Genomic Adaptations of Saccharomyces Genus to Wine Niche
Abstract
1. Introduction
2. Sugar Transporters (HXT3 and FSY1)
3. Cysteine-S-β-lyase IRC7
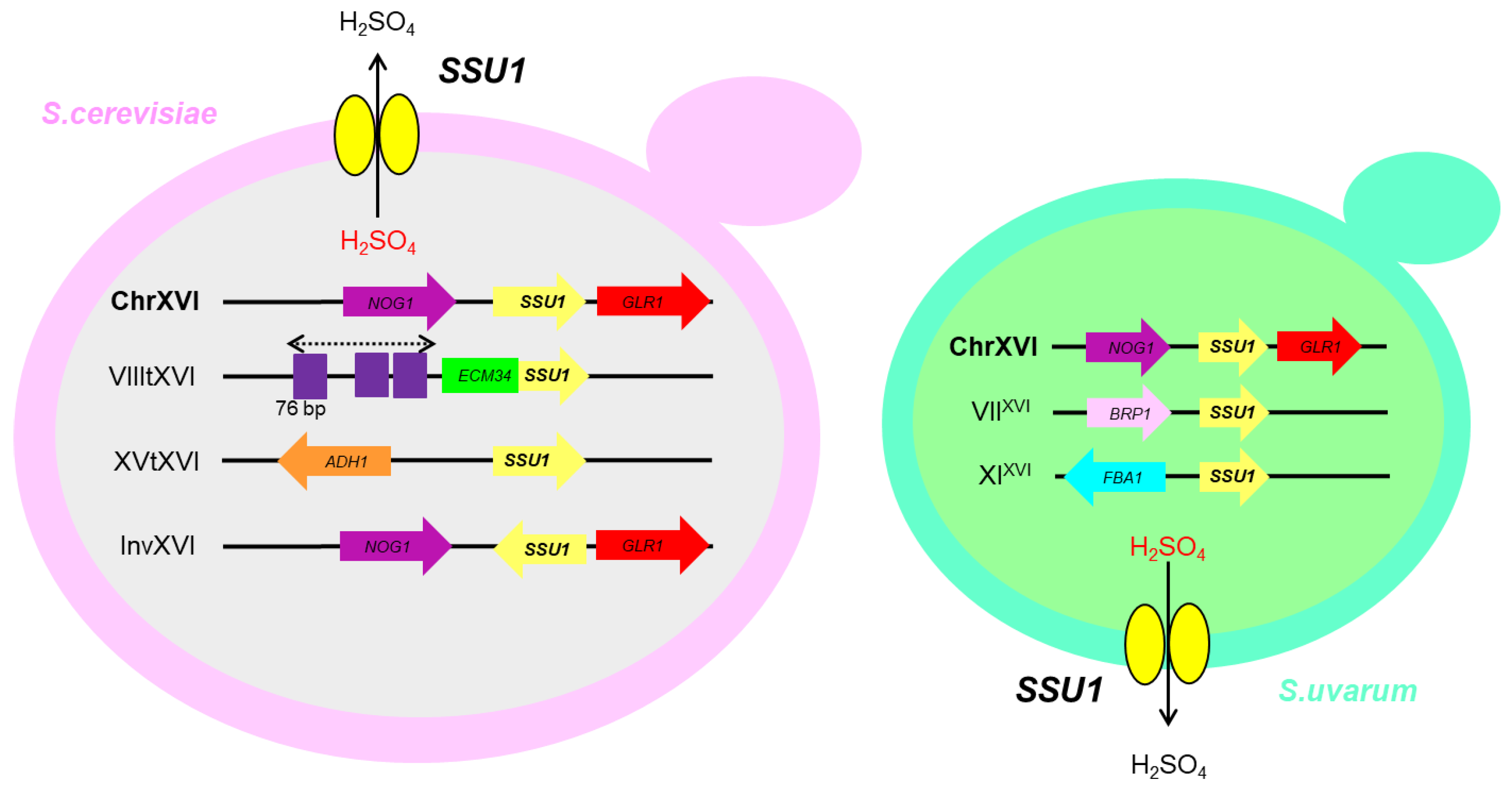
4. Sulphite Resistance
5. Copper Tolerance
6. Oligopeptide Uptake: FOT Genes
7. Biofilm Formation: FLO Genes
8. Inactivation of Aquaporin Genes
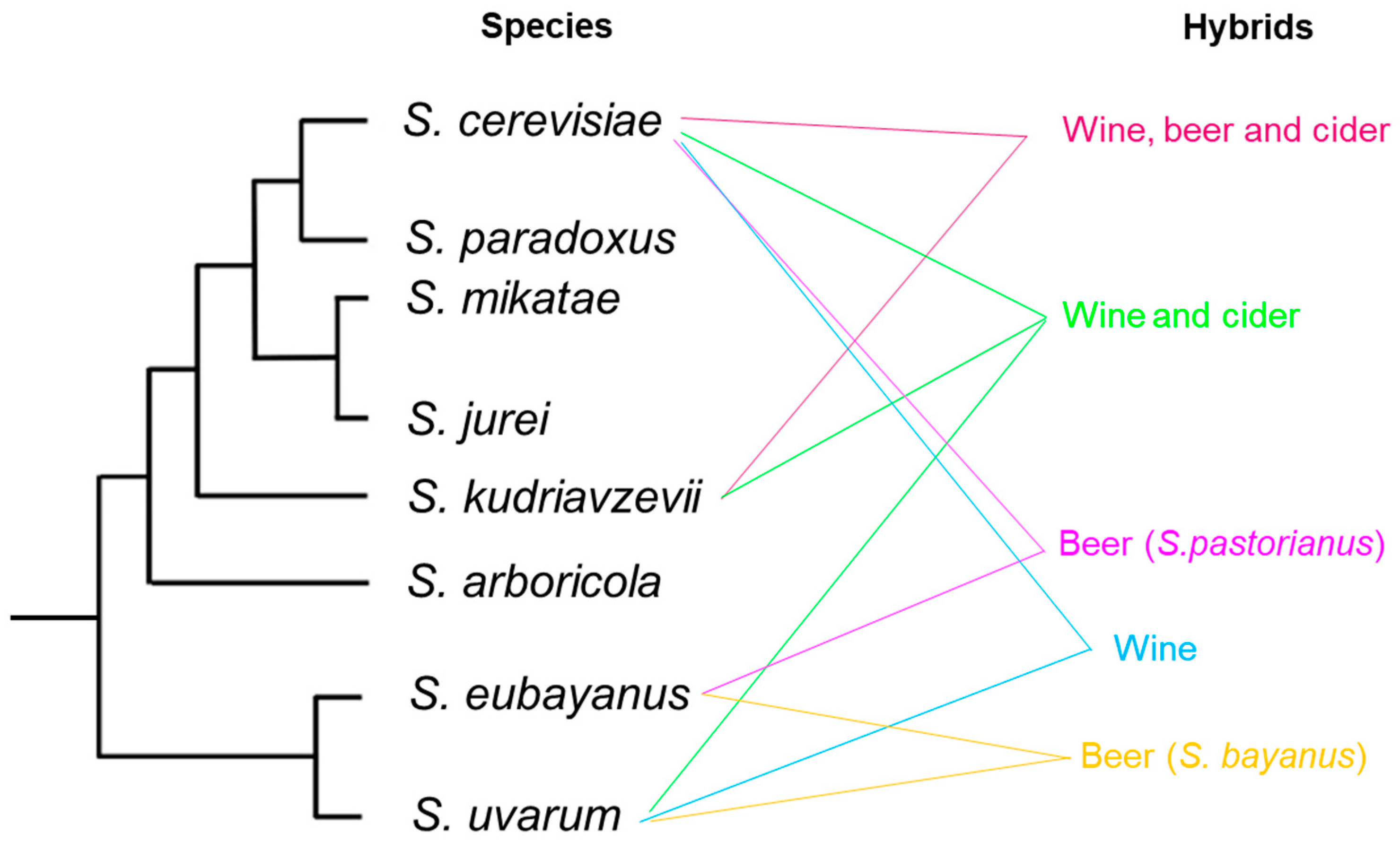
9. Interspecific Hybridization
10. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
References
- Steensels, J.L.; Meersman, E.; Snoek, T.; Saels, V.; Verstrepen, K.J. Large-scale selection and breeding to generate industrial yeasts with superior aroma production. Appl. Environ. Microbiol. 2014, 80, 6965–6975. [Google Scholar] [CrossRef] [PubMed]
- Parapouli, M.; Vasileiadis, A.; Afendra, A.S.; Hatziloukas, E. Saccharomyces cerevisiae and its industrial applications. AIMS Microbiol. 2020, 6, 1–31. [Google Scholar] [CrossRef]
- Legras, J.; Galeote, V.; Bigey, F.; Camarasa, C.; Marsit, S.; Nidelet, T.; Sanchez, I.; Couloux, A.; Guy, J.; Franco-duarte, R.; et al. Adaptation of S. cerevisiae to fermented food environments reveals remarkable genome plasticity and the footprints of domestication. Mol. Biol. Evol. 2018, 35, 1712–1727. [Google Scholar] [CrossRef] [PubMed]
- Alsammar, H.; Delneri, D. An update on the diversity, ecology and biogeography of the Saccharomyces genus. FEMS Yeast Res. 2020, 20, foaa013. [Google Scholar] [CrossRef] [PubMed]
- Molinet, J.; Cubillos, F.A. Wild yeast for the future: Exploring the use of wild strains for wine and beer fermentation. Front. Genet. 2020, 11, 589350. [Google Scholar] [CrossRef]
- García-Ríos, E.; López-Malo, M.; Guillamón, J.M. Global phenotypic and genomic comparison of two Saccharomyces cerevisiae wine strains reveals a novel role of the sulfur assimilation pathway in adaptation at low temperature fermentations. BMC Genom. 2014, 15, 1059. [Google Scholar] [CrossRef]
- García-Ríos, E.; Ramos-Alonso, L.; Guillamón, J.M. Correlation between low temperature adaptation and oxidative stress in Saccharomyces cerevisiae. Front. Microbiol. 2016, 7, 1199. [Google Scholar] [CrossRef]
- García-Ríos, E.; Guillamón, J.M. Mechanisms of yeast adaptation to wine fermentations. Prog. Mol. Subcell. Biol. 2019, 58, 37–59. [Google Scholar] [CrossRef]
- Brice, C.; Sanchez, I.; Bigey, F.; Legras, J.-L.; Blondin, B. A genetic approach of wine yeast fermentation capacity in nitrogen-starvation reveals the key role of nitrogen signaling. BMC Genom. 2014, 15, 495. [Google Scholar] [CrossRef]
- Warringer, J.; Zörgö, E.; Cubillos, F.A.; Zia, A.; Gjuvsland, A.; Simpson, J.T.; Forsmark, A.; Durbin, R.; Omholt, S.W.; Louis, E.J.; et al. Trait variation in yeast is defined by population history. PLoS Genet. 2011, 7, e1002111. [Google Scholar] [CrossRef]
- Taymaz-Nikerel, H.; Cankorur-Cetinkaya, A.; Kirdar, B. Genome-wide transcriptional response of Saccharomyces cerevisiae to stress-induced perturbations. Front. Bioeng. Biotechnol. 2016, 4, 17. [Google Scholar] [CrossRef] [PubMed]
- Steensels, J.; Gallone, B.; Voordeckers, K.; Verstrepen, K.J. Domestication of Industrial Microbes. Curr. Biol. 2019, 29, R381–R393. [Google Scholar] [CrossRef] [PubMed]
- Pronk, J.T.; Steensma, H.Y.; Van Dijken, J.P. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast 1996, 12, 1607–1633. [Google Scholar] [CrossRef]
- Belda, I.; Ruiz, J.; Santos, A.; Van Wyk, N.; Pretorius, I.S. Saccharomyces cerevisiae. Trends Genet. 2019, 35, 956–957. [Google Scholar] [CrossRef] [PubMed]
- Piskur, J.; Rozpedowska, E.; Polakova, S.; Merico, A.; Compagno, C. How did Saccharomyces evolve to become a good brewer? Trends Genet. 2006, 22, 183–186. [Google Scholar] [CrossRef] [PubMed]
- Hagman, A.; Säll, T.; Compagno, C.; Piskur, J. Yeast “Make-Accumulate-Consume” life strategy evolved as a multi-step process that predates the whole genome duplication. PLoS ONE 2013, 8, e68734. [Google Scholar] [CrossRef]
- Pfeiffer, T.; Schuster, S.; Bonhoeffer, S. Cooperation and competition in the evolution of ATP-producing pathways. Science 2001, 292, 504–507. [Google Scholar] [CrossRef]
- Albergaria, H.; Arneborg, N. Dominance of Saccharomyces cerevisiae in alcoholic fermentation processes: Role of physiological fitness and microbial interactions. Appl. Microbiol. Biotechnol. 2016, 100, 2035–2046. [Google Scholar] [CrossRef]
- Yue, J.-X.; Li, J.; Aigrain, L.; Hallin, J.; Persson, K.; Oliver, K.; Bergström, A.; Coupland, P.; Warringer, J.; Cosentino Lagomarsino, M.; et al. Contrasting genome dynamics between domesticated and wild yeasts. Nat. Genet. 2017, 49, 913–924. [Google Scholar] [CrossRef]
- Gallone, B.; Steensels, J.; Prahl, T.; Soriaga, L.; Saels, V.; Herrera-Malaver, B.; Merlevede, A.; Roncoroni, M.; Voordeckers, K.; Miraglia, L.; et al. Domestication and divergence of Saccharomyces cerevisiae beer yeasts. Cell 2016, 166, 1397–1410.e16. [Google Scholar] [CrossRef]
- Liti, G.; Carter, D.M.; Moses, A.M.; Warringer, J.; Parts, L.; James, S.A.; Davey, R.P.; Roberts, I.N.; Burt, A.; Koufopanou, V.; et al. Population genomics of domestic and wild yeasts. Nature 2009, 458, 337–341. [Google Scholar] [CrossRef] [PubMed]
