurillac); Marc FELLER (CH Blois); Amine BOURIGA (CH Calais); Moulni HAMROUNI (CH Chartres); Florent BAVOZET (CH Dreux); Guillaume SCHNELL (CH Le Havre); Danièle GOLDGRAN TOLEDANO (GHI Montfermeil); Thomas BAUDRY (CH Nord-Ouest Villefranche); Pierre FILLATRE (CH Saint-Brieuc); Eric DIEYE (CH St-Gaudens); Gilles TROCHE (CH Versailles); Serge ALFANDARI (CH Tourcoing); Hakim SLIMANI (CH Trevenans); Malcie MESNIL (Hôpital Fondation de Rothschild, Paris). Infection control: Yasmina BERROUANE, Patricia VEYRES (CHU Nice); Maryvonne DEMASURE (CHR Orléans); Christine LAWRENCE (HU Paris Site R. Poincaré); Virginie MORANGE (CHRU Tours); Joël DELHOMME (CHIC Alencon-Mamers); Catherine GUIGNABERT (CH Aurillac); Valérie BARRY-PERDEREAU (CH Blois); Sylvie JORON (CH Calais); Oana ZAMFIR (CH Chartres); Valérie BREAN (CH Dreux); Sandra BOURDON, Elise BENARD (CH Le Havre); Laurence MARTY (GHEF Marne-la-Vallée); Agnès CECILLE (GHI Montfermeil); Anne PEREZ (CH Nord-Ouest Villefranche); Véronique MARIE (CH St-Brieuc); Claudia DOUATBEYRIES (CH St-Gaudens); Séverine GALLAIS (CH St-Nazaire); Alexandra ALLAIRE (CH St-Lo); Caroline NEULIER (CH Versailles); Serge ALFANDARI (CH Tourcoing); Anne-Marie DETULLIO (CH Trevenans); Marion LECURU (Hôpital Foch, Suresnes); Malcie MESNIL (Hôpital Fondation de Rothschild, Paris); Fabienne GUASP (Hôpital privé de Provence, Aix-en-Provence).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Costerton, J.W.; Stewart, P.S.; Greenberg, E.P. Bacterial Biofilms: A Common Cause of Persistent Infections. Science 1999, 284, 1318–1322. [Google Scholar] [CrossRef]
- Mah, T.-F.C.; O’Toole, G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef]
- O’Neill, E.; Pozzi, C.; Houston, P.; Smyth, D.; Humphreys, H.; Robinson, D.A.; O’Gara, J.P. Association between Methicillin Susceptibility and Biofilm Regulation in Staphylococcus aureus Isolates from Device-Related Infections. J. Clin. Microbiol. 2007, 45, 1379–1388. [Google Scholar] [CrossRef]
- van Belkum, A.; Melles, D.C.; Peeters, J.K.; van Leeuwen, W.B.; van Duijkeren, E.; Huijsdens, X.W.; Spalburg, E.; de Neeling, A.J.; Verbrugh, H.A. Methicillin-Resistant and -Susceptible Staphylococcus aureus Sequence Type 398 in Pigs and Humans. Emerg. Infect. Dis. 2008, 14, 479–483. [Google Scholar] [CrossRef] [PubMed]
- Valentin-Domelier, A.-S.; Girard, M.; Bertrand, X.; Violette, J.; Francois, P.; Donnio, P.-Y.; Talon, D.; Quentin, R.; Schrenzel, J.; Van Der Mee-Marquet, N.; et al. Methicillin-Susceptible ST398 Staphylococcus aureus Responsible for Bloodstream Infections: An Emerging Human-Adapted Subclone? PLoS ONE 2011, 6, e28369. [Google Scholar] [CrossRef]
- Valour, F.; Tasse, J.; Trouillet-Assant, S.; Rasigade, J.-P.; Lamy, B.; Chanard, E.; Verhoeven, P.; Decousser, J.-W.; Marchandin, H.; Bés, M.; et al. Methicillin-susceptible Staphylococcus aureus clonal complex 398: High prevalence and geographical heterogeneity in bone and joint infection and nasal carriage. Clin. Microbiol. Infect. 2014, 20, O772–O775. [Google Scholar] [CrossRef] [PubMed]
- Brunel, A.-S.; Bañuls, A.-L.; Marchandin, H.; Bouzinbi, N.; Morquin, D.; Jumas-Bilak, E.; Corne, P. Methicillin-Sensitive Staphylococcus aureus CC398 in Intensive Care Unit, France. Emerg. Infect. Dis. 2014, 20, 1511–1515. [Google Scholar] [CrossRef]
