Abstract
Furfural and hydroxy-methyl-furfural (HMF) are produced by lignocellulosic biomass during heat or acid pretreatment and are toxic to yeast. Aldehyde reductase is the main enzyme to reduce furfural and HMF. To improve the conversion efficiency of lignocellulosic biomass into ethanol, we constructed Saccharomyces cerevisiae with overexpression of aldehyde reductase (encoded by ari1). The gene of aldehyde reductase (encoded by ari1) was cloned via polymerase chain reaction (PCR) and ligated with the expression vector pGAPZαC. Western blot coupled with anti-His tag confirmed overexpression of the ari1 gene. The growth curves of the wild and ari1-overexpressed strain in the YPD medium were found to be almost identical. Compare to the ari1-overexpressed strain, the wild strain showed a longer doubling time and lag phase in the presence of 20 mM furfural and 60 mM HMF, respectively. The real-time PCR results showed that furfural was much more potent than HMF in stimulating ari1 expression, but the cell growth patterns showed that 60 mM HMF was more toxic to yeast than 20 mM furfural. S. cerevisiae with ari1 overexpression appeared to confer higher tolerance to aldehyde inhibitors, thereby increasing the growth rate and ethanol production capacity of S. cerevisiae in an aldehyde-containing environment.
1. Introduction
Based on the concept of low-cost waste utilization, lignocellulosic biomass and agricultural residues are ideal raw materials for bioethanol production [1,2]. The production of ethanol from lignocellulosic biomass involves the main operations of pretreatment, hydrolysis, fermentation and distillation [3]. Acid hydrolysis or alkali swelling combined with heating is the key technology used to destroy the cellulose crystal structure [4]. Pretreatment of the biomass with acid hydrolysis will produce furfural or hydroxymethyl furfural (HMF), which is the most effective and representative inhibitory compound that interferes with subsequent fermentation [5,6]. After acid/heat treatment, furfural and HMF are formed from the dehydration of pentose and hexose sugars, respectively [7,8]. Research has shown that ethanol productivity in S. cerevisiae can be reduced under 30 mM furfural or 60 mM HMF [9]. These inhibitors hinder yeast growth by disrupting cell walls and membranes, reducing enzyme activity, damaging DNA and inhibiting protein and RNA synthesis [9].
Saccharomyces cerevisiae, a species of yeast, has been traditionally used for brewing and fuel ethanol production. Additionally, the aldehyde reductase originally present in S. cerevisiae can reduce furfural and HMF to furfuryl alcohol [10,11] and 2,5-bis-hydroxymethylfuran [5], respectively. However, in high furfural or HMF environments, S. cerevisiae growth is inhibited and, when furfural and HMF are reduced to sufficiently low concentrations, cell growth recovers and ethanol production resumes until fermentation is complete [12,13]. Overexpression of the dehydrogenase/reductase genes adh6 (encodes for NADPH-dependent Alcohol Dehydrogenase 6), adh7 (encodes for NADPH-dependent Alcohol Dehydrogenase 7), ari1 (encodes for NADPH-dependent aldehyde reductase) and ald6 (encodes for Magnesium-activated aldehyde dehydrogenase) increase furfural and/or HMF reduction and host S. cerevisiae strain tolerance to inhibitors [14,15,16,17].
Liu and Moon [11] discovered a new aldehyde reductase gene, ari1/YGL157W, from S. cerevisiae, NRRL Y-12632, which is a NADPH-dependent aldehyde reduction enzyme (Saccharomyces Genome Database http://www.yeastgenome.org/; accessed on 10 October 2021) with reduction activity for 14 aldehydes. The optimum temperature for this enzyme’s activity is 25 °C, the optimum pH is 7.0 and the protein molecular weight is 38 kDa. This enzyme is an intermediate member of the short-chain dehydrogenase/reductase superfamily and catalyzes Tyr169-XXX-Lys173, whose catalytic position requires four amino acids: Asn106, Ser131, Tyr169 and Lys173. The cofactor binding site for this enzyme is Gly11-XX-Gly14-XX-Ala17 near the N-terminal position [10,11].