- Strope, P.K.; Skelly, D.A.; Kozmin, S.G.; Mahadevan, G.; Stone, E.A.; Magwene, P.M.; Dietrich, F.S.; McCusker, J.H. The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome Res. 2015, 125, 762–774. [Google Scholar] [CrossRef] [PubMed]
- Fay, J.C.; Benavides, J.A. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS Genet. 2005, 1, e5. [Google Scholar] [CrossRef] [PubMed]
- Legras, J.L.; Merdinoglu, D.; Cornuet, J.M.; Karst, F. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol. Ecol. 2007, 16, 2091–2102. [Google Scholar] [CrossRef]
- Barbosa, R.; Pontes, A.; Santos, R.O.; Montandon, G.G.; De Ponzzes-Gomes, C.M.; Morais, P.B.; Gonçalves, P.; Rosa, C.A.; Sampaio, J.D.S.P. Multiple rounds of artificial selection promote microbe secondary domestication- the case of cachaça yeasts. Genome Biol. Evol. 2018, 10, 1939–1955. [Google Scholar] [CrossRef]
- Pontes, A.; Hutzler, M.; Brito, P.H.; Sampaio, J.P. Revisiting the taxonomic synonyms and populations of Saccharomyces cerevisiae—phylogeny, phenotypes, ecology and domestication. Microorganisms 2020, 8, 903. [Google Scholar] [CrossRef]
- Peter, J.; De Chiara, M.; Friedrich, A.; Yue, J.-X.; Pflieger, D.; Bergstrom, A.; Sigwalt, A.; Barré, B.; Freel, K.; Llored, A.; et al. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature 2018, 556, 339–344. [Google Scholar] [CrossRef]
- Schacherer, J.; Shapiro, J.A.; Ruderfer, D.M.; Kruglyak, L. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 2009, 458, 342–345. [Google Scholar] [CrossRef]
- Giannakou, K.; Cotterrell, M.; Delneri, D. Genomic adaptation of Saccharomyces species to industrial environments. Front. Genet. 2020, 11, 916. [Google Scholar] [CrossRef]
- Guillamón, J.M.; Barrio, E. Genetic polymorphism in wine yeasts: Mechanisms and methods for its detection. Front. Microbiol. 2017, 8, 806. [Google Scholar] [CrossRef]
- Gonçalves, M.; Pontes, A.; Almeida, P.; Barbosa, R.; Serra, M.; Libkind, D.; Hutzler, M.; Gonçalves, P.; Sampaio, J.P. Distinct domestication trajectories in top-fermenting beer yeasts and wine yeasts. Curr. Biol. 2016, 26, 2750–2761. [Google Scholar] [CrossRef] [PubMed]
- Marsit, S.; Dequin, S. Diversity and adaptive evolution of Saccharomyces wine yeast: A review. FEMS Yeast Res. 2015, 15, fov067. [Google Scholar] [CrossRef] [PubMed]
- Mendes, I.; Sanchez, I.; Franco-duarte, R.; Camarasa, C.; Schuller, D.; Dequin, S.; Sousa, M.J. Integrating transcriptomics and metabolomics for the analysis of the aroma profiles of Saccharomyces cerevisiae strains from diverse origins. BMC Genom. 2017, 18, 455. [Google Scholar] [CrossRef]
- Franco-Duarte, R.; Mendes, I.; Umek, L.; Drumonde-Neves, J.; Zupan, B.; Schuller, D. Computational models reveal genotype-phenotype associations in Saccharomyces cerevisiae. Yeast 2014, 31, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Franco-Duarte, R.; Bigey, F.; Carreto, L.; Mendes, I.; Dequin, S.; Santos, M.A.S.; Pais, C.; Schuller, D. Intrastrain genomic and phenotypic variability of the commercial Saccharomyces cerevisiae strain Zymaflore VL1 reveals microevolutionary adaptation to vineyard environments. FEMS Yeast Res. 2015, 15, fov063. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liti, G.; Louis, E.J. Yeast Evolution and Comparative Genomics. Annu. Rev. Microbiol 2005, 59, 135–153. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Bergström, A.; Simpson, J.T.; Salinas, F.; Barré, B.; Parts, L.; Zia, A.; Nguyen Ba, A.N.; Moses, A.M.; Louis, E.J.; Mustonen, V.; et al. A high-definition view of functional genetic variation from natural yeast genomes. Mol. Biol. Evol. 2014, 31, 872–888. [Google Scholar] [CrossRef]
- Boynton, P.J.; Greig, D. The ecology and evolution of non-domesticated Saccharomyces species. Yeast 2014, 31, 449–462. [Google Scholar] [CrossRef]
- Kruckeberg, A.L. The hexose transporter family of Saccharomyces cerevisiae. Arch. Microbiol. 1996, 166, 283–292. [Google Scholar] [CrossRef]
- Luyten, K.; Riou, C.; Blondin, B. The hexose transporters of Saccharomyces cerevisiae play different roles during enological fermentation. Yeast 2002, 19, 713–726. [Google Scholar] [CrossRef]
- Guillaume, C.; Delobel, P.; Sablayrolles, J.M.; Blondin, B. Molecular basis of fructose utilization by the wine yeast Saccharomyces cerevisiae: A mutated HXT3 allele enhances fructose fermentation. Appl. Environ. Microbiol. 2007, 73, 2432–2439. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Hellín, P.; Naranjo, V.; Úbeda, J.; Briones, A. Saccharomyces cerevisiae and metabolic activators: HXT3 gene expression and fructose/glucose discrepancy in sluggish fermentation conditions. World J. Microbiol. Biotechnol. 2016, 32, 196. [Google Scholar] [CrossRef] [PubMed]
- Zuchowska, M.; Jaenicke, E.; König, H.; Claus, H. Allelic variants of hexose transporter Hxt3p and hexokinases Hxk1p/Hxk2p in strains of Saccharomyces cerevisiae and interspecies hybrids. Yeast 2015, 32, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Carrascosa, A.; Muñoz, R.; González, R. Molecular Wine Microbiology; Carrascosa, A., Muñoz, R., González, R., Eds.; Academic Press: Amsterdam, The Netherlands, 2011; ISBN 978-0-12-375021-1. [Google Scholar]
