Molecular Epidemiology of Penicillin-Susceptible Staphylococcus aureus Bacteremia in Australia and Reliability of Diagnostic Phenotypic Susceptibility Methods to Detect Penicillin Susceptibility
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolate Collection
2.2. Whole Genome Sequencing
2.3. Genomic Assembly and Phylogenetic Reconstruction
2.4. Bioinformatics Analyses
2.5. β-Lactamase Detection
2.6. Penicillin Susceptibility Testing
2.7. Comparison of Proportions
3. Results
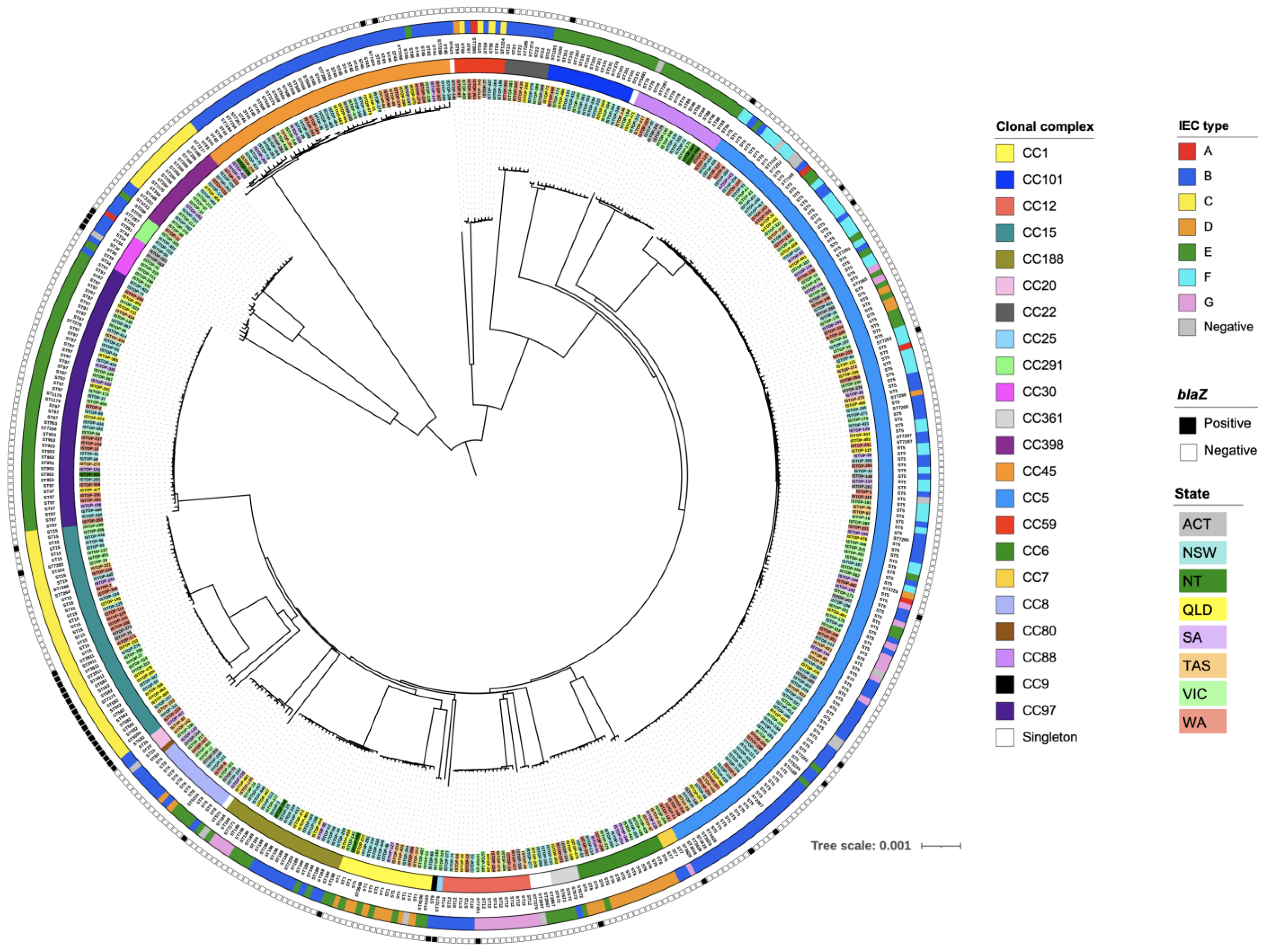
3.1. Population Structure of PSSA
3.2. Distribution of Virulence Genes
3.3. Distribution of AMR Genes
3.4. Clonal Complexes
3.4.1. Clonal Complex 1
3.4.2. Clonal Complex 5
3.4.3. Clonal Complex 6
3.4.4. Clonal Complex 7
3.4.5. Clonal Complex 8
3.4.6. Clonal Complex 9
3.4.7. Clonal Complex 12
3.4.8. Clonal Complex 15
3.4.9. Clonal Complex 20
3.4.10. Clonal Complex 22
3.4.11. Clonal Complex 25
3.4.12. Clonal Complex 30
3.4.13. Clonal Complex 45
3.4.14. Clonal Complex 59
3.4.15. Clonal Complex 80
3.4.16. Clonal Complex 88
3.4.17. Clonal Complex 97
3.4.18. Clonal Complex 101
3.4.19. Clonal Complex 188
3.4.20. Clonal Complex 291
3.4.21. Clonal Complex 361
3.4.22. Clonal Complex 398
3.4.23. Singletons
- Three agr group II/capsule type 5 ST2867 isolates with three closely related spa types. Two of the three isolates harbored a type E IEC, while in one isolate the IEC was not detected. The three isolates harbored selx and edinB, and one type E IEC isolate harbored ant(4’)-Ia and aadD.
- One agr group II/capsule type 5 ST425 isolate with a type B IEC harboring selx.
