Inhibitors of Nucleotide Biosynthesis as Candidates for a Wide Spectrum of Antiviral Chemotherapy
Abstract
1. Introduction
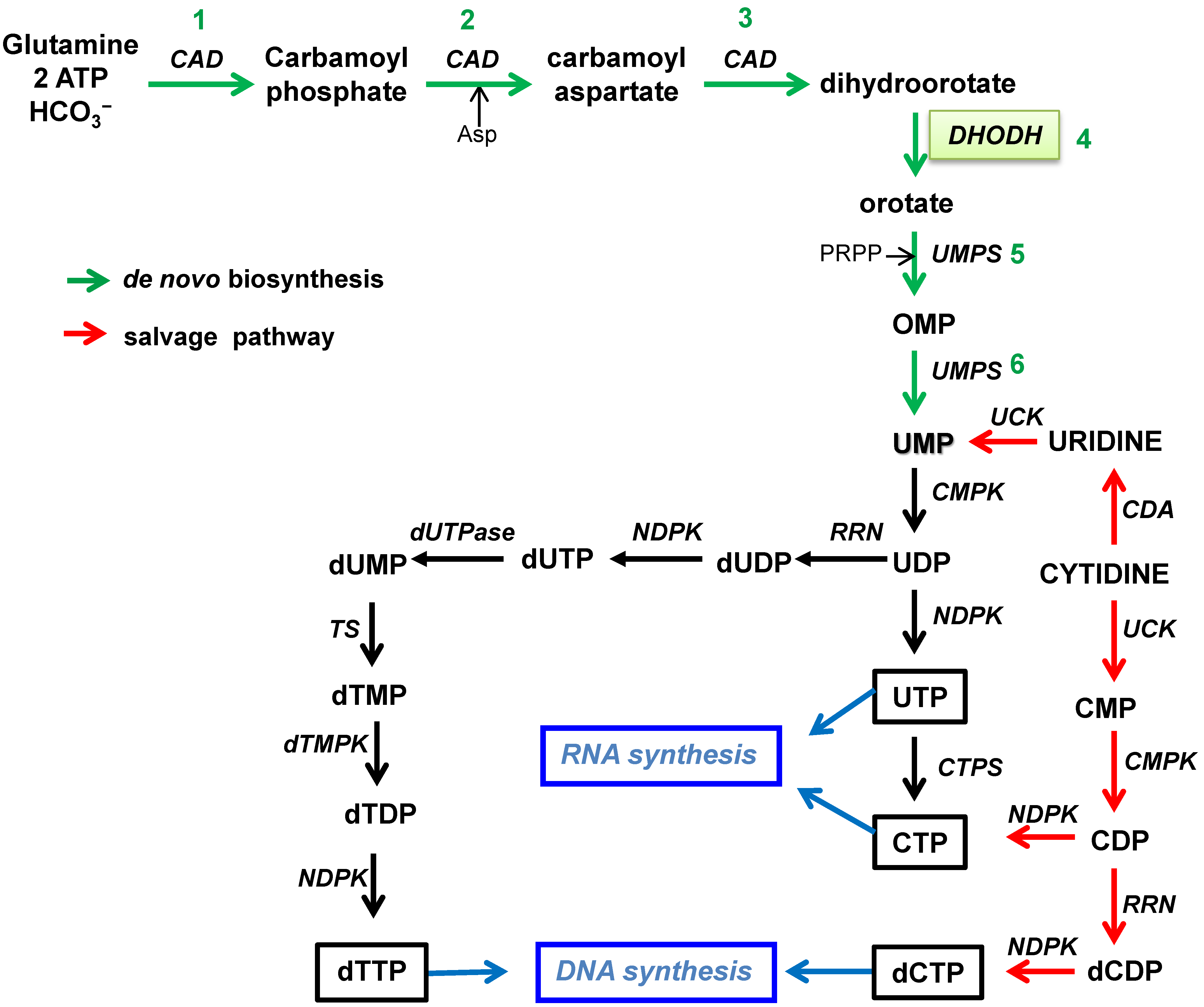
2. Nucleotide Biosynthesis Pathways
2.1. Biosynthesis of Pyrimidines
2.2. Biosynthesis of Purines
3. Inhibition of Nucleotide Biosynthesis and the Potential for Therapy
4. Pyrimidine Inhibitors as Antivirals
4.1. Leflunomide and Teriflunomide
4.2. Brequinar (DuP-785)
4.3. S312 and S416
4.4. Emvododstat (PTC299)
4.5. Vidofludimus Calcium (IMU-838)
4.6. A3
4.7. FA-613
4.8. BAY2402234
4.9. MEDS433
4.10. RYL-634
4.11. GSK 983
4.12. AR-12 Derivatives
5. Purine Inhibitors as Antivirals
5.1. Ribavirin
5.2. Mizoribine
5.3. EICAR
5.4. C-Nucleosides
5.5. Mycophenolic Acid
5.6. Merimepodib
5.7. New IMPDH Inhibitors
6. Concluding Remarks and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Cheng, V.C.C.; To, K.K.W.; Tse, H.; Hung, I.F.N.; Yuen, K.-Y. Two years after pandemic Influenza A/2009/H1N1: What have we learned? Clin. Microbiol. Rev. 2012, 25, 223–263. [Google Scholar] [CrossRef]
- Gubler, D.J.; Vasilakis, N.; Musso, D. History and emergence of Zika virus. J. Infect. Dis. 2017, 216, S860–S867. [Google Scholar] [CrossRef]
- Guarner, J. Three emerging coronaviruses in two decades. Am. J. Clin. Pathol. 2020, 153, 420–421. [Google Scholar] [CrossRef]
- Roychoudhury, S.; Das, A.; Sengupta, P.; Dutta, S.; Roychoudhury, S.; Choudhury, A.P.; Fuzayel Ahmed, A.B.; Bhattacharjee, S.; Slama, P. Viral pandemics of the last four decades: Pathophysiology, health impacts and perspectives. Int. J. Environ. Res. Public Health 2020, 17, 9411. [Google Scholar] [CrossRef]