- Coi, A.L.; Bigey, F.; Mallet, S.; Marsit, S.; Zara, G.; Gladieux, P.; Galeote, V.; Budroni, M.; Dequin, S.; Legras, J.L. Genomic signatures of adaptation to wine biological ageing conditions in biofilm-forming flor yeasts. Mol. Ecol. 2017, 26, 2150–2166. [Google Scholar] [CrossRef] [PubMed]
- Novo, M.; Dé, F.; Bigey, R.; Beyne, E.; Galeote, V.; Gavory, R.; Mallet, S.; Cambon, B.; Legras, J.-L.J.-L.; Wincker, P.; et al. Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118. Proc. Natl. Acad. Sci. USA 2009, 106, 16333–16338. [Google Scholar] [CrossRef]
- Marsit, S.; Mena, A.; Bigey, F.; Sauvage, F.X.; Couloux, A.; Guy, J.; Legras, J.L.; Barrio, E.; Dequin, S.; Galeote, V. Evolutionary advantage conferred by an eukaryote-to-eukaryote gene transfer event in wine yeasts. Mol. Biol. Evol. 2015, 32, 1695–1707. [Google Scholar] [CrossRef]
- Galeote, V.; Novo, M.; Salema-Oom, M.; Brion, C.; Valério, E.; Gonçalves, P.; Dequin, S. FSY1, a horizontally transferred gene in the Saccharomyces cerevisiae EC1118 wine yeast strain, encodes a high-affinity fructose/H+ symporter. Microbiology 2010, 156, 3754–3761. [Google Scholar] [CrossRef]
- Charpentier, C.; Colin, A.; Alais, A.; Legras, J.L. French Jura flor yeasts: Genotype and technological diversity. Antonie Van Leeuwenhoek 2009, 95, 263–273. [Google Scholar] [CrossRef]
- Roncoroni, M.; Santiago, M.; Hooks, D.O.; Moroney, S.; Harsch, M.J.; Lee, S.A.; Richards, K.D.; Nicolau, L.; Gardner, R.C. The yeast IRC7 gene encodes a β-lyase responsible for production of the varietal thiol 4-mercapto-4-methylpentan-2-one in wine. Food Microbiol. 2011, 28, 926–935. [Google Scholar] [CrossRef]
- Santiago, M.; Gardner, R.C. Yeast genes required for conversion of grape precursors to varietal thiols in wine. FEMS Yeast Res. 2015, 15, fov034. [Google Scholar] [CrossRef]
- Belda, I.; Ruiz, J.; Navascués, E.; Marquina, D.; Santos, A. Improvement of aromatic thiol release through the selection of yeasts with increased β-lyase activity. Int. J. Food Microbiol. 2016, 225, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, J.; de Celis, M.; Martín-Santamaría, M.; Benito-Vázquez, I.; Pontes, A.; Lanza, V.F.; Sampaio, J.P.; Santos, A.; Belda, I. Global distribution of IRC7 alleles in Saccharomyces cerevisiae populations: A genomic and phenotypic survey within the wine clade. Environ. Microbiol. 2021, 23, 3182–3195. [Google Scholar] [CrossRef] [PubMed]
- Cordente, A.G.; Borneman, A.R.; Bartel, C.; Capone, D.; Solomon, M.; Roach, M.; Curtin, C.D. Inactivating mutations in Irc7p are common in wine yeasts, attenuating carbon-sulfur beta-lyase activity and volatile sulfur compound production. Appl. Environ. Microbiol. 2019, 85, e02684-18. [Google Scholar] [CrossRef]
- Dunn, B.; Richter, C.; Kvitek, D.J.; Pugh, T.; Sherlock, G. Analysis of the Saccharomyces cerevisiae pan-genome reveals a pool of copy number variants distributed in diverse yeast strains from differing industrial environments. Genome Res. 2012, 22, 908–924. [Google Scholar] [CrossRef] [PubMed]
- Santiago, M.; Gardner, R.C. The IRC7 gene encodes cysteine desulphydrase activity and confers on yeast the ability to grow on cysteine as a nitrogen source. Yeast 2015, 32, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Zara, G.; Nardi, T. Yeast metabolism and its exploitation in emerging winemaking trends: From sulfite tolerance to sulfite reduction. Fermentation 2021, 7, 57. [Google Scholar] [CrossRef]
- Pérez-Ortín, J.E.; Querol, A.; Puig, S.; Barrio, E. Molecular characterization of a chromosomal rearrangement involved in the adaptive evolution of yeast strains. Genome Res. 2002, 12, 1533–1539. [Google Scholar] [CrossRef]
- Zimmer, A.; Durand, C.; Loira, N.; Durrens, P.; Sherman, D.J.; Marullo, P. QTL dissection of lag phase in wine fermentation reveals a new translocation responsible for Saccharomyces cerevisiae adaptation to sulfite. PLoS ONE 2014, 9, e86298. [Google Scholar] [CrossRef]
- García-Ríos, E.; Nuévalos, M.; Barrio, E.; Puig, S.; Guillamón, J.M. A new chromosomal rearrangement improves the adaptation of wine yeasts to sulfite. Environ. Microbiol. 2019, 21, 1771–1781. [Google Scholar] [CrossRef]
- Yuasa, N.; Nakagawa, Y.; Hayakawa, M.; Iimura, Y. Two alleles of the sulfite resistance genes are differentially regulated in Saccharomyces cerevisiae. Biosci. Biotechnol. Biochem. 2005, 69, 1584–1588. [Google Scholar] [CrossRef][Green Version]
- Marullo, P.; Claisse, O.; Raymond Eder, M.L.; Börlin, M.; Feghali, N.; Bernard, M.; Legras, J.L.; Albertin, W.; Rosa, A.L.; Masneuf-Pomarede, I. SSU1 checkup, a rapid tool for detecting chromosomal rearrangements related to the SSU1 promoter in Saccharomyces cerevisiae: An ecological and technological study on wine yeast. Front. Microbiol. 2020, 11, 1331. [Google Scholar] [CrossRef] [PubMed]
- Nardi, T.; Corich, V.; Giacomini, A.; Blondin, B. A sulphite-inducible form of the sulphite efflux gene SSU1 in a Saccharomyces cerevisiae wine yeast. Microbiology 2010, 156, 1686–1696. [Google Scholar] [CrossRef] [PubMed]