- One agr group II/capsule type 5 ST573 t1839 isolate with an unidentified IEC (only scn identified) harboring selx, selz, sel27, sel28, the egc-cluster, and sec2 + sel.
- One agr group III/capsule type 8 ST5491 t5925 isolate with a type E IEC harboring selx.
- One agr group I/capsule type 5 ST7270 isolate with a type G IEC harboring selx and seb.
3.5. Phylogenetic Analyses of PSSA Isolates
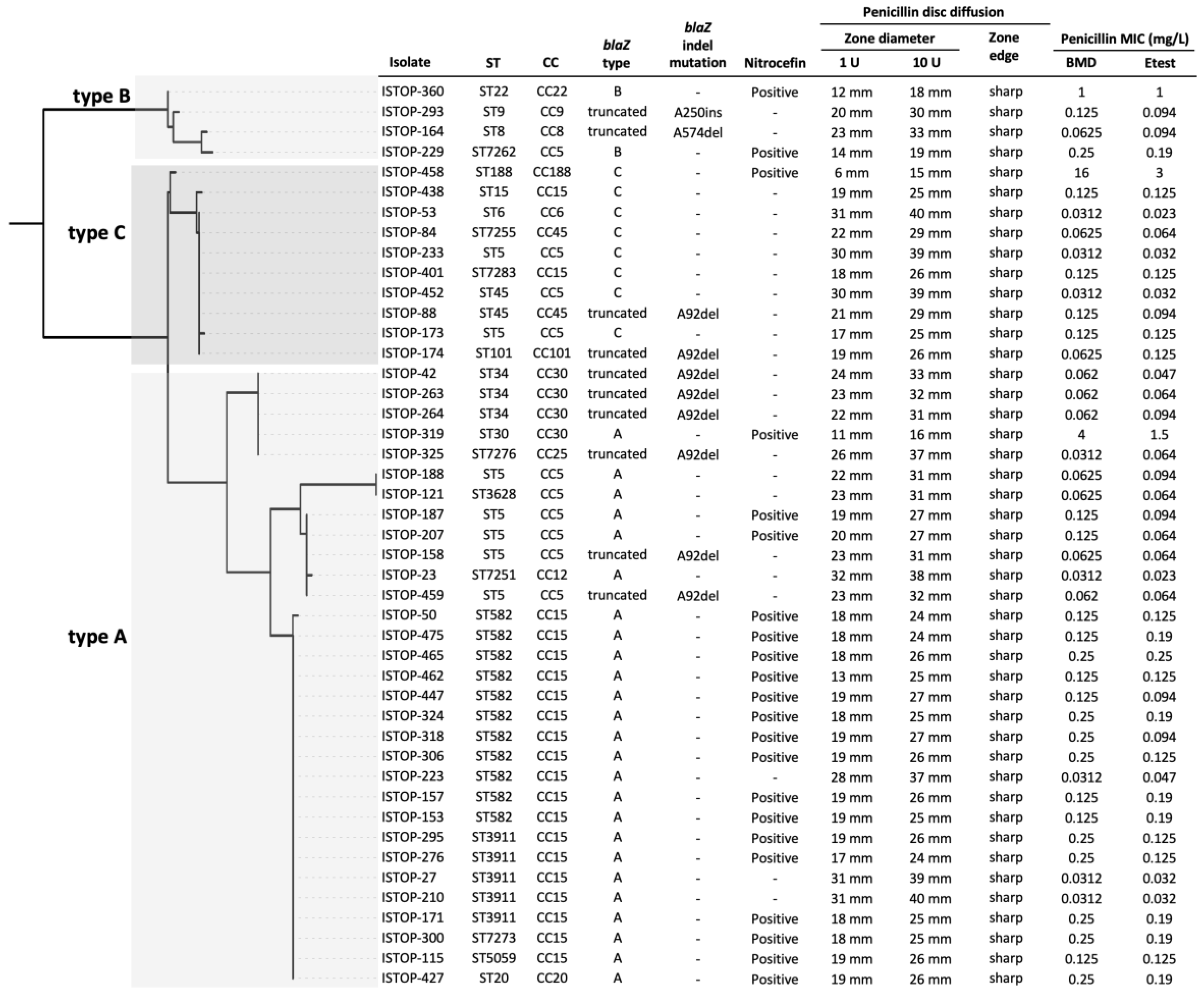
3.6. Phenotypic Antimicrobial Susceptibility Testing of blaZ-Positive Isolates Classified as Penicillin-Susceptible by Vitek® 2
3.7. Genotypic Characterisation of blaZ-Positive Isolates Classified as Penicillin-Susceptible by Vitek® 2
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tong, S.Y.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed]
- Jensen, S.O.; Lyon, B.R. Genetics of antimicrobial resistance in Staphylococcus aureus. Future Microbiol. 2009, 4, 565–582. [Google Scholar] [CrossRef] [PubMed]
- Kirby, W.M. Extraction of a Highly Potent Penicillin Inactivator from Penicillin-resistant Staphylococci. Science 1944, 99, 452–453. [Google Scholar] [CrossRef] [PubMed]
- Barber, M.; Rozwadowska-Dowzenko, M. Infection by penicillin-resistant staphylococci. Lancet 1948, 2, 641–644. [Google Scholar] [CrossRef]
- Chambers, H.F. The changing epidemiology of Staphylococcus aureus? Emerg. Infect. Dis. 2001, 7, 178–182. [Google Scholar] [CrossRef]
- Cheng, M.P.; Rene, P.; Cheng, A.P.; Lee, T.C. Back to the Future: Penicillin-Susceptible Staphylococcus aureus. Am. J. Med. 2016, 129, 1331–1333. [Google Scholar] [CrossRef] [PubMed]