In the present study, ari1 was overexpressed in S. cerevisiae BCRC 21685. The engineered strain exhibited higher tolerance to aldehyde inhibitors, thereby enhancing the growth rate of S. cerevisiae and its ethanol production capacity. We investigated the growth patterns and degradation capacity of wild and engineered strains in YPD broth supplemented with furfural (20 mM) and HMF (60 mM). The engineered strains constructed in this study exhibited better responses at higher concentrations of HMF (60 mM).
2. Materials and Methods
2.1. Strains, Vectors and Media
S. cerevisiae (BCRC 21685) was purchased from the Bioresource Collection and Research Center, Food Industry Research and Development Institution, Shinchu, Taiwan. Escherichia coli TOP10F’ and the expression vector pGAPZαC were purchased from Novagen Inc. (Madison, WI, USA) and Invitrogen (Carlsbad, CA, USA) and served as the cloning host cell and expression vector, respectively.
Parent E. coli TOP10F’ and the Zeocin-resistant transformant were cultured, respectively, in a Luria–Bertani (LB) medium (10 g/L peptone, 10 g/L NaCl and 5 g/L yeast extract) and low salt Zeocin-LB plates (10 g/L peptone, 5 g/L NaCl, 5 g/L yeast extract and 25 mg/L Zeocin) at 37 °C. S. cerevisiae was maintained in a YPD medium (10 g/L yeast extract, 20 g/L peptone, 20 g/L dextrose) at 28 °C and 100 mg/L of Zeocin (Invitrogen Corp., Carlsbad, CA, USA) was used to select the engineered yeast. Vectors in the E. coli TOP10F’ and yeast genomic DNA were extracted using a Gene-spin miniprep plasmid purification kit (Protech Technology, Taipei, Taiwan) and Genomic DNA purification kit (BioKit, Miaoli, Taiwan).
2.2. Primers
Table 1 lists the primers used for ari1 cloning, verification of the ari1 gene insertion and quantitative real-time PCR in this study. The nucleotide sequences of the ari1 gene (GenBank ID: NM_001181022.3) and taf10 (GenBank ID: NM_001180474.3) (housekeeping genes used as the internal control) from S. cerevisiae were acquired from the NCBI website (National Center for Biotechnology Information, Bethesda, MD, USA).

Table 1.
Oligonucleotides used in this study.
2.3. Genetic Manipulation
The expression vector pGAPZαC carrying the ari1 gene (pGAPZC-ari1) was constructed according to our previous study [12]. After being linearized with the AvrII enzyme, pGAPZC-ari1 was transformed via electroporation according to the manufacturer’s instructions (MicroPulser electroporation apparatus, Bio-Rad Laboratories, Hercules, CA, USA). Zeocin-resistant colonies were selected and confirmed with PCR. The yeast carrying pGAPZC-ari1 was named SCA, while the parent S. cerevisiae was named SC.
2.4. Expression of Recombinant Protein
Yeast was incubated in a YPD broth with shaking (150 rpm) at 30 °C for 9 h and pellets were collected for protein extraction [13]. Proteins were separated using SDS-PAGE according to Laemmli [14] and the target protein (aldehyde reductase) probed with the Anti-His tag was visualized with Western blotting [12].
2.5. Quantification of Gene Expression via Real-Time Reverse Transcription PCR (RT-qPCR)
To evaluate the ari1 expression level under the pressure of furfural or HMF, yeast was cultured in fresh YPD broth (100 mL) containing 20 mM furfural or 60 mM HMF at 30 °C with shaking (150 rpm).
At the indicated time intervals, cells were collected and centrifuged at 10,000× g for 5 min. TRIzol reagent was applied to extract the total RNA (Life Technologies, Inc., Grand Island, NY, USA). An iScriptTM cDNA synthesis kit (BioRad, Hercules, CA, USA) was used to reverse transcribe RNA to DNA. Real-time PCR was performed with iQ SYBR Green Supermix (BioRad, Hercules, CA, USA) and quantified using a MiniOpticonTM system (BioRad, Hercules, CA, USA). Amplifications were performed under the following thermo-cycle conditions: pre-denaturation at 95 °C/3 min, 40 cycles at 95 °C/10 s and 57.8 °C/30 s and final extension at 95 °C/10 s. The gene expression level was analyzed using the BioRad CFX manager 2.1 software (BioRad, Hercules, CA, USA) and presented as the ratio of ari1 to taf10 [15].