- Macías, L.G.; Flores, M.G.; Adam, A.C.; Rodríguez, M.E.; Querol, A.; Barrio, E.; Lopes, C.A.; Pérez-Torrado, R. Convergent adaptation of Saccharomyces uvarum to sulfite, an antimicrobial preservative widely used in human-driven fermentations. PLoS Genet. 2021, 17, e1009872. [Google Scholar] [CrossRef]
- Martins, V.; Teixeira, A.; Bassil, E.; Hanana, M.; Blumwald, E.; Gerós, H. Copper-based fungicide Bordeaux mixture regulates the expression of Vitis vinifera copper transporters. Aust. J. Grape Wine Res. 2014, 20, 451–458. [Google Scholar] [CrossRef]
- Carpenè, E.; Andreani, G.; Isani, G. Metallothionein functions and structural characteristics. J. Trace Elem. Med. Biol. 2007, 21, 35–39. [Google Scholar] [CrossRef] [PubMed]
- Fogel, S.; Welch, J.W. Tandem gene amplification mediates copper resistance in yeast. Proc. Natl. Acad. Sci. USA 1982, 79, 5342–5346. [Google Scholar] [CrossRef]
- Adamo, G.M.; Brocca, S.; Passolungui, S.; Salvato, B.; Lotti, M. Laboratory evolution of copper tolerant yeast strains. Microb. Cell Fact. 2012, 11, 1. [Google Scholar] [CrossRef]
- Crosato, G.; Nadai, C.; Carlot, M.; Garavaglia, J.; Ziegler, D.R.; Rossi, R.C.; De Castilhos, J.; Campanaro, S.; Treu, L.; Giacomini, A.; et al. The impact of CUP1 gene copy-number and XVI-VIII/XV-XVI translocations on copper and sulfite tolerance in vineyard Saccharomyces cerevisiae strain populations. FEMS Yeast Res. 2020, 20, foaa028. [Google Scholar] [CrossRef]
- Hull, R.M.; Cruz, C.; Jack, C.V.; Houseley, J. Environmental change drives accelerated adaptation through stimulated copy number variation. PLoS Biol. 2017, 15, e2001333. [Google Scholar] [CrossRef]
- Whale, A.J.; King, M.; Hull, R.M.; Krueger, F.; Houseley, J. Stimulation of adaptive gene amplification by origin firing under replication fork constraint. Nucleic Acids Res. 2022, 50, 915–936. [Google Scholar] [CrossRef]
- Liu, J.; Martin-Yken, H.; Bigey, F.; Dequin, S.; François, J.M.; Capp, J.P. Natural yeast promoter variants reveal epistasis in the generation of transcriptional-mediated noise and its potential benefit in stressful conditions. Genome Biol. Evol. 2015, 7, 969–984. [Google Scholar] [CrossRef] [PubMed]
- Gerstein, A.C.; Ono, J.; Lo, D.S.; Campbell, M.L.; Kuzmin, A.; Otto, S.P. Too much of a good thing: The unique and repeated paths toward copper adaptation. Genetics 2014, 199, 555–571. [Google Scholar] [CrossRef] [PubMed]
- Henschke, P.A.; Jiranek, V. Yeast: Metabolism of nitrogen compounds. In Wine Microbiology and Biotechnology; Fleet, G.H., Ed.; Harwood Academic Publishers: Cambridge, UK, 1993; pp. 77–164. ISBN 0-415-27850-3. [Google Scholar]
- Perry, J.R.; Basrai, M.A.; Steiner, H.Y.; Naider, F.; Becker, J.M. Isolation and characterization of a Saccharomyces cerevisiae peptide transport gene. Mol. Cell. Biol. 1994, 14, 104–115. [Google Scholar] [CrossRef]
- Nelissen, B.; De Wachter, R.; Goffeau, A. Classification of all putative permeases and other membrane plurispanners of the major facilitator superfamily encoded by the complete genome of Saccharomyces cerevisiae. FEMS Microbiol. Rev. 1997, 21, 113–134. [Google Scholar] [CrossRef]
- Hauser, M.; Donhardt, A.M.; Barnes, D.; Naider, F.; Becker, J.M. Enkephalins are transported by a novel eukaryotic peptide uptake system. J. Biol. Chem. 2000, 275, 3037–3041. [Google Scholar] [CrossRef] [PubMed]
- Wiles, A.M.; Cai, H.; Naider, F.; Becker, J.M. Nutrient regulation of oligopeptide transport in Saccharomyces cerevisiae. Microbiology 2006, 152, 3133–3145. [Google Scholar] [CrossRef]
- Damon, C.; Vallon, L.; Zimmermann, S.; Haider, M.Z.; Galeote, V.; Dequin, S.; Luis, P.; Fraissinet-Tachet, L.; Marmeisse, R. A novel fungal family of oligopeptide transporters identified by functional metatranscriptomics of soil eukaryotes. ISME J. 2011, 5, 1871–1880. [Google Scholar] [CrossRef] [PubMed]
- Becerra-Rodríguez, C.; Marsit, S.; Galeote, V. Diversity of oligopeptide transport in yeast and its impact on adaptation to winemaking conditions. Front. Genet. 2020, 11, 602. [Google Scholar] [CrossRef]
- Marsit, S.; Sanchez, I.; Galeote, V.; Dequin, S. Horizontally acquired oligopeptide transporters favour adaptation of Saccharomyces cerevisiae wine yeast to oenological environment. Environ. Microbiol. 2016, 18, 1148–1161. [Google Scholar] [CrossRef]
- Duc, C.; Maçna, F.; Sanchez, I.; Galeote, V.; Delpech, S.; Silvano, A.; Mouret, J.R. Large-scale screening of thiol and fermentative aroma production during wine alcoholic fermentation: Exploring the effects of assimilable nitrogen and peptides. Fermentation 2020, 6, 98. [Google Scholar] [CrossRef]
- Becerra-Rodríguez, C.; Taghouti, G.; Portier, P.; Dequin, S.; Casal, M.; Paiva, S.; Galeote, V. Yeast plasma membrane fungal oligopeptide transporters display distinct substrate preferences despite their high sequence identity. J. Fungi 2021, 7, 963. [Google Scholar] [CrossRef]