- Resman, F.; Thegerstrom, J.; Mansson, F.; Ahl, J.; Tham, J.; Riesbeck, K. The prevalence, population structure and screening test specificity of penicillin-susceptible Staphylococcus aureus bacteremia isolates in Malmo, Sweden. J. Infect. 2016, 73, 129–135. [Google Scholar] [CrossRef] [PubMed]
- Butler-Laporte, G.; Lee, T.C.; Cheng, M.P. Increasing Rates of Penicillin Sensitivity in Staphylococcus aureus. Antimicrob. Agents Chemother. 2018, 62, e00680-18. [Google Scholar] [CrossRef]
- Chabot, M.R.; Stefan, M.S.; Friderici, J.; Schimmel, J.; Larioza, J. Reappearance and treatment of penicillin-susceptible Staphylococcus aureus in a tertiary medical centre. J. Antimicrob. Chemother. 2015, 70, 3353–3356. [Google Scholar]
- Jin, Y.; Zhou, W.; Zhan, Q.; Chen, Y.; Luo, Q.; Shen, P.; Xiao, Y. Genomic epidemiology and characterisation of penicillin-sensitive Staphylococcus aureus isolates from invasive bloodstream infections in China: An increasing prevalence and higher diversity in genetic typing be revealed. Emerg. Microbes. Infect. 2022, 11, 326–336. [Google Scholar] [CrossRef]
- Coombs, G.W.; Daley, D.A.; Yee, N.W.T.; Shoby, P.; Mowlaboccus, S. Australian Group on Antimicrobial Resistance (AGAR) Australian Staphylococcus aureus Sepsis Outcome Programme (ASSOP) Annual Report 2020. Commun. Dis. Intell. 2022, 46, 2018. [Google Scholar] [CrossRef] [PubMed]
- Henderson, A.; Harris, P.; Hartel, G.; Paterson, D.; Turnidge, J.; Davis, J.S.; Tong, S.Y.C. Benzylpenicillin versus flucloxacillin for penicillin-susceptible Staphylococcus aureus bloodstream infections from a large retrospective cohort study. Int. J. Antimicrob. Agents 2019, 54, 491–495. [Google Scholar] [CrossRef] [PubMed]
