The Effects of Secondary Growth of Spartina alterniflora after Treatment on Sediment Microorganisms in the Yellow River Delta
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area and Soil Sampling
2.2. Soil Physicochemical Properties Analysis
2.3. Soil DNA Extraction, Amplification, and Sequencing
2.4. Sequence Processing and Statistical Analysis
3. Results
3.1. Soil Physicochemical Properties
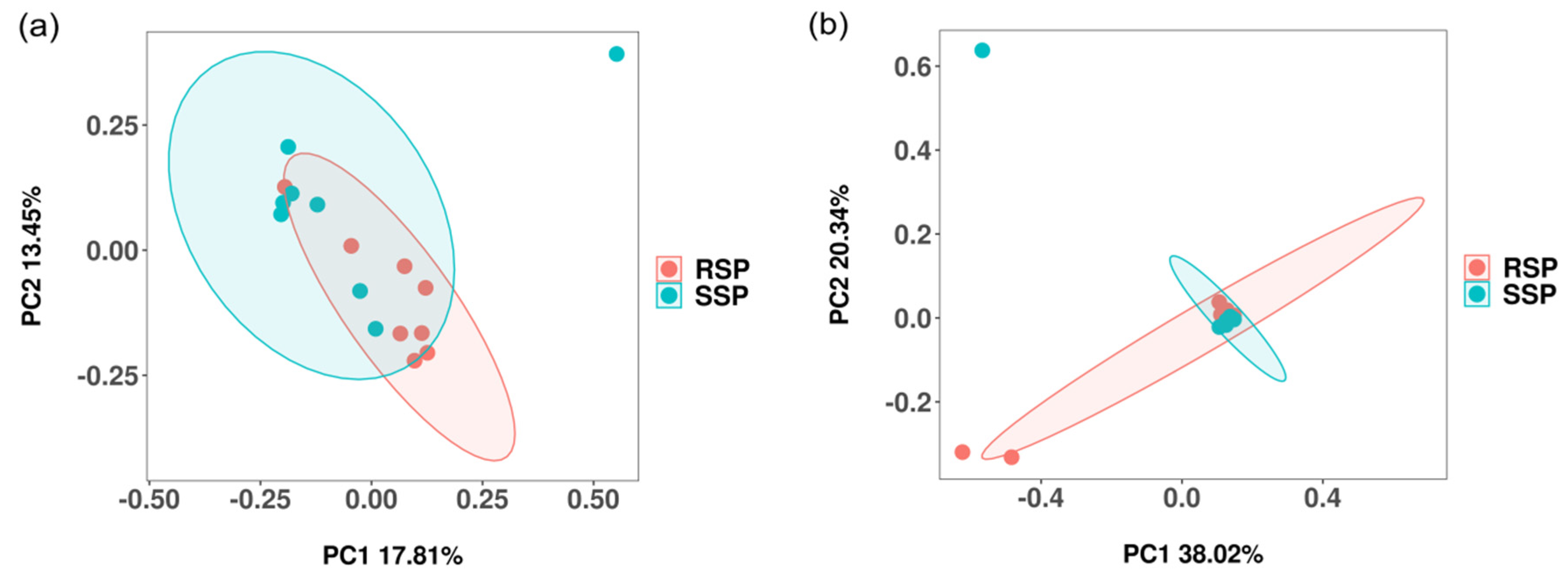
3.2. Bacterial and Fungal Community Diversity
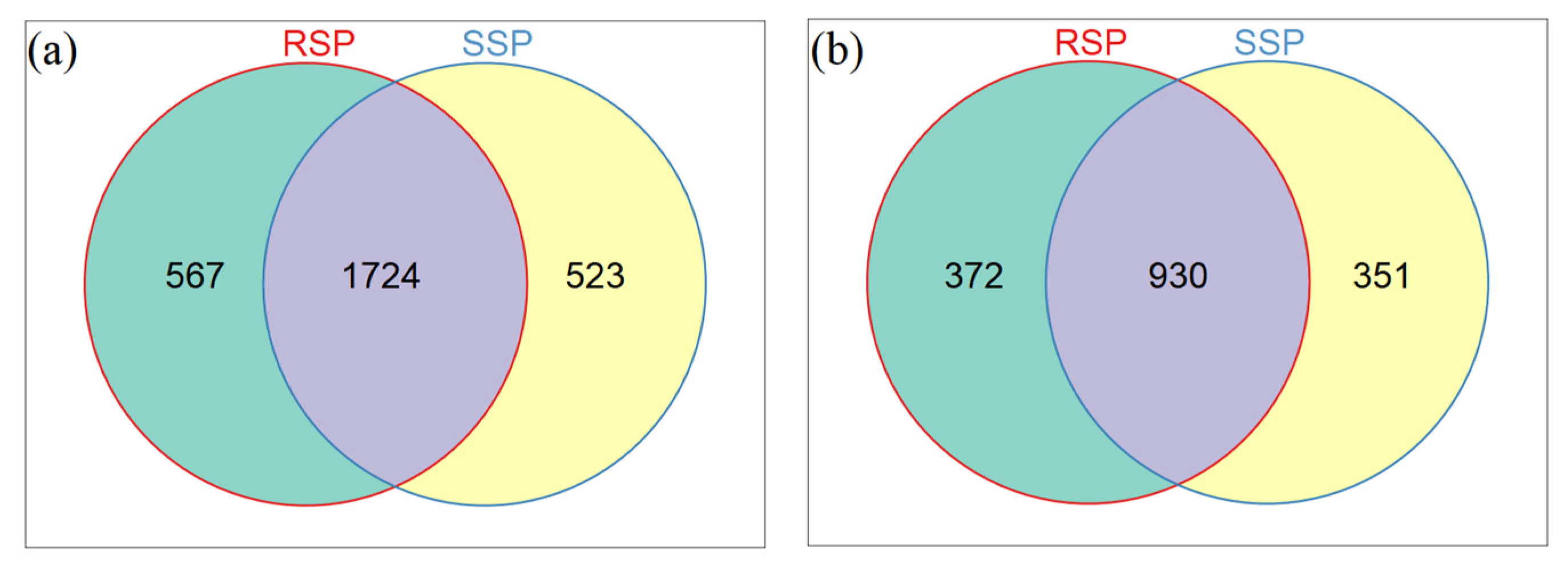
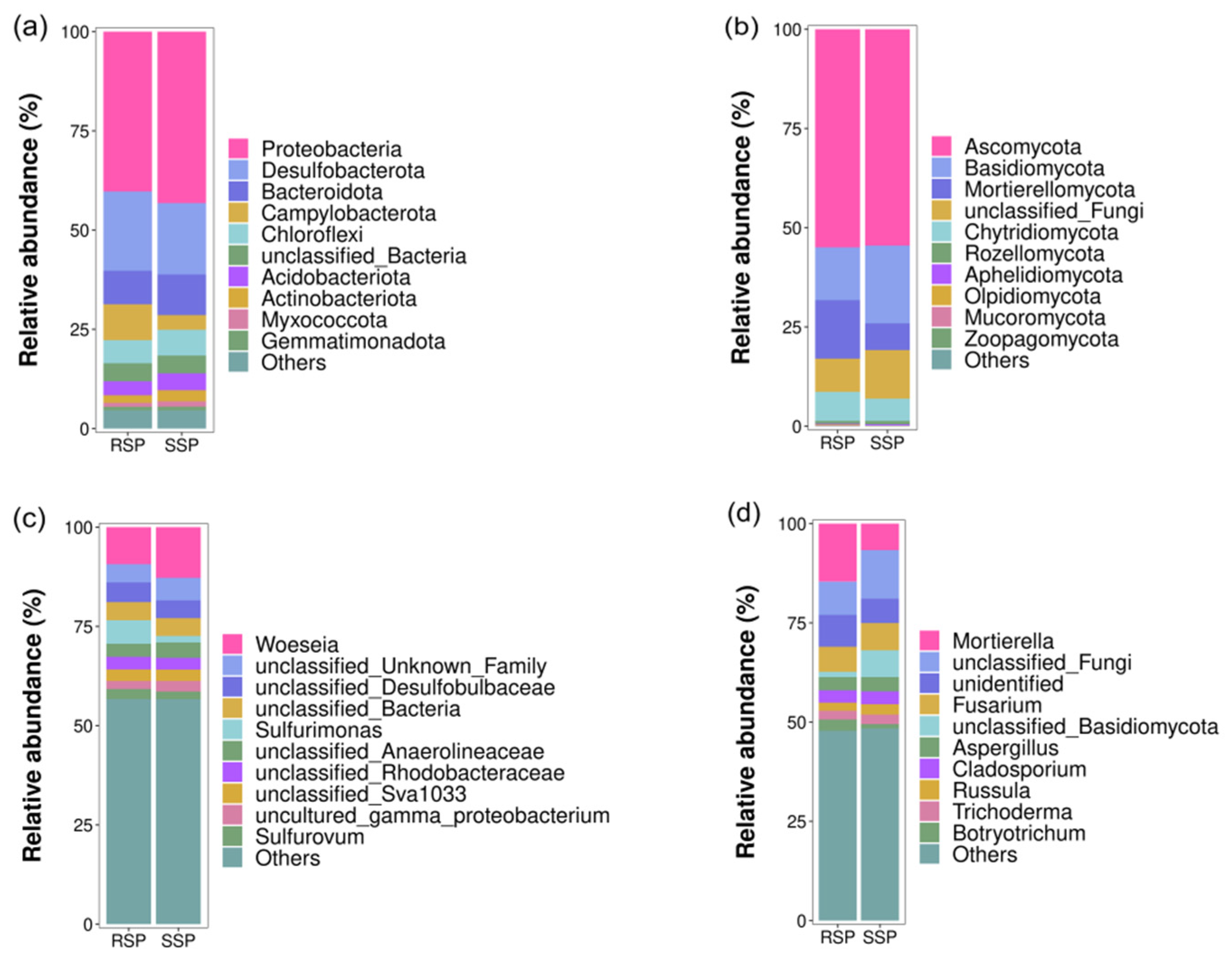
3.3. Bacterial and Fungal Community Composition
3.4. LefSe Analysis
3.5. Redundancy Analysis (RDA) Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Alp, M.; Pichon, C.L. Getting from Sea to Nurseries: Considering Tidal Dynamics of Juvenile Habitat Distribution and Connectivity in a Highly Modified Estuarine Riverscape. Ecosystems 2020, 24, 583–601. [Google Scholar] [CrossRef]
- Wigginton, R.D.; Pearson, J.; Whitcraft, C.R. Invasive plant ecosystem engineer facilitates community and trophic level alteration for brackish marsh invertebrates. Ecosphere 2014, 5, 40. [Google Scholar] [CrossRef]
- Blanco, A.; Larrinaga, A.R.; Neto, J.M.; Troncoso, J.; Mendez, G.; Dominguez-Lapido, P.; Ovejero, A.; Pereira, L.; Mouga, T.M.; Gaspar, R.; et al. Spotting intruders: Species distribution models for managing invasive intertidal macroalgae. J. Environ. Manag. 2021, 281, 111861. [Google Scholar] [CrossRef]
- Flood, P.J.; Duran, A.; Barton, M.; Mercado-Molina, A.E.; Trexler, J.C. Invasion impacts on functions and services of aquatic ecosystems. Hydrobiologia 2020, 847, 1571–1586. [Google Scholar] [CrossRef]