- Almeida, P.; Gonçalves, C.; Teixeira, S.; Libkind, D.; Bontrager, M.; Masneuf-Pomarède, I.; Albertin, W.; Durrens, P.; Sherman, D.J.; Marullo, P.; et al. A Gondwanan imprint on global diversity and domestication of wine and cider yeast Saccharomyces uvarum. Nat. Commun. 2014, 5, 4044. [Google Scholar] [CrossRef]
- Marullo, P.; Aigle, M.; Bely, M.; Masneuf-Pomarède, I.; Durrens, P.; Dubourdieu, D.; Yvert, G. Single QTL mapping and nucleotide-level resolution of a physiologic trait in wine Saccharomyces cerevisiae strains. FEMS Yeast Res. 2007, 7, 941–952. [Google Scholar] [CrossRef][Green Version]
- Ibstedt, S.; Stenberg, S.; Bages, S.; Gjuvsland, A.B.; Salinas, F.; Kourtchenko, O.; Samy, J.K.A.; Blomberg, A.; Omholt, S.W.; Liti, G.; et al. Concerted evolution of life stage performances signals recent selection on yeast nitrogen use. Mol. Biol. Evol. 2015, 32, 153–161. [Google Scholar] [CrossRef]
- MacLean, R.C. The tragedy of the commons in microbial populations: Insights from theoretical, comparative and experimental studies. Heredity 2008, 100, 233–239. [Google Scholar] [CrossRef]
- Alexandre, H. Flor yeasts of Saccharomyces cerevisiae-Their ecology, genetics and metabolism. Int. J. Food Microbiol. 2013, 167, 269–275. [Google Scholar] [CrossRef]
- Avdanina, D.; Zghun, A. Sherry wines: Worldwide production, chemical composition and screening conception for flor yeasts. Fermentation 2022, 8, 381. [Google Scholar] [CrossRef]
- Legras, J.-L.; Erny, C.; Charpentier, C. Population structure and comparative genome hybridization of European flor yeast reveal a unique group of Saccharomyces cerevisiae strains with few gene duplications in their genome. PLoS ONE 2014, 9, e108089. [Google Scholar] [CrossRef]
- Govender, P.; Bester, M.; Bauer, F.F. FLO gene-dependent phenotypes in industrial wine yeast strains. Appl. Microbiol. Biotechnol. 2010, 86, 931–945. [Google Scholar] [CrossRef]
- Verstrepen, K.J.; Klis, F.M. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 2006, 60, 5–15. [Google Scholar] [CrossRef]
- Fidalgo, M.; Barrales, R.R.; Ibeas, J.I.; Jimenez, J. Adaptive evolution by mutations in the FLO11 gene. Proc. Natl. Acad. Sci. USA 2006, 103, 11228–11233. [Google Scholar] [CrossRef]
- Barré, B.P.; Hallin, J.; Yue, J.X.; Persson, K.; Mikhalev, E.; Irizar, A.; Holt, S.; Thompson, D.; Molin, M.; Warringer, J.; et al. Intragenic repeat expansion in the cell wall protein gene HPF1 controls yeast chronological aging. Genome Res. 2020, 30, 697–710. [Google Scholar] [CrossRef]
- Váchová, L.; Štoví, V.; Hlaváček, O.; Chernyavskiy, O.; Štěpánek, L.; Kubínová, L.; Palková, Z. Flo11p, drug efflux pumps, and the extracellular matrix cooperate to form biofilm yeast colonies. J. Cell Biol. 2011, 194, 679–687. [Google Scholar] [CrossRef]
- Douglas, L.M.; Li, L.; Yang, Y.; Dranginis, A.M. Expression and characterization of the flocculin Flo11/Muc1, a Saccharomyces cerevisiae mannoprotein with homotypic properties of adhesion. Eukaryot. Cell 2007, 6, 2214–2221. [Google Scholar] [CrossRef]
- Bumgarner, S.L.; Dowell, R.D.; Grisafi, P.; Gifford, D.K.; Fink, G.R. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc. Natl. Acad. Sci. USA 2009, 106, 18321–18326. [Google Scholar] [CrossRef]
- Verstrepen, K.J.; Jansen, A.; Lewitter, F.; Fink, G.R. Intragenic tandem repeats generate functional variability. Nat. Genet. 2005, 37, 986–990. [Google Scholar] [CrossRef]
- Fidalgo, M.; Barrales, R.R.; Jimenez, J. Coding repeat instability in the FLO11 gene of Saccharomyces cerevisiae. Yeast 2008, 25, 879–889. [Google Scholar] [CrossRef]
- Legras, J.L.; Moreno-Garcia, J.; Zara, S.; Zara, G.; Garcia-Martinez, T.; Mauricio, J.C.; Mannazzu, I.; Coi, A.L.; Zeidan, M.B.; Dequin, S.; et al. Flor yeast: New perspectives beyond wine aging. Front. Microbiol. 2016, 7, 503. [Google Scholar] [CrossRef]
- Sabir, F.; Loureiro-Dias, M.C.; Prista, C. Comparative analysis of sequences, polymorphisms and topology of yeasts aquaporins and aquaglyceroporins. FEMS Yeast Res. 2016, 16, fow025. [Google Scholar] [CrossRef]
- Will, J.L.; Kim, H.S.; Clarke, J.; Painter, J.C.; Fay, J.C.; Gasch, A.P. Incipient balancing selection through adaptive loss of aquaporins in natural Saccharomyces cerevisiae populations. PLoS Genet. 2010, 6, e1000893. [Google Scholar] [CrossRef]
- Pettersson, N.; Filipsson, C.; Becit, E.; Brive, L.; Hohmann, S. Aquaporins in yeasts and filamentous fungi. Biol. Cell 2005, 97, 487–500. [Google Scholar] [CrossRef]
- Schluter, D. Evidence for ecological speciation and its alternative. Science 2009, 323, 737–741. [Google Scholar] [CrossRef]
- Sabir, F.; Loureiro-Dias, M.C.; Soveral, G.; Prista, C. Functional relevance of water and glycerol channels in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 2017, 364, fnx080. [Google Scholar] [CrossRef]
- Morales, L.; Dujon, B. Evolutionary role of interspecies hybridization and genetic exchanges in yeasts. Microbiol. Mol. Biol. Rev. 2012, 76, 721–739. [Google Scholar] [CrossRef]
- Stelkens, R.; Bendixsen, D.P. The evolutionary and ecological potential of yeast hybrids. Curr. Opin. Genet. Dev. 2022, 76, 101958. [Google Scholar] [CrossRef]
- Bendixsen, D.P.; Frazão, J.G.; Stelkens, R. Saccharomyces yeast hybrids on the rise. Yeast 2022, 39, 40–54. [Google Scholar] [CrossRef]