- Baddour, L.M.; Wilson, W.R.; Bayer, A.S.; Fowler, V.G., Jr.; Tleyjeh, I.M.; Rybak, M.J.; Barsic, B.; Lockhart, P.B.; Gewitz, M.H.; Levison, M.E.; et al. Infective Endocarditis in Adults: Diagnosis, Antimicrobial Therapy, and Management of Complications: A Scientific Statement for Healthcare Professionals From the American Heart Association. Circulation 2015, 132, 1435–1486. [Google Scholar] [CrossRef]
- Papanicolas, L.E.; Bell, J.M.; Bastian, I. Performance of phenotypic tests for detection of penicillinase in Staphylococcus aureus isolates from Australia. J. Clin. Microbiol. 2014, 52, 1136–1138. [Google Scholar] [CrossRef] [PubMed]
- Banic, S.; Dragas, A.Z.; Fiser, J. Comparison of the efficiency of three methods for the determination of staphylococcal beta-lactamase. J. Chemother. 1990, 2, 17–19. [Google Scholar] [CrossRef]
- Kaase, M.; Lenga, S.; Friedrich, S.; Szabados, F.; Sakinc, T.; Kleine, B.; Gatermann, S.G. Comparison of phenotypic methods for penicillinase detection in Staphylococcus aureus. Clin. Microbiol. Infect. 2008, 14, 614–616. [Google Scholar] [CrossRef]
- James, S.L., II; PharmD; FIDSA. M100-ED31:2021. In Performance Standards for Antimicrobial Susceptibility Testing, 31st ed.; Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2022; Available online: http://em100.edaptivedocs.net/GetDoc.aspx?doc=CLSI%20M100%20ED31:2021&xormat=SPDF&src=BB (accessed on 1 August 2021).
- Breakpoint Tables for Interpretation of MICs and Zone Diameters Version 11.0. Available online: http://www.eucast.org (accessed on 1 August 2021).