- Williams, E.P.; Spruill-Harrell, B.M.; Taylor, M.K.; Lee, J.; Nywening, A.V.; Yang, Z.; Nichols, J.H.; Camp, J.V.; Owen, R.D.; Jonsson, C.B. Common themes in zoonotic spillover and disease emergence: Lessons learned from bat- and rodent-borne RNA viruses. Viruses 2021, 13, 1509. [Google Scholar] [CrossRef]
- Rosenberg, R. Detecting the emergence of novel, zoonotic viruses pathogenic to humans. Cell. Mol. Life Sci. 2015, 72, 1115–1125. [Google Scholar] [CrossRef]
- De Clercq, E.; Li, G. Approved antiviral drugs over the past 50 years. Clin. Microbiol. Rev. 2016, 29, 695–747. [Google Scholar] [CrossRef]
- Debing, Y.; Neyts, J.; Delang, L. The future of antivirals: Broad-spectrum inhibitors. Curr. Opin. Infect. Dis. 2015, 28, 596–602. [Google Scholar] [CrossRef]
- Linero, F.N.; Sepúlveda, C.S.; Giovannoni, F.; Castilla, V.; García, C.C.; Scolaro, L.A.; Damonte, E.B. Host cell factors as antiviral targets in arenavirus infection. Viruses 2012, 4, 1569–1591. [Google Scholar] [CrossRef]
- Zakaria, M.K.; Carletti, T.; Marcello, A. Cellular targets for the treatment of flavivirus infections. Front. Cell. Infect. Microbiol. 2018, 8, 398. [Google Scholar] [CrossRef]
- Wong, K.Z.; Hann, J.J. The interplay of viral and host Factors in Chikungunya virus infection: Targets for antiviral strategies. Viruses 2018, 10, 294. [Google Scholar] [CrossRef] [PubMed]
- Mercorelli, B.; Palù, G.; Loregian, A. Drug repurposing for viral infectious diseases: How far are we? Trends Microbiol. 2018, 26, 865–876. [Google Scholar] [CrossRef]
- Evans, D.R.; Guy