- Chen, J.; Wang, L.; Li, Y.; Zhang, W.; Fu, X.; Le, Y. Effect of Spartina alterniflora invasion and its controlling technologies on soil microbial respiration of a tidal wetland in Chongming Dongtan, China. Ecol. Eng. 2012, 41, 52–59. [Google Scholar] [CrossRef]
- Li, H.; Zhang, L. An experimental study on physical controls of an exotic plant Spartina alterniflora in Shanghai, China. Ecol. Eng. 2008, 32, 11–21. [Google Scholar] [CrossRef]
- Yuan, J.; Ding, W.; Liu, D.; Kang, H.; Freeman, C.; Xiang, J.; Lin, Y. Exotic Spartina alterniflora invasion alters ecosystem-atmosphere exchange of CH4 and N2O and carbon sequestration in a coastal salt marsh in China. Glob. Chang. Biol. 2015, 21, 1567–1580. [Google Scholar] [CrossRef]
- Yuan, L.; Zhang, L.; Xiao, D.; Huang, H. The application of cutting plus waterlogging to control Spartina alterniflora on saltmarshes in the Yangtze Estuary, China. Estuar. Coast. Shelf Sci. 2011, 92, 103–110. [Google Scholar] [CrossRef]
- Zhang, G.; Bai, J.; Jia, J.; Wang, W.; Wang, X.; Zhao, Q.; Lu, Q. Shifts of soil microbial community composition along a short-term invasion chronosequence of Spartina alterniflora in a Chinese estuary. Sci. Total Environ. 2019, 657, 222–233. [Google Scholar] [CrossRef]
- Lin, H.-J.; Hsu, C.-B.; Liao, S.-H.; Chen, C.-P.; Hsieh, H.-L. Effects of Spartina alterniflora Invasion on the Abundance and Community of Meiofauna in a Subtropical Wetland. Wetlands 2015, 35, 547–556. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Grinyer, J.; Reich, P.B.; Singh, B.K.; Allen, E. Relative importance of soil properties and microbial community for soil functionality: Insights from a microbial swap experiment. Funct. Ecol. 2016, 30, 1862–1873. [Google Scholar] [CrossRef]
- Kowalchuk, G.A.; Buma, D.S.; de Boer, W.; Klinkhamer, P.G.; van Veen, J.A. Effects of above-ground plant species composition and diversity on the diversity of soil-borne microorganisms. Antonie Van Leeuwenhoek 2002, 81, 509–520. [Google Scholar] [CrossRef]