- Gill, V.J.; Manning, C.B.; Ingalls, C.M. Correlation of penicillin minimum inhibitory concentrations and penicillin zone edge appearance with staphylococcal beta-lactamase production. J. Clin. Microbiol. 1981, 14, 437–440. [Google Scholar] [CrossRef]
- Mama, O.M.; Aspiroz, C.; Lozano, C.; Ruiz-Ripa, L.; Azcona, J.M.; Seral, C.; Cercenado, E.; Lopez-Cerero, L.; Palacian, P.; Belles-Belles, A.; et al. Penicillin susceptibility among invasive MSSA infections: A multicentre study in 16 Spanish hospitals. J. Antimicrob. Chemother. 2021, 76, 2519–2527. [Google Scholar] [CrossRef]
- Campbell, A.J.; Mowlaboccus, S.; Coombs, G.W.; Daley, D.A.; Al Yazidi, L.S.; Phuong, L.K.; Leung, C.; Best, E.J.; Webb, R.H.; Voss, L.; et al. Whole genome sequencing and molecular epidemiology of pediatric Staphylococcus aureus bacteremia. J. Glob. Antimicrob. Resist. 2022, 29, 197–206. [Google Scholar] [CrossRef]
- Prjibelski, A.; Antipov, D.; Meleshko, D.; Lapidus, A.; Korobeynikov, A. Using SPAdes De Novo Assembler. Curr. Protoc. Bioinform. 2020, 70, e102. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Mlst, Github. Available online: https://github.com/tseemann/mlst (accessed on 1 February 2022).
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Alikhan, N.F.; Sergeant, M.J.; Luhmann, N.; Vaz, C.; Francisco, A.P.; Carriço, J.A.; Achtman, M. GrapeTree: Visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018, 28, 1395–1404. [Google Scholar] [CrossRef] [PubMed]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.-H.; McDermott, P.F.; et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019, 63, e00483-19. [Google Scholar] [CrossRef]
- Zankari, E.; Allesoe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2017, 72, 2764–2768. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Carvajal, L.P.; Rincon, S.; Echeverri, A.M.; Porras, J.; Rios, R.; Ordonez, K.M.; Seas, C.; Gomez-Villegas, S.I.; Diaz, L.; Arias, C.A.; et al. Novel Insights into the Classification of Staphylococcal beta-Lactamases in Relation to the Cefazolin Inoculum Effect. Antimicrob. Agents Chemother. 2020, 64, e02511-19. [Google Scholar] [CrossRef]
- BD BBL™ Cefinase™ Paper Discs. Available online: https://www.bd.com/en-ca/offerings/capabilities/microbiology-solutions/identification-and-susceptibility-testing/bd-bbl-sensi-disc-products/bd-bbl-cefinase-paper-discs (accessed on 1 August 2021).
- Grinberg, A.; Biggs, P.J.; Zhang, J.; Ritchie, S.; Oneroa, Z.; O’Neill, C.; Karkaba, A.; Velathanthiri, N.S.; Coombs, G.W. Genomic epidemiology of methicillin-susceptible Staphylococcus aureus across colonisation and skin and soft tissue infection. J. Infect. 2017, 75, 326–335. [Google Scholar] [CrossRef]
- Coombs, G.W.; Nimmo, G.R.; Daly, D.A.; Le, T.T.; Pearson, J.C.; Tan, H.L.; Robinson, J.O.; Collignon, P.J.; McLaws, M.L.; Turnidge, J.D.; et al. Australian Staphylococcus aureus Sepsis Outcome Programme annual report, 2013. Commun. Dis. Intell. Q. Rep. 2014, 38, E309–E319. [Google Scholar]