2.6. Microorganism Growth
The growth of yeast was determined by measuring the OD600 or Colony Forming Units with YPD broth or YPD agar at the relevant time intervals.
2.7. Furfural and HMF Reduction
In a 500 mL baffled Erlenmeyer flask, one mL of log-growth culture was adjusted to an OD600 value of 0.3 and added to 100 mL of YPD broth containing 20 mM furfural or 60 mM HMF, and incubated at 30 °C with shaking at 150 rpm. After centrifugation at 10,000× g for 10 min, the supernatant was collected and filtered through a 0.45 mm membrane. To determine furfural and HMF concentrations, the HPLC system was equipped with a refractive index detector and ICSep ICE-COREGEL-87H3 column (Transgenomic, Omaha, NB, USA). The mobile phase was 5 mM H2SO4 with a flow rate of 0.8 mL/min [16]. The reduction of furfural and HMF was expressed as the decrease in furfural and HMF concentrations in the growth broth over time.
2.8. Ethanol Productivity
The log-growth yeast (1 mL, 3 × 107 cells/mL) was inoculated into 100 mL YP broth (10 g/L yeast extract, 20 g/L peptone) containing 10% glucose. The cells were incubated statically at 30 °C and 1 mL of liquid was withdrawn at each indicated time interval. Gas chromatography was performed according to the procedure in [17].
3. Results
3.1. Molecular Characteristics
An ari1 gene with 1044 bp was cloned in this study. This gene showed 99% similarity to the ari1 open reading frame of S. cerevisiae S288c (NC-001139.8) listed in the NCBI gene bank (Figure 1). The aldehyde reductase expressed by the genetically recombined S. cerevisiae was found to have a total of six amino acids which were different from the protein sequence of the aldehyde reductase expressed by S. cerevisiae S288c (Figure 2). Fortunately, none of these amino acids were located in the enzyme active region (Tyr169-Lys173) or cofactor binding site (Gly11-Ala17).

Figure 1.
Comparison of the ari1 DNA sequence for ari1 obtained in this study with that published by Liu et al., 2009. Query: The ari1 DNA sequence published by Liu et al., 2009. Subject: The ari1 DNA sequence from this study. It is shown that the DNA sequence of this study is different from that of Liu et al.

Figure 2.
Comparison of the amino acid sequence for the ARI protein obtained in this study with that published by Liu et al., 2009. Query: The ARI amino acid sequence published by Liu et al., 2009. Subject: The ARI amino acid sequence in this study. 1 Cofactor binding site, 2 Active site, s DNA sequences were different from those of Liu et al. but their amino acids were the same. d Both DNA sequences and amino acids were different from those of Liu et al., 2009.
3.2. Expression of Aldehyde Reductase
A DNA fragment appeared in the position at about 1.0 kb, which corresponds to the ari1 gene (1044 bp was expected) in SCA (Figure 3A), whereas no PCR product was generated in the parental strain (SC). The result of Western blotting probed with an anti-His label (Figure 3B) confirmed that the SCA-expressed protein was in the range of 35~48 kDa, which matches the molecular weight of aldehyde reductase reported by Moon and Liu [18]. Again, SC did not produce the target protein band. The results of PCR and Western blot confirmed that ari1 was successfully recombined and expressed in SCA.

Figure 3.
(A) PCR confirmation of the recombinant expression vector pGAPZC-ari1 in SC and SCA. (B) Overexpression of ARI in SC and SCA. SC (S. cerevisiae) and SCA (S. cerevisiae with ari1 gene overexpression).