- Cao, M.; Cui, L.; Sun, H.; Zhang, X.; Zheng, X.; Jiang, J. Effects of Spartina alterniflora Invasion on Soil Microbial Community Structure and Ecological Functions. Microorganisms 2021, 9, 138. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Wang, S.; Li, K.; Qiao, J.; Guo, Y.; Liu, Z.; Guo, X. Responses of soil bacterial and fungal communities to the long-term monoculture of grapevine. Appl. Microbiol. Biotechnol. 2021, 105, 7035–7050. [Google Scholar] [CrossRef] [PubMed]
- Shang, S.; Hu, S.; Liu, X.; Zang, Y.; Chen, J.; Gao, N.; Li, L.; Wang, J.; Liu, L.; Xu, J.; et al. Effects of Spartina alterniflora invasion on the community structure and diversity of wetland soil bacteria in the Yellow River Delta. Ecol. Evol. 2022, 12, e8905. [Google Scholar] [CrossRef]
- Su, Z.; Qiu, G.; Fan, H.; Li, M.; Fang, C. Changes in carbon storage and macrobenthic communities in a mangrove-seagrass ecosystem after the invasion of smooth cordgrass in southern China. Mar. Pollut Bull. 2020, 152, 110887. [Google Scholar] [CrossRef]
- Jiang, S.; Zhang, C.; Chen, L.; Liu, C.; Yan, L.; Li, B. Effects of Smooth Cordgrass Spartina alterniflora Invasion on Macrobenthic Fauna in the Yellow River Delta. Wetlands 2022, 42, 1–12. [Google Scholar] [CrossRef]
- Li, L.; Jiang, X.; Zhou, Q.; Chen, J.; Zang, Y.; Zhang, Z.; Gao, C.; Tang, X.; Shang, S. Responses of Soil Microbiota to Different Control Methods of the Spartina alterniflora in the Yellow River Delta. Microorganisms 2022, 10, 1122. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, H.; Liu, J.; Wang, Q.; Shen, T.; Guo, W.; Wang, R. Shifts in microbial community function and structure along the successional gradient of coastal wetlands in Yellow River Estuary. Eur. J. Soil Biol. 2012, 49, 12–21. [Google Scholar] [CrossRef]