- Levinson, G.; Gutman, G.A. Slipped-strand mispairing: A major mechanism for DNA sequence evolution. Mol. Biol. Evol. 1987, 4, 203–221. [Google Scholar]
- Skov, R.; Lonsway, D.R.; Larsen, J.; Larsen, A.R.; Samulioniene, J.; Limbago, B.M. Evaluation of methods for detection of beta-lactamase production in MSSA. J. Antimicrob. Chemother. 2021, 76, 1487–1494. [Google Scholar] [CrossRef]

| Clone | N | PVL | tst | edinB | eta | sea | seb | sec + sel | sed + selj + ser | sek + seq | sep | see | seh | egc- cluster | selx | sely | selz | sel28 | sel31 | sel32 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC1 | 17 | 1 | 1 | 9 | 1 | 1 | 14 | 15 | 17 | |||||||||||
| CC5 | 150 | 3 | 8 | 1 | 2 | 17 | 49 | 150 | 150 | 1 | 1 | |||||||||
| CC6 | 16 | 15 | 3 | 16 | ||||||||||||||||
| CC7 | 3 | 1 | 3 | |||||||||||||||||
| CC8 | 14 | 2 | 1 | 1 | 1 | 4 | 14 | |||||||||||||
| CC9 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||
| CC12 | 16 | 10 | 1 | 10 | 16 | 16 | ||||||||||||||
| CC15 | 41 | 2 | 2 | 40 | ||||||||||||||||
| CC20 | 3 | 1 | 3 | 3 | 3 | |||||||||||||||
| CC22 | 8 | 2 | 1 | 8 | 8 | |||||||||||||||
| CC25 | 1 | 1 | 1 | 1 | ||||||||||||||||
| CC30 | 7 | 1 | 6 | 1 | 2 | 4 | 6 | |||||||||||||
| CC45 | 47 | 10 | 1 | 28 | 46 | 47 | ||||||||||||||
| CC59 | 9 | 2 | 5 | 5 | 9 | 9 | ||||||||||||||
| CC80 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||||||||||||
| CC88 | 17 | 1 | 3 | 17 | ||||||||||||||||
| CC97 | 48 | 2 | 2 | 48 | ||||||||||||||||
| CC101 | 16 | 2 | 16 | |||||||||||||||||
| CC188 | 23 | 1 | 4 | 23 | ||||||||||||||||
| CC291 | 4 | 4 | 1 | |||||||||||||||||
| CC361 | 5 | 1 | 5 | 5 | ||||||||||||||||
| CC398 | 16 | |||||||||||||||||||
| ST425 | 1 | 1 | ||||||||||||||||||
| ST573 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||||||||||
| ST2867 | 3 | 3 | 3 | |||||||||||||||||
| ST5491 | 1 | 1 | ||||||||||||||||||
| ST7270 | 1 | 1 | 1 | 1 | ||||||||||||||||
| Total | 470 | 3 | 23 | 9 | 4 | 37 | 29 | 44 | 18 | 24 | 65 | 1 | 20 | 221 | 442 | 14 | 17 | 2 | 1 | 1 |
| Clone | N | blaZ | ant(4′)-Ia | aadD | ant(9)-Ia | aph(3′)-IIIa | dfrG | ermA | ermB | ermC | ermT | fexA | fusC | lnuA | mphC | tet(K) | tet(M) | mdfA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CC1 | 17 | 12 | ||||||||||||||||
| CC5 | 150 | 10 | 1 | 4 | 1 | 1 | 8 | 2 | 1 | 6 | 1 | |||||||
| CC6 | 16 | 1 | ||||||||||||||||
| CC7 | 3 | |||||||||||||||||
| CC8 | 14 | 1 | 1 | |||||||||||||||
| CC9 | 1 | 1 | ||||||||||||||||
| CC12 | 16 | 1 | ||||||||||||||||
| CC15 | 41 | 20 | 1 | 1 | 1 | |||||||||||||
| CC20 | 3 | 1 | ||||||||||||||||
| CC22 | 8 | 1 | ||||||||||||||||
| CC25 | 1 | 1 | ||||||||||||||||
| CC30 | 7 | 4 | 1 | 1 | 1 | 1 | ||||||||||||
| CC45 | 47 | 2 | 1 | 1 | 1 | 1 | ||||||||||||
| CC59 | 9 | 1 | 1 | |||||||||||||||