3.3. Furfural Tolerance
Both SC and SCA were inhibited by furfural and HMF. The maximum OD600 values of SC and SCA were not affected, while their lag phases were extended in a dose-dependent manner (Figure 4 and Figure 5). The level of growth inhibition was greater in SC than in SCA.
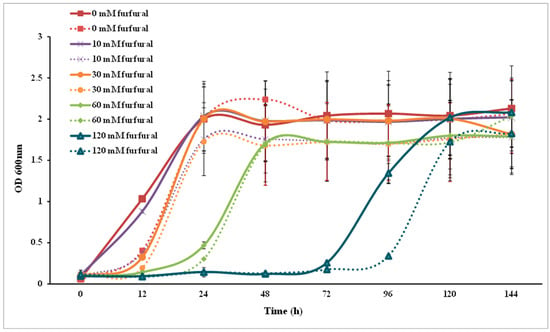
Figure 4.
Furfural tolerance analysis of Saccharomyces cerevisiae. Solid lines represent engineered strains; dashed lines represent wild strains.
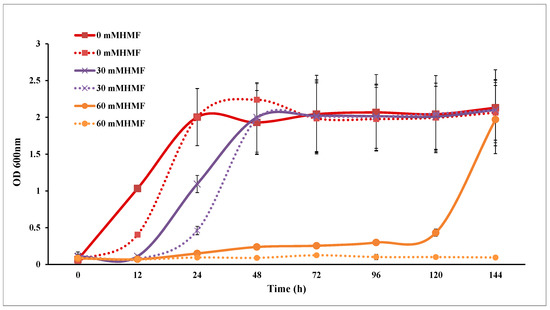
Figure 5.
HMF tolerance analysis of Saccharomyces cerevisiae. Solid lines represent engineered strains; dashed lines represent wild strains.
3.4. ari1 Gene Expression
SC did not show ari1 gene expression within 144 h of culturing in the YPD broth, while SCA showed ari1 gene expression after 24 h of culturing. The expression of ari1 in SCA was highest at 120 h-4.4-fold higher than that in SC. Although the growth patterns of SC and SCA were not significantly different, the expression of the ari1 gene was higher in SCA than that in SC (Figure 6A).
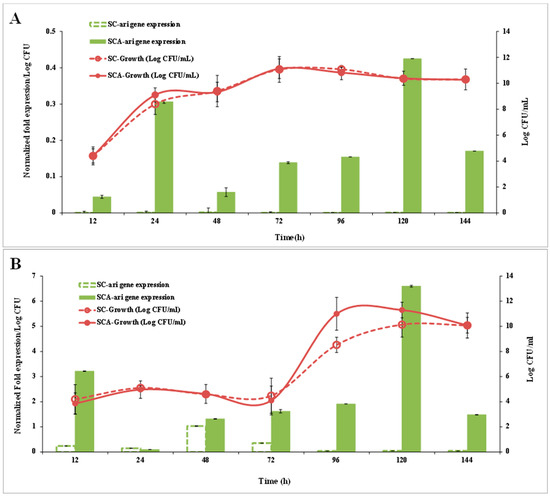
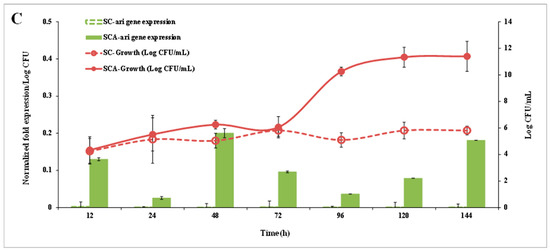
Figure 6.
Ari gene expression (bar chart) and cell growth (graph) of SCA (filled symbol) and SC (open symbol) on the YPD medium (A), YPD + 20 mM furfural (B) and YPD + 60 mM HMF (C). SC (S. cerevisiae) and SCA (S. cerevisiae with ari1 gene overexpression).