- Liu, F.; Mo, X.; Kong, W.; Song, Y. Soil bacterial diversity, structure, and function of Suaeda salsa in rhizosphere and non-rhizosphere soils in various habitats in the Yellow River Delta, China. Sci. Total Environ. 2020, 740, 140144. [Google Scholar] [CrossRef]
- Martin, M. Cut adapt removes adapter sequences from high-throughput sequencing reads, EMBnet. EMBnet 17:10-12. Embnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glockner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Callaway, R.M.; Thelen, G.C.; Rodriguez, A.; Holben, W.E. Soil biota and exotic plant invasion. Nature 2004, 427, 731–733. [Google Scholar] [CrossRef]
- Zhang, P.; Nie, M.; Li, B.; Wu, J. The transfer and allocation of newly fixed C by invasive Spartina alterniflora and native Phragmites australis to soil microbiota. Soil Biol. Biochem. 2017, 113, 231–239. [Google Scholar] [CrossRef]
- Trivedi, P.; Batista, B.D.; Bazany, K.E.; Singh, B.K. Plant–microbiome interactions under a changing world: Responses, consequences and perspectives. New Phytol. 2022, 6, 1951–1959. [Google Scholar] [CrossRef]
- Li, Z.; Zu, C.; Wang, C.; Yang, J.; Yu, H.; Wu, H. Different responses of rhizosphere and non-rhizosphere soil microbial communities to consecutive Piper nigrum L. monoculture. Sci. Rep. 2016, 6, 35825. [Google Scholar] [CrossRef] [PubMed]
- Qu, Z.; Liu, B.; Ma, Y.; Xu, J.; Sun, H.; Gallery, R. The response of the soil bacterial community and function to forest succession caused by forest disease. Funct. Ecol. 2020, 34, 2548–2559. [Google Scholar] [CrossRef]
- Zheng, J.; Li, J.; Lan, Y.; Liu, S.; Zhou, L.; Luo, Y.; Liu, J.; Wu, Z. Effects of Spartina alterniflora invasion on Kandelia candel rhizospheric bacterial community as determined by high-throughput sequencing analysis. J. Soils Sediments 2018, 19, 332–344. [Google Scholar] [CrossRef]