- Langdon, Q.K.; Peris, D.; Baker, E.P.C.P.; Opulente, D.A.; Bond, U.; Gonçalves, P.; Sampaio, J.P.; Libkind, D.; Nguyen, H.V.; Bond, U.; et al. Fermentation innovation through complex hybridization of wild and domesticated yeasts. Nat. Ecol. Evol. 2019, 3, 1576–1586. [Google Scholar] [CrossRef]
- González, S.S.; Gallo, L.; Climent, D.; Barrio, E.; Querol, A.; Climent, M.A.D.; Barrio, E.; Querol, A. Enological characterization of natural hybrids from Saccharomyces cerevisiae and S. kudriavzevii. Int. J. Food Microbiol. 2007, 116, 11–18. [Google Scholar] [CrossRef]
- Tronchoni, J.; Gamero, A.; Arroyo-López, F.N.; Barrio, E.; Querol, A. Differences in the glucose and fructose consumption profiles in diverse Saccharomyces wine species and their hybrids during grape juice fermentation. Int. J. Food Microbiol. 2009, 134, 237–243. [Google Scholar] [CrossRef]
- Arroyo-López, F.N.; Orlic, S.; Querol, A.; Barrio, E. Effects of temperature, pH and sugar concentration on the growth parameters of Saccharomyces cerevisiae, S. kudriavzevii and their interspecific hybrid. Int. J. Food Microbiol. 2009, 131, 120–127. [Google Scholar] [CrossRef]
- Belloch, C.; Orlic, S.; Barrio, E.; Querol, A. Fermentative stress adaptation of hybrids within the Saccharomyces sensu stricto complex. Int. J. Food Microbiol. 2008, 122, 188–195. [Google Scholar] [CrossRef]
- Peris, D.; Pérez-Torrado, R.; Hittinger, C.T.; Barrio, E.; Querol, A. On the origins and industrial applications of Saccharomyces cerevisiae × Saccharomyces kudriavzevii hybrids. Yeast 2009, 26, 545–551. [Google Scholar] [CrossRef]
- Origone, A.C.; del Mónaco, S.M.; Ávila, J.R.; González Flores, M.; Rodríguez, M.E.; Lopes, C.A. Tolerance to winemaking stress conditions of Patagonian strains of Saccharomyces eubayanus and Saccharomyces uvarum. J. Appl. Microbiol. 2017, 123, 450–463. [Google Scholar] [CrossRef]
- Querol, A.; Pérez-Torrado, R.; Alonso-del-Real, J.; Minebois, R.; Stribny, J.; Oliveira, B.M.; Barrio, E. New Trends in the Uses of Yeasts in Oenology, 1st ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2018; Volume 85, ISBN 1043-4526. [Google Scholar]
- García-Ríos, E.; Querol, A.; Guillamón, J.M. iTRAQ-based proteome profiling of Saccharomyces cerevisiae and cryotolerant species S. uvarum and S. kudriavzevii during low-temperature wine fermentation. J. Proteom. 2016, 146, 70–79. [Google Scholar] [CrossRef]
- Alonso-del-Real, J.; Pérez-Torrado, R.; Querol, A.; Barrio, E. Dominance of wine Saccharomyces cerevisiae strains over S. kudriavzevii in industrial fermentation competitions is related to an acceleration of nutrient uptake and utilization. Environ. Microbiol. 2019, 21, 1627–1644. [Google Scholar] [CrossRef]
- Minebois, R.; Pérez-Torrado, R.; Querol, A. Metabolome segregation of four strains of Saccharomyces cerevisiae, Saccharomyces uvarum and Saccharomyces kudriavzevii conducted under low temperature oenological conditions. Environ. Microbiol. 2020, 22, 3700–3721. [Google Scholar] [CrossRef]
- Navarro, D.P. Genome characterization of natural Saccharomyces hybrids of biotechnological interest. Ph.D. Thesis, Universitat de València, València, Spain, 2012. [Google Scholar]
- Gamero, A.; Tronchoni, J.; Querol, A.; Belloch, C. Production of aroma compounds by cryotolerant Saccharomyces species and hybrids at low and moderate fermentation temperatures. J. Appl. Microbiol. 2013, 114, 1405–1414. [Google Scholar] [CrossRef]
- Stribny, J.; Gamero, A.; Pérez-Torrado, R.; Querol, A. Saccharomyces kudriavzevii and Saccharomyces uvarum differ from Saccharomyces cerevisiae during the production of aroma-active higher alcohols and acetate esters using their amino acidic precursors. Int. J. Food Microbiol. 2015, 205, 41–46. [Google Scholar] [CrossRef]
- Replansky, T.; Koufopanou, V.; Greig, D.; Bell, G. Saccharomyces sensu stricto as a model system for evolution and ecology. Trends Ecol. Evol. 2008, 23, 494–501. [Google Scholar] [CrossRef]
- Erny, C.; Raoult, P.; Alais, A.; Butterlin, G.; Delobel, P.; Matei-Radoi, F.; Casaregola, S.; Legras, J.L. Ecological success of a group of Saccharomyces cerevisiae/Saccharomyces kudriavzevii hybrids in the northern European wine-making environment. Appl. Environ. Microbiol. 2012, 78, 3256–3265. [Google Scholar] [CrossRef]
- Lopandic, K.; Gangl, H.; Wallner, E.; Tscheik, G.; Leitner, G.; Querol, A.; Borth, N.; Breitenbach, M.; Prillinger, H.; Tiefenbrunner, W. Genetically different wine yeasts isolated from Austrian vine-growing regions influence wine aroma differently and contain putative hybrids between Saccharomyces cerevisiae and Saccharomyces kudriavzevii. FEMS Yeast Res. 2007, 7, 953–965. [Google Scholar] [CrossRef] [PubMed]
- Naumova, E.S.; Naumov, G.I.; Masneuf-Pomarède, I.; Aigle, M.; Dubourdieu, D. Molecular genetic study of introgression between Saccharomyces bayanus and S. cerevisiae. Yeast 2005, 22, 1099–1115. [Google Scholar] [CrossRef] [PubMed]
- Groth, C.; Hansen, J.; Piskur, J. A natural chimeric yeast containing genetic material from three species. Int. J. Syst. Bacteriol. 1999, 49, 1933–1938. [Google Scholar] [CrossRef]