Figure 6B,C show the ari1 gene expression and growth performance of SC and SCA in the YPD broth supplemented with 20 mM furfural or 60 mM HMF, respectively. When YPD contained 20 mM furfural, the expression of ari1 in SCA was higher than that in SC after 12 h of culturing and the expression of ari1 in SCA was the highest after 120 h of culturing (74 times higher than that of SC). Both SC and SCA entered the logarithmic growth phase after 72 h of incubation; however, SCA showed the highest growth at 96 h of incubation (Figure 6B). In the presence of 60 mM HMF, the expression of ari1 in SCA increased from 1.2-fold to 2-fold over an incubation period of 48~144 h. However, expression of the ari1 gene in the parental strain remained below 0.008-fold. On the other hand, SCA entered the logarithmic growth phase after 72 h of culturing, but when there was a large amount of HMF (60 mM) in the medium, the SC did not show significant growth (Figure 6C).
3.5. Furfural and HMF Reduction Capacities
SC and SCA were independently incubated in the YPD broth and challenged with furfural (20 mM) or HMF (60 mM) to compare their abilities to reduce these compounds (Figure 7). SCA completely reduced furfural (20 mM) within 96 h; however, 4.3 mM furfural remained after SC was incubated for 144 h. SCA reduced HMF (60 mM) to 27 mM and 15 mM after 72 h and 144 h of incubation, respectively, whereas SC was unable to degrade HMF.
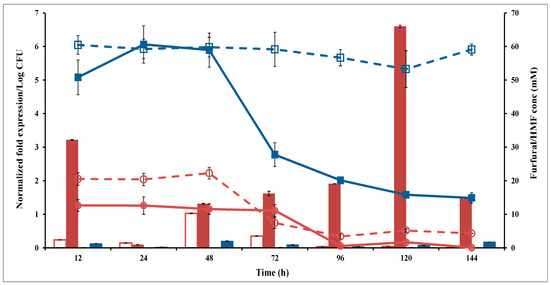
Figure 7.
Ari gene expression (bar chart) and aldehyde degradation (line chart) of SCA (filled symbol) and SC (open symbol) in the presence of 60 mM HMF (Blue color) or 20 mM Furfural (Red color) on the YPD medium. SC (S. cerevisiae) and SCA (S. cerevisiae with ari1 gene overexpression).
3.6. Ethanol Production Capacities
To evaluate the ethanol-producing capacity of the engineered strain, SCA was incubated in a high-glucose YPD broth (containing 10% glucose). After 48 h of incubation, both SC and SCA produced the largest amount of ethanol, with ethanol concentrations of 40.21 and 43.03 mg/mL, respectively (Figure 8); the conversion rates were 78.69 ± 1.88% and 84.21 ± 2.13%, respectively. SCA can more efficiently produce ethanol than its parent strain (SC).
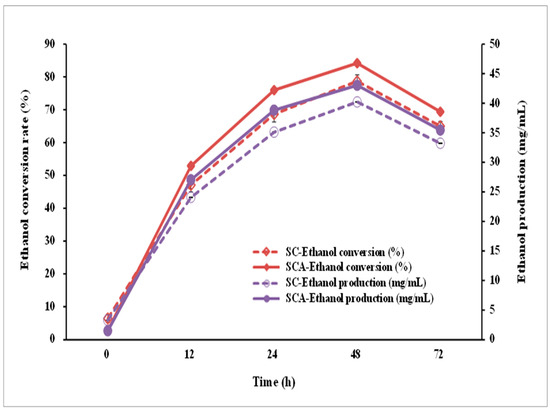
Figure 8.
Ethanol conversion rate and production using SCA (filled symbol) and SC (open symbol) in the presence of 10% glucose on the YPD medium. SC (S. cerevisiae) and SCA (S. cerevisiae with ari1 gene overexpression).