- Price, L.B.; Stegger, M.; Hasman, H.; Aziz, M.; Larsen, J.; Andersen, P.S.; Pearson, T.; Waters, A.E.; Foster, J.T.; Schupp, J.; et al. Staphylococcus aureus CC398: Host Adaptation and Emergence of Methicillin Resistance in Livestock. mBio 2012, 3, e00305–e00311. [Google Scholar] [CrossRef]
- van der Mee-Marquet, N.; Corvaglia, A.-R.; Valentin, A.-S.; Hernandez, D.; Bertrand, X.; Girard, M.; Kluytmans, J.; Donnio, P.-Y.; Quentin, R.; François, P. Analysis of prophages harbored by the human-adapted subpopulation of Staphylococcus aureus CC398. Infect. Genet. Evol. 2013, 18, 299–308. [Google Scholar] [CrossRef]
- Uhlemann, A.-C.; Porcella, S.F.; Trivedi, S.; Sullivan, S.B.; Hafer, C.; Kennedy, A.D.; Barbian, K.D.; McCarthy, A.; Street, C.; Hirschberg, D.L.; et al. Identification of a highly transmissible animal-independent Staphylococcus aureus ST398 clone with distinct genomic and cell adhesion properties. mBio 2012, 3, e00027-12. [Google Scholar] [CrossRef]
- Lu, H.; Zhao, L.; Si, Y.; Jian, Y.; Wang, Y.; Li, T.; Dai, Y.; Huang, Q.; Ma, X.; He, L.; et al. The Surge of Hypervirulent ST398 MRSA Lineage with higher biofilm-forming ability is a critical threat to clinics. Front. Microbiol. 2021, 12, 636788. [Google Scholar] [CrossRef] [PubMed]
- Von Dach, E.; Diene, S.M.; Fankhauser, C.; Schrenzel, J.; Harbarth, S.; François, P. Comparative Genomics of Community-Associated Methicillin-Resistant Staphylococcus aureus shows the emergence of clone ST8-USA300 in Geneva, Switzerland. J. Infect. Dis. 2016, 213, 1370–1379. [Google Scholar] [CrossRef] [PubMed]
- Christensen, G.D.; Simpson, W.A.; Younger, J.J.; Baddour, L.M.; Barrett, F.F.; Melton, D.M.; Beachey, E.H. Adherence of coagulase-negative staphylococci to plastic tissue culture plates: A quantitative model for the adherence of staphylococci to medical devices. J. Clin. Microbiol. 1985, 22, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Stepanović, S.; Vuković, D.; Hola, V.; DI Bonaventura, G.D.; Djukić, S.; Ćirković, I.; Ruzicka, F. Quantification of biofilm in microtiter plates: Overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 2007, 115, 891–899. [Google Scholar] [CrossRef] [PubMed]
- Ceri, H.; Olson, M.E.; Stremick, C.; Read, R.R.; Morck, D.; Buret, A. The Calgary Biofilm Device: New Technology for Rapid Determination of Antibiotic Susceptibilities of Bacterial Biofilms. J. Clin. Microbiol. 1999, 37, 1771–1776. [Google Scholar] [CrossRef]
- Diene, S.M.; Corvaglia, A.R.; François, P.; Van Der Mee-Marquet, N.L. Prophages and adaptation of Staphylococcus aureus ST398 to the human clinic. BMC Genom. 2017, 18, 133. [Google Scholar] [CrossRef]
- Cheng, A.G.; Missiakas, D.; Schneewind, O. The Giant Protein Ebh Is a Determinant of Staphylococcus aureus Cell Size and Complement Resistance. J. Bacteriol. 2013, 196, 971–981. [Google Scholar] [CrossRef]
- Mama, O.M.; Aspiroz, C.; Ruiz-Ripa, L.; Ceballos, S.; Iñiguez-Barrio, M.; Cercenado, E.; Azcona, J.M.; López-Cerero, L.; Seral, C.; López-Calleja, A.I.; et al. Prevalence and Genetic Characteristics of Staphylococcus aureus CC398 Isolates From Invasive Infections in Spanish Hospitals, Focusing on the Livestock-Independent CC398-MSSA Clade. Front. Microbiol. 2021, 12, 623108. [Google Scholar] [CrossRef]