- Naumov, G.I.; James, S.A.; Naumova, E.S.; Louis, E.J.; Roberts, I.N. Three new species in the Saccharomyces sensu stricto complex: Saccharomyces cariocanus, Saccharomyces kudriavzevii and Saccharomyces mikatae. Int. J. Syst. Evol. Microbiol. 2000, 50, 1931–1942. [Google Scholar] [CrossRef]
- Sampaio, J.P.; Gonçalves, P. Natural populations of Saccharomyces kudriavzevii in Portugal are associated with oak bark and are sympatric with S. cerevisiae and S. paradoxus. Appl. Environ. Microbiol. 2008, 74, 2144–2152. [Google Scholar] [CrossRef] [PubMed]
- Antunovics, Z.; Nguyen, H.V.; Gaillardin, C.; Sipiczki, M. Gradual genome stabilisation by progressive reduction of the Saccharomyces uvarum genome in an interspecific hybrid with Saccharomyces cerevisiae. FEMS Yeast Res. 2005, 5, 1141–1150. [Google Scholar] [CrossRef] [PubMed]
- Morard, M.; Ibáñez, C.; Adam, A.C.; Querol, A.; Barrio, E.; Toft, C. Genomic instability in an interspecific hybrid of the genus Saccharomyces: A matter of adaptability. Microb. Genom. 2020, 6, mgen000448. [Google Scholar] [CrossRef]
- Sipiczki, M. Interspecies hybridization and recombination in Saccharomyces wine yeasts. FEMS Yeast Res. 2008, 8, 996–1007. [Google Scholar] [CrossRef]
- Gilchrist, C.; Stelkens, R. Aneuploidy in yeast: Segregation error or adaptation mechanism? Yeast 2019, 36, 525–539. [Google Scholar] [CrossRef]
- Ortiz-Tovar, G.; Pérez-Torrado, R.; Adam, A.C.; Barrio, E.; Querol, A. A comparison of the performance of natural hybrids Saccharomyces cerevisiae × Saccharomyces kudriavzevii at low temperatures reveals the crucial role of their S. kudriavzevii genomic contribution. Int. J. Food Microbiol. 2018, 274, 12–19. [Google Scholar] [CrossRef]
- Peris, D.; Lopes, C.A.; Belloch, C.; Querol, A.; Barrio, E. Comparative genomics among Saccharomyces cerevisiae × Saccharomyces kudriavzevii natural hybrid strains isolated from wine and beer reveals different origins. BMC Genom. 2012, 13, 407. [Google Scholar] [CrossRef]
- Borneman, A.R.; Desany, B.A.; Riches, D.; Affourtit, J.P.; Forgan, A.H.; Pretorius, I.S.; Egholm, M.; Chambers, P.J. The genome sequence of the wine yeast VIN7 reveals an allotriploid hybrid genome with Saccharomyces cerevisiae and Saccharomyces kudriavzevii origins. FEMS Yeast Res. 2012, 12, 88–96. [Google Scholar] [CrossRef]
- Peris, D.; Alexander, W.G.; Fisher, K.J.; Moriarty, R.V.; Basuino, M.G.; Ubbelohde, E.J.; Wrobel, R.L.; Hittinger, C.T. Synthetic hybrids of six yeast species. Nat. Commun. 2020, 11, 2085. [Google Scholar] [CrossRef]
- Gallone, B.; Steensels, J.; Mertens, S.; Dzialo, M.C.; Gordon, J.L.; Wauters, R.; Theßeling, F.A.; Bellinazzo, F.; Saels, V.; Herrera-malaver, B.; et al. Interspecific hybridization facilitates niche adaptation in beer yeast. Nat. Ecol. Evol. 2019, 3, 1562–1575. [Google Scholar] [CrossRef]
- Steensels, J.; Snoek, T.; Meersman, E.; Nicolino, M.P.; Voordeckers, K.; Verstrepen, K.J. Improving industrial yeast strains: Exploiting natural and artificial diversity. FEMS Microbiol. Rev. 2014, 38, 947–995. [Google Scholar] [CrossRef]
- Langdon, Q.K.; Peris, D.; Kyle, B.; Hittinger, C.T. Sppider: A species identification tool to investigate hybrid genomes with high-throughput sequencing. Mol. Biol. Evol. 2018, 35, 2835–2849. [Google Scholar] [CrossRef]
- Li, J.; Llorente, B.; Liti, G.; Yue, J.-X. RecombineX: A generalized computational framework for automatic high-throughput gamete genotyping and tetrad-based recombination analysis. PLOS Genet. 2022, 18, e1010047. [Google Scholar] [CrossRef]
- Tattini, L.; Tellini, N.; Mozzachiodi, S.; D’Angiolo, M.; Loeillet, S.; Nicolas, A.; Liti, G.; D’Angiolo, M.; Loeillet, S.; Nicolas, A.; et al. Accurate tracking of the mutational landscape of diploid hybrid genomes. Mol. Biol. Evol. 2019, 36, 2861–2877. [Google Scholar] [CrossRef]
- Smukowski Heil, C.S.; Large, C.R.L.; Patterson, K.; Hickey, A.S.M.; Yeh, C.L.C.; Dunham, M.J. Temperature preference can bias parental genome retention during hybrid evolution. PLoS Genet. 2019, 15, e1008383. [Google Scholar] [CrossRef]
- Gorkosvskiy, A.; Verstrepen, K.J. The role of structural variation in adaptation and evolution of yeast and other fungi. Genes 2021, 12, 699. [Google Scholar] [CrossRef]
- Ohno, S.; Wolf, U.; Atkin, N.B. Evolution from fish to mammals. Hereditas 1967, 59, 169–187. [Google Scholar] [CrossRef] [PubMed]
- García-Ríos, E.; Lairón-Peris, M.; Muñiz-Calvo, S.; Heras, J.M.; Ortiz-Julien, A.; Poirot, P.; Rozes, N.; Querol, A.; Guillamón, J.M.; García-Ríos, E.; et al. Thermo-adaptive evolution to generate improved Saccharomyces cerevisiae strains for cocoa pulp fermentations. Int. J. Food Microbiol. 2021, 342, 109177. [Google Scholar] [CrossRef] [PubMed]
- Dunham, M.J.; Badrane, H.; Ferea, T.; Adams, J.; Brown, P.O.; Rosenzweig, F.; Botstein, D. Characteristic genome rearrangements in experimental evolution of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2002, 99, 16144–16149. [Google Scholar] [CrossRef]