4. Discussion
The bioconversion of lignocellulosic material to ethanol is a feasible technology based on inexpensive raw materials [1]; the main operations involve pretreatment, hydrolysis, fermentation and distillation [3]. The major components of lignocellulosic biomass are cellulose, hemicellulose and lignin. The relative amounts of these components vary among the different sources of lignocellulosic biomass. For example, plant cell walls contain 30–45% cellulose, 20–30% hemicellulose and 20–35% lignin [19]. Pretreatments, such as acid and alkaline hydrolysis, are commonly used to release hemicellulose (major) and lignin (minor) to facilitate enzyme access and ultimately increase ethanol production. Acid-pretreated hemicellulose is easily degraded into its constituent sugar units (mainly xylose, mannose, arabinose and glucose [20]), while fructose, arabinose, rhamnose, galactose, glucose and xylose were detected in the lignin hydrolysate [21]. However, monosaccharides can be converted to furan derivatives upon heating in acidic solutions, in which pentoses are dehydrated into furfural and hexoses are dehydrated into HMF [7].
Furfural and HMF are the most representative inhibitors of yeast growth [5,7]. Researchers have reported that these aldehydes can damage DNA, hinder RNA and protein synthesis and reduce enzymatic activity, thereby inhibiting cell growth [6,19,20,21,22,23].
Likewise, the multiple dehydrogenases/reductases in S. cerevisiae are capable of reducing furfural and HMF to their corresponding, less toxic alcohols [11,24]. Studies have shown that the overexpression of dehydrogenase/reductase genes (ari1, adh7, ald6 and adh6) increases enzyme activities for furfural and/or HMF reduction while increasing the tolerance of yeast to inhibitors [14,15,16,17]. Liu and Moon [11] showed that S. cerevisiae NRRL Y-12632, which overexpressed the aldehyde reductase gene, was not only more tolerant to furfural (20 mM) and HMF (40 mM) than the wild type but also more easily recovered and subject to better growth. Strain improvement played important roles in cell viability and ethanol production under several different stress conditions.
Consistent with the above-mentioned factors, the engineered strain SCA (overexpression of ari1 gene) constructed in this study showed greater tolerance to HMF and furfural and consequently increased ethanol production. SCA was more tolerant to furfural and HM than SC and increased resistance to HMF to a greater extent than furfural. Even though the air1 expression in SCA incubated with 60 mM HMF was not as good as that in 20 mM furfural, the level of expression was sufficient to remove almost 100% of HMF.
Without the presence of HMF or furfural, the growth of SC and SCA was almost the same. Studies have shown that HMF is less toxic to yeast than furfural [25], so we compared the effects of higher HMF concentrations on yeast to those of lower furfural concentrations. According to Fenske et al. [23], the concentrations of furfural and HMF are approximately 0.01 g/L (0.1 mM) in corn stover, switchgrass and poplar pre-hydrolysates. Almeida et al. [22] reported that the HMF concentrations in spruce hydrolysate can vary from 2.0 to 5.9 g/L (15.8 and 46.6 mM), depending on whether one-step- or two-step-dilution acid hydrolysis is performed. The HMF concentrations are around 1 g/L (10 mM). A recent study reported by Erkan et al. [24] showed that 1 mM furfural decreased the ethanol yield by 10% when using Saccharomyces cerevisiae. Furfural is more toxic to S. cerevisiae. Based on the above literature, in this study 20 mM furfural and 60 mM HMF were used to analyze the air1 gene expression levels when simulating the conversion process of lignocellulosic biomass.
SC was found to overcome the toxicity of 20 mM furfural after 72 h incubation and subsequently showed growth. However, the growth rate was slower compared to that of SCA. On the other hand, 60 mM HMF was toxic to SC and caused SC to cease growth for 144 h. Inhibition with HMF and furfural exhibited distinct patterns on SC: 20 mM furfural increased the doubling time during logarithmic growth and decreased the amount of total biomass, while 60 mM HMF increased the lag phase to 144 h, the end of the experiment time. Both decreased the biomass or prolonged the lag phase under the stress of furfural and HMF, respectively, suggesting that the cells had difficulty adapting to the stress conditions. In terms of furfural and HMF’s reduction capacities, SCA significantly expressed ari1 and reduced HMF from 60 to 29 mM under 72 h incubation, which allowed SCA to overcome the inhibitory effect of HMF. SC exhibited the ability to reduce furfural when cultured for 72 h, which was why SC did not show a delay in the lag phase but did present a decrease in biomass.