| CC80 | 1 | |||||||||||||||||
| CC88 | 17 | 5 | 5 | |||||||||||||||
| CC97 | 48 | 1 | 1 | 1 | 1 | |||||||||||||
| CC101 | 16 | 1 | ||||||||||||||||
| CC188 | 23 | 1 | 1 | |||||||||||||||
| CC291 | 4 | |||||||||||||||||
| CC361 | 5 | |||||||||||||||||
| CC398 | 16 | 1 | 12 | |||||||||||||||
| ST425 | 1 | |||||||||||||||||
| ST573 | 1 | |||||||||||||||||
| ST2867 | 3 | 1 | 1 | |||||||||||||||
| ST5491 | 1 | |||||||||||||||||
| ST7270 | 1 | |||||||||||||||||
| Total | 470 | 45 | 2 | 2 | 9 | 1 | 6 | 9 | 1 | 13 | 12 | 2 | 13 | 1 | 1 | 2 | 6 | 1 |
| Method | Criteria | Interpretive Guidelines | Resistance | Resistant n (%) | Sensitive n (%) |
|---|---|---|---|---|---|
| Disc Diffusion | Zone Diameter | EUCAST P1 | <26 mm | 37 (82.2) | 8 (17.8) |
| CLSI P10 | <29 mm | 26 (57.8) | 19 (42.2) | ||
| Disc Diffusion | Zone Edge | EUCAST P1 | Sharp Zone Edge | 45 (100) | 0 (0) |
| CLSI P10 | Sharp Zone Edge | 45 (100) | 0 (0) | ||
| Broth Microdilution | Breakpoint | EUCAST | >0.125 mg/L | 13 (28.8) | 32 (71.1) |
| CLSI | >0.125 mg/L | ||||
| Etest® | Breakpoint | EUCAST | >0.125 mg/L | 12 (26.7) | 33 (73.3) |
| CLSI | >0.125 mg/L | ||||
| Positive | Negative | ||||
| Nitrocefin Test | Colour Change | Red | 22 (48.9) | 23 (51.1) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Coombs, G.W.; Yee, N.W.T.; Daley, D.; Bennett, C.M.; Robinson, J.O.; Stegger, M.; Shoby, P.; Mowlaboccus, S. Molecular Epidemiology of Penicillin-Susceptible Staphylococcus aureus Bacteremia in Australia and Reliability of Diagnostic Phenotypic Susceptibility Methods to Detect Penicillin Susceptibility. Microorganisms 2022, 10, 1650. https://doi.org/10.3390/microorganisms10081650
Coombs GW, Yee NWT, Daley D, Bennett CM, Robinson JO, Stegger M, Shoby P, Mowlaboccus S. Molecular Epidemiology of Penicillin-Susceptible Staphylococcus aureus Bacteremia in Australia and Reliability of Diagnostic Phenotypic Susceptibility Methods to Detect Penicillin Susceptibility. Microorganisms. 2022; 10(8):1650. https://doi.org/10.3390/microorganisms10081650
Chicago/Turabian StyleCoombs, Geoffrey W., Nicholas W. T. Yee, Denise Daley, Catherine M. Bennett, James O. Robinson, Marc Stegger, Princy Shoby, and Shakeel Mowlaboccus. 2022. "Molecular Epidemiology of Penicillin-Susceptible Staphylococcus aureus Bacteremia in Australia and Reliability of Diagnostic Phenotypic Susceptibility Methods to Detect Penicillin Susceptibility" Microorganisms 10, no. 8: 1650. https://doi.org/10.3390/microorganisms10081650
APA StyleCoombs, G. W., Yee, N. W. T., Daley, D., Bennett, C. M., Robinson, J. O., Stegger, M., Shoby, P., & Mowlaboccus, S. (2022). Molecular Epidemiology of Penicillin-Susceptible Staphylococcus aureus Bacteremia in Australia and Reliability of Diagnostic Phenotypic Susceptibility Methods to Detect Penicillin Susceptibility. Microorganisms, 10(8), 1650. https://doi.org/10.3390/microorganisms10081650