- Voordeckers, K.; Kominek, J.; Das, A.; Espinosa-Cantú, A.; De Maeyer, D.; Arslan, A.; Van Pee, M.; van der Zande, E.; Meert, W.; Yang, Y.; et al. Adaptation to high ethanol reveals complex evolutionary pathways. PLoS Genet. 2015, 11, e1005635. [Google Scholar] [CrossRef] [PubMed]
- Morard, M.; Macías, L.G.; Adam, A.C.; Lairón-Peris, M.; Pérez-Torrado, R.; Toft, C.; Barrio, E. Aneuploidy and ethanol tolerance in Saccharomyces cerevisiae. Front. Genet. 2019, 10, 82. [Google Scholar] [CrossRef]
- Chen, G.; Rubinstein, B.; Li, R. Whole chromosome aneuploidy: Big mutations drive adaptation by phenotypic leap. Bioessays 2012, 34, 893–900. [Google Scholar] [CrossRef]
- Selmecki, A.M.; Maruvka, Y.E.; Richmond, P.A.; Guillet, M.; Shoresh, N.; Sorenson, A.L.; De, S.; Kishony, R.; Michor, F.; Dowell, R.; et al. Polyploidy can drive rapid adaptation in yeast. Nature 2015, 519, 349–352. [Google Scholar] [CrossRef]
- Todd, R.T.; Forche, A.; Selmecki, A. Ploidy variation in Fungi: Polyploidy, aneuploidy, and genome evolution. Microbiol. Spectr. 2017, 5, FUNK-0051-2016. [Google Scholar] [CrossRef]
- Liti, G.; Warringer, J.; Blomberg, A. Budding yeast strains and genotype—phenotype mapping. Cold Spring Harb. Protoc. 2017, 8, 606–611. [Google Scholar] [CrossRef]
- García-Ríos, E.; Morard, M.; Parts, L.; Liti, G.; Guillamón, J.M. The genetic architecture of low-temperature adaptation in the wine yeast Saccharomyces cerevisiae. BMC Genom. 2017, 18, 159. [Google Scholar] [CrossRef]
- García-Ríos; Ying, K.; Lip, F.; Pinheiro, T.; Teixeira, J.; van Gulik, W.; Domingues, L.; Querol, A.; Guillamón, J.M. Genome-wide effect of non-optimal temperatures under anaerobic conditions on gene expression in Saccharomyces cerevisiae. Genomics 2022, 114, 110386. [Google Scholar] [CrossRef] [PubMed]
- Marullo, P.; Durrens, P.; Peltier, E.; Bernard, M.; Mansour, C.; Dubourdieu, D. Natural allelic variations of Saccharomyces cerevisiae impact stuck fermentation due to the combined effect of ethanol and temperature; a QTL-mapping study. BMC Genom. 2019, 20, 680. [Google Scholar] [CrossRef] [PubMed]
- Peltier, E.; Friedrich, A.; Schacherer, J.; Marullo, P. Quantitative trait nucleotides impacting the technological performances of industrial Saccharomyces cerevisiae strains. Front. Genet. 2019, 10, 683. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Ji, B.; Nielsen, J. The pan-genome of Saccharomyces cerevisiae. FEMS Yeast Res. 2019, 19, foz064. [Google Scholar] [CrossRef] [PubMed]
- Peltier, E.; Sharma, V.; Martí Raga, M.; Roncoroni, M.; Bernard, M.; Jiranek, V.; Gibon, Y.; Marullo, P. Dissection of the molecular bases of genotype x environment interactions: A study of phenotypic plasticity of Saccharomyces cerevisiae in grape juices. BMC Genom. 2018, 19, 772. [Google Scholar] [CrossRef]
- Liti, G.; Louis, E.J. Advances in Quantitative Trait Analysis in Yeast. PLoS Genet. 2012, 8, e1002912. [Google Scholar] [CrossRef]
- Kessi-Pérez, E.I.; Araos, S.; García, V.; Salinas, F.; Abarca, V.; Larrondo, L.F.; Martínez, C.; Cubillos, F.A. RIM15 antagonistic pleiotropy is responsible for differences in fermentation and stress response kinetics in budding yeast. FEMS Yeast Res. 2016, 16, fow021. [Google Scholar] [CrossRef][Green Version]
- Cubillos, F.A.; Brice, C.; Molinet, J.; Tisné, S.; Abarca, V.; Tapia, S.M.; Oporto, C.; García, V.; Liti, G.; Martínez, C. Identification of nitrogen consumption genetic variants in yeast through QTL mapping and bulk segregant RNA-seq analyses. G3 Genes Genomes Genet. 2017, 7, 1693–1705. [Google Scholar] [CrossRef]
- Yeung, C.H.L.; Sahin, N.; Andrews, B. Phenomics approaches to understand genetic networks and gene function in yeast. Biochem. Soc. Trans. 2022, 50, 713–721. [Google Scholar] [CrossRef]
- Vizeacoumar, F.J.; Chong, Y.; Boone, C.; Andrews, B.J. A picture is worth a thousand words: Genomics to phenomics in the yeast Saccharomyces cerevisiae. FEBS Lett. 2009, 583, 1656–1661. [Google Scholar] [CrossRef]
- Costanzo, M.; Kuzmin, E.; van Leeuwen, J.; Mair, B.; Moffat, J.; Boone, C.; Andrews, B. Global genetic networks and the genotype-to-phenotype relationship. Cell 2019, 177, 85–100. [Google Scholar] [CrossRef] [PubMed]
- Ohya, Y.; Kimori, Y.; Okada, H.; Ohnuki, S. Single-cell phenomics in budding yeast. Mol. Biol. Cell 2015, 26, 3920–3925. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
García-Ríos, E.; Guillamón, J.M. Genomic Adaptations of Saccharomyces Genus to Wine Niche. Microorganisms 2022, 10, 1811. https://doi.org/10.3390/microorganisms10091811
García-Ríos E, Guillamón JM. Genomic Adaptations of Saccharomyces Genus to Wine Niche. Microorganisms. 2022; 10(9):1811. https://doi.org/10.3390/microorganisms10091811
Chicago/Turabian StyleGarcía-Ríos, Estéfani, and José Manuel Guillamón. 2022. "Genomic Adaptations of Saccharomyces Genus to Wine Niche" Microorganisms 10, no. 9: 1811. https://doi.org/10.3390/microorganisms10091811
APA StyleGarcía-Ríos, E., & Guillamón, J. M. (2022). Genomic Adaptations of Saccharomyces Genus to Wine Niche. Microorganisms, 10(9), 1811. https://doi.org/10.3390/microorganisms10091811