At the same concentration levels, yeast strains are more sensitive to inhibition with furfural than that with HMF [25]. The effects of furfural and 5-hydroxymethylfurfural (5-HMF) on oxidative metabolism and fermentation were also investigated using Candida guilliermondii and S. cerevisiae, respectively. When added to the medium at a concentration of 0.2%, furfural was found to be a strong inhibitor of both functions, whereas a smaller dose of furoic acid was detected in the supernatant of the C. guillermondii medium. The inhibitory effect of 5-HMF on fermentation and growth was weak and no metabolite was detected in the supernatant. The results showed that the metabolic pathways of furfural and 5-HMF are different depending on whether they enter the fermentation pathway or the oxidative pathway in the yeast strains studied [6].
5. Conclusions
In this study, an inhibitor-tolerant yeast strain was constructed. This strain showed enhanced abilities to reduce furfural and/or HMF through overexpression of the ari1 gene, thus indicating higher cell viability in the environment containing aldehydes.
Author Contributions
N.R.D.: manuscript preparation and experiment executor. P.-J.H.: experiment executor. G.-H.C.: technique and instrumentation support. Y.-C.C.: experiment supervisor, project IP and manuscript preparation. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Ministry of Science and Technology, R.O.C. Taiwan (MOST110-2313-B-126-003-MY3). This financial support is greatly appreciated.
Institutional Review Board Statement
The study protocol was approved by the Biological Experiment Safety Committee of Providence University (27 December 2010).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Bothast, R.J.; Saha, B.C. Ethanol production from agricultural biomass substrates. Adv. Appl. Microbiol. 1997, 44, 261–286. [Google Scholar] [CrossRef]
- Saha, B.C. Hemicellulose bioconversion. J. Ind. Microbiol. Biotechnol. 2003, 30, 279–291. [Google Scholar] [CrossRef] [PubMed]
- Galbe, M.; Zacchi, G. Pretreatment of lignocellulosic materials for efficient bioethanol production. Adv. Biochem. Eng. Biotechnol. 2007, 108, 41–65. [Google Scholar] [CrossRef]
- Sun, Y.; Cheng, J. Hydrolysis of lignocellulosic materials for ethanol production: A review. Bioresour. Technol. 2002, 83, 1–11. [Google Scholar] [CrossRef]
- Elander, M.; Myrback, K. Isolation of crystalline trehalose after fermentation of glucose by maceration juice. Arch. Biochem. 1949, 21, 249–255. [Google Scholar] [PubMed]
- Sanchez, B.; Bautista, J. Effects of furfural and 5-hydroxymethylfurfural on the fermentation of Saccharomyces cerevisiae and biomass production from Candida guilliermondii. Enzyme Microb. Technol. 1988, 10, 315–318. [Google Scholar] [CrossRef]
- Boopathy, R.; Bokang, H.; Daniels, L. Biotransformation of furfural and 5-hydroxymethyl furfural by enteric bacteria. J. Ind. Microbiol. 1993, 11, 147–150. [Google Scholar] [CrossRef]
- Larsson, S.; Palmqvist, E.; Hahn-Hgerdal, B.; Tengborg, C.; Stenberg, K.; Zacchi, G.; Nilvebrant, N.O. The generation of fermentation inhibitors during dilute acid hydrolysis of softwood. Enzyme Microb. Technol. 1999, 24, 151–159. [Google Scholar] [CrossRef]
- Liu, Z.L.; Blaschek, H.P. Biomass Conversion Inhibitors and In Situ Detoxification. Biomass Biofuels Strateg. Glob. Ind. 2010, 233–259. [Google Scholar] [CrossRef]
- Lewis Liu, Z.; Moon, J.; Andersh, B.J.; Slininger, P.J.; Weber, S. Multiple gene-mediated NAD(P)H-dependent aldehyde reduction is a mechanism of in situ detoxification of furfural and 5-hydroxymethylfurfural by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2008, 81, 743–753. [Google Scholar] [CrossRef]