- Yang, W.; An, S.; Zhao, H.; Xu, L.; Qiao, Y.; Cheng, X. Impacts of Spartina alterniflora invasion on soil organic carbon and nitrogen pools sizes, stability, and turnover in a coastal salt marsh of eastern China. Ecol. Eng. 2016, 86, 174–182. [Google Scholar] [CrossRef]
- Huang, X.; Feng, J.; Dong, J.; Zhang, J.; Yang, Q.; Yu, C.; Wu, M.; Zhang, W.; Ling, J. Spartina alterniflora invasion and mangrove restoration alter diversity and composition of sediment diazotrophic community. Appl. Soil Ecol. 2022, 177, 104519. [Google Scholar] [CrossRef]
- Takai, K. Sulfurimonas paralvinellae sp. nov., a novel mesophilic, hydrogen- and sulfur-oxidizing chemolithoautotroph within the Epsilonproteobacteria isolated from a deep-sea hydrothermal vent polychaete nest, reclassification of Thiomicrospira denitrificans as Sul. Int. J. Syst. Evol. Microbiol. 2006, 56, 1725–1733. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, S.; Jiang, Z.; Wu, Y.; Huang, X. Gradient of microbial communities around seagrass roots was mediated by sediment grain size. Ecosphere 2022, 13, e3942. [Google Scholar] [CrossRef]
- Liu, D.; Liu, G.; Chen, L.; Wang, J.; Zhang, L. Soil pH determines fungal diversity along an elevation gradient in Southwestern China. Sci. China Life Sci. 2018, 61, 718–726. [Google Scholar] [CrossRef]
- Li, J.; Wang, Y.; Qiu, Y.; Xie, Z.; Zhang, Y.; Hua, C. Decomposition effects of Lanzhou lily (Lilium davidii var. unicolor) flowers on soil physical and chemical properties and microbial community diversity. Sci. Cold Arid. Reg. 2022, 14, 212–222. [Google Scholar] [CrossRef]


| Groups | Sulfur (mg/kg) | TN (mg/kg) | pH | EC (ms/cm) |
|---|---|---|---|---|
| SSP | 1.39 ± 0.11 a | 232.88 ± 40.20 a | 8.26 ± 0.49 a | 2.49 ± 0.21 a |