- Bonnet, I.; Millon, B.; Meugnier, H.; Vandenesch, F.; Maurin, M.; Pavese, P.; Boisset, S. High prevalence of spa type t571 among methicillin-susceptible Staphylococcus aureus from bacteremic patients in a French University Hospital. PLoS ONE 2018, 13, e0204977. [Google Scholar] [CrossRef]
- He, L.; Zheng, H.-X.; Wang, Y.; Le, K.Y.; Liu, Q.; Shang, J.; Dai, Y.; Meng, H.; Wang, X.; Li, T.; et al. Detection and analysis of methicillin-resistant human-adapted sequence type 398 allows insight into community-associated methicillin-resistant Staphylococcus aureus evolution. Genome Med. 2018, 10, 5. [Google Scholar] [CrossRef]
- Sun, L.; Chen, Y.; Wang, D.; Wang, H.; Wu, D.; Shi, K.; Yan, P.; Yu, Y. Surgical Site Infections Caused by Highly Virulent Methicillin-Resistant Staphylococcus aureus Sequence Type 398, China. Emerg. Infect. Dis. 2019, 25, 157–160. [Google Scholar] [CrossRef]
- Bouiller, K.; Bertrand, X.; Hocquet, D.; Chirouze, C. Human Infection of Methicillin-Susceptible Staphylococcus aureus CC398: A Review. Microorganisms 2020, 8, 1737. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Chen, Z.-J.; Sun, Z.; Feng, X.; Zou, M.; Cao, W.; Wang, S.; Zeng, J.; Wang, Y.; Sun, M. Molecular characteristics and virulence factors in methicillin-susceptible, resistant, and heterogeneous vancomycin-intermediate Staphylococcus aureus from central-southern China. J. Microbiol. Immunol. Infect. 2015, 48, 490–496. [Google Scholar] [CrossRef]
- Coombs, G.W.; Daley, D.; Shoby, P.; Yee, N.W.; Robinson, J.O.; Murray, R.; Korman, T.M.; Warner, M.S.; Papanaoum, K.; Derrington, P.; et al. Genomic characterisation of CC398 MRSA causing severe disease in Australia. Int. J. Antimicrob. Agents 2022, 59, 106577. [Google Scholar] [CrossRef] [PubMed]
- Goerke, C.; Wirtz, C.; Fluckiger, U.; Wolz, C. Extensive phage dynamics in Staphylococcus aureus contributes to adaptation to the human host during infection. Mol. Microbiol. 2006, 61, 1673–1685. [Google Scholar] [CrossRef]
- Rooijakkers, S.H.M.; Ruyken, M.; van Roon, J.; van Kessel, K.P.M.; van Strijp, J.A.G.; van Wamel, W.J.B. Early expression of SCIN and CHIPS drives instant immune evasion by Staphylococcus aureus. Cell. Microbiol. 2006, 8, 1282–1293. [Google Scholar] [CrossRef] [PubMed]
- Messad, N.; Landraud, L.; Canivet, B.; Lina, G.; Richard, J.-L.; Sotto, A.; Lavigne, J.-P.; Lemichez, E. Distribution of edin in Staphylococcus aureus isolated from diabetic foot ulcers. Clin. Microbiol. Infect. 2013, 19, 875–880. [Google Scholar] [CrossRef]
- Del Rio, A.; Cervera, C.; Moreno, A.; Moreillon, P.; Miró, J.M. Patients at Risk of Complications of Staphylococcus aureus Bloodstream Infection. Clin. Infect. Dis. 2009, 48, S246–S253. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.-S.; Lee, D.Y.; Rayamajhi, N.; Kang, M.L.; Yoo, H.S. Biofilm-forming associated genotypic and phenotypic characteristics of Staphylococcus spp. isolated from animals and air. Res. Veter-Sci. 2008, 85, 433–438. [Google Scholar] [CrossRef]