- Liu, Z.L.; Moon, J. A novel NADPH-dependent aldehyde reductase gene from Saccharomyces cerevisiae NRRL Y-12632 involved in the detoxification of aldehyde inhibitors derived from lignocellulosic biomass conversion. Gene 2009, 446, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Divate, N.R.; Chen, G.H.; Divate, R.D.; Ou, B.R.; Chung, Y.C. Metabolic engineering of Saccharomyces cerevisiae for improvement in stresses tolerance. Bioengineered 2017, 8, 524–535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horvath, A.; Riezman, H. Rapid protein extraction from Saccharomyces cerevisiae. Yeast 1994, 10, 1305–1310. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Teste, M.A.; Duquenne, M.; François, J.M.; Parrou, J.L. Validation of reference genes for quantitative expression analysis by real-time RT-PCR in Saccharomyces cerevisiae. BMC Mol. Biol. 2009, 10, 99. [Google Scholar] [CrossRef] [Green Version]
- Kupiainen, L.; Ahola, J.; Tanskanen, J. Kinetics of glucose decomposition in formic acid. Chem. Eng. Res. Des. 2011, 89, 2706–2713. [Google Scholar] [CrossRef]
- Divate, N.R.; Chen, G.H.; Wang, P.M.; Ou, B.R.; Chung, Y.C. Engineering Saccharomyces cerevisiae for improvement in ethanol tolerance by accumulation of trehalose. Bioengineered 2016, 7, 445–458. [Google Scholar] [CrossRef] [Green Version]
- Moon, J.; Liu, Z.L. Direct enzyme assay evidence confirms aldehyde reductase function of Ydr541cp and Ygl039wp from Saccharomyces cerevisiae. Yeast 2015, 32, 399–407. [Google Scholar] [CrossRef]
- Carrasco, J.E.; Sáiz, M.C.; Navarro, A.; Soriano, P.; Sáez, F.; Martinez, J.M. Effects of dilute acid and steam explosion pretreatments on the cellulose structure and kinetics of cellulosic fraction hydrolysis by dilute acids in lignocellulosic materials. Appl. Biochem. Biotechnol. 1994, 45, 23–34. [Google Scholar] [CrossRef]
- Sjöström, E. Wood Chemistry: Fundamentals and Applications; Academic Press: San Diego, CA, USA, 1993. [Google Scholar]
- Mirpoor, S.F.; Restaino, O.F.; Schiraldi, C.; Giosafatto, C.V.L.; Ruffo, F.; Porta, R. Lignin/carbohydrate complex isolated from posidonia oceanica sea balls (Egagropili): Characterization and antioxidant reinforcement of protein-based films. Int. J. Mol. Sci. 2021, 22, 9147. [Google Scholar] [CrossRef]
- Almeida, J.R.M.; Modig, T.; Petersson, A.; Hähn-Hägerdal, B.; Lidén, G.; Gorwa-Grauslund, M.F. Increased tolerance and conversion of inhibitors in lignocellulosic hydrolysates by Saccharomyces cerevisiae. J. Chem. Technol. Biotechnol. 2007, 82, 340–349. [Google Scholar] [CrossRef]
- Fenske, J.J.; Griffin, D.A.; Penner, M.H. Comparison of aromatic monomers in lignocellulosic biomass prehydrolysates. J. Ind. Microbiol. Biotechnol. 1998, 20, 364–368. [Google Scholar] [CrossRef]
- Erkan, S.B.; Yatmaz, E.; Germec, M.; Turhan, I. Effect of furfural concentration on ethanol production using Saccharomyces cerevisiae in an immobilized cells stirred-tank bioreactor with glucose-based medium and mathematical modeling. J. Food Process. Preserv. 2021, 45, e14635. [Google Scholar] [CrossRef]
- Liu, Z.L.; Slininger, P.J.; Dien, B.S.; Berhow, M.A.; Kurtzman, C.P.; Gorsich, S.W. Adaptive response of yeasts to furfural and 5-hydroxymethylfurfural and new chemical evidence for HMF conversion to 2,5-bis-hydroxymethylfuran. J. Ind. Microbiol. Biotechnol. 2004, 31, 345–352. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).