| RSP | 1.33 ± 0.15 a | 169.13 ± 29.38 a | 7.89 ± 0.49 b | 1.98 ± 0.15 a |
| Groups | ACE | Chao1 | Simpson | Shannon |
|---|---|---|---|---|
| SSP | 707.73 ± 35.83 | 712.83 ± 36.53 | 0.9965 ± 0.0004 | 8.79 ± 0.11 |
| RSP | 708.90 ± 20.64 | 709.83 ± 20.79 | 0.9955 ± 0.0005 | 8.68 ± 0.08 |
| Groups | ACE | Chao1 | Simpson | Shannon |
|---|---|---|---|---|
| SSP | 421.61 ± 45.59 | 421.69 ± 45.61 | 0.9809 ± 0.0057 | 7.24 ± 0.28 |
| RSP | 409.13 ± 28.15 | 409.84 ± 28.19 | 0.9586 ± 0.0121 | 6.81 ± 0.27 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shang, S.; Li, L.; Zhang, Z.; Zang, Y.; Chen, J.; Wang, J.; Wu, T.; Xia, J.; Tang, X. The Effects of Secondary Growth of Spartina alterniflora after Treatment on Sediment Microorganisms in the Yellow River Delta. Microorganisms 2022, 10, 1722. https://doi.org/10.3390/microorganisms10091722
Shang S, Li L, Zhang Z, Zang Y, Chen J, Wang J, Wu T, Xia J, Tang X. The Effects of Secondary Growth of Spartina alterniflora after Treatment on Sediment Microorganisms in the Yellow River Delta. Microorganisms. 2022; 10(9):1722. https://doi.org/10.3390/microorganisms10091722
Chicago/Turabian StyleShang, Shuai, Liangyu Li, Zaiwang Zhang, Yu Zang, Jun Chen, Jun Wang, Tao Wu, Jiangbao Xia, and Xuexi Tang. 2022. "The Effects of Secondary Growth of Spartina alterniflora after Treatment on Sediment Microorganisms in the Yellow River Delta" Microorganisms 10, no. 9: 1722. https://doi.org/10.3390/microorganisms10091722
APA StyleShang, S., Li, L., Zhang, Z., Zang, Y., Chen, J., Wang, J., Wu, T., Xia, J., & Tang, X. (2022). The Effects of Secondary Growth of Spartina alterniflora after Treatment on Sediment Microorganisms in the Yellow River Delta. Microorganisms, 10(9), 1722. https://doi.org/10.3390/microorganisms10091722