- Ammendolia, M.G.; Di Rosa, R.; Montanaro, L.; Arciola, C.R.; Baldassarri, L. Slime Production and Expression of the Slime-Associated Antigen by Staphylococcal Clinical Isolates. J. Clin. Microbiol. 1999, 37, 3235–3238. [Google Scholar] [CrossRef] [PubMed]
- Croes, S.; Deurenberg, R.H.; Boumans, M.-L.L.; Beisser, P.S.; Neef, C.; Stobberingh, E.E. Staphylococcus aureus biofilm formation at the physiologic glucose concentration depends on the S. aureus lineage. BMC Microbiol. 2009, 9, 229. [Google Scholar] [CrossRef]
- Pérez-Montarelo, D.; Viedma, E.; Larrosa, M.N.; Gómez-González, C.; de Gopegui, E.R.; Muñoz-Gallego, I.; Juan, R.S.; Fernández-Hidalgo, N.; Almirante, B.; Chaves, F. Molecular Epidemiology of Staphylococcus aureus Bacteremia: Association of Molecular Factors with the Source of Infection. Front. Microbiol. 2018, 9, 2210. [Google Scholar] [CrossRef] [PubMed]
- Lebeaux, D.; Ghigo, J.-M.; Beloin, C. Biofilm-Related Infections: Bridging the Gap between Clinical Management and Fundamental Aspects of Recalcitrance toward Antibiotics. Microbiol. Mol. Biol. Rev. 2014, 78, 510–543. [Google Scholar] [CrossRef] [PubMed]
- Sritharadol, R.; Hamada, M.; Kimura, S.; Ishii, Y.; Srichana, T.; Tateda, K. Mupirocin at Subinhibitory Concentrations Induces Biofilm Formation in Staphylococcus aureus. Microb. Drug Resist. 2018, 24, 1249–1258. [Google Scholar] [CrossRef] [PubMed]
- Jo, A.; Ahn, J. Phenotypic and genotypic characterisation of multiple antibiotic-resistant Staphylococcus aureus exposed to subinhibitory levels of oxacillin and levofloxacin. BMC Microbiol. 2016, 16, 170. [Google Scholar] [CrossRef] [PubMed]
- Madsen, J.S.; Burmølle, M.; Hansen, L.H.; Sørensen, S.J. The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 2012, 65, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Oie, S.; Hosokawa, I.; Kamiya, A. Contamination of room door handles by methicillin-sensitive/methicillin-resistant Staphylococcus aureus. J. Hosp. Infect. 2002, 51, 140–143. [Google Scholar] [CrossRef]
- Weinstein, R.A.; Hota, B. Contamination, Disinfection, and Cross-Colonization: Are Hospital Surfaces Reservoirs for Nosocomial Infection? Clin. Infect. Dis. 2004, 39, 1182–1189. [Google Scholar] [CrossRef]
- Sexton, T.; Clarke, P.; O’Neill, E.; Dillane, T.; Humphreys, H. Environmental reservoirs of methicillin-resistant Staphylococcus aureus in isolation rooms: Correlation with patient isolates and implications for hospital hygiene. J. Hosp. Infect. 2006, 62, 187–194. [Google Scholar] [CrossRef]
- Stewart, P.S.; Costerton, J.W. Antibiotic resistance of bacteria in biofilms. Lancet 2001, 358, 135–138. [Google Scholar] [CrossRef]
- Chuard, C.; Vaudaux, P.; Waldvogel, F.A.; Lew, D.P. Susceptibility of Staphylococcus aureus growing on fibronectin-coated surfaces to bactericidal antibiotics. Antimicrob. Agents Chemother. 1993, 37, 625–632. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Gilbert, P.; Das, J.; Foley, I. Biofilm Susceptibility to Antimicrobials. Adv. Dent. Res. 1997, 11, 160–167. [Google Scholar] [CrossRef] [PubMed]
- Macia, M.; Rojo-Molinero, E.; Oliver, A. Antimicrobial susceptibility testing in biofilm-growing bacteria. Clin. Microbiol. Infect. 2014, 20, 981–990. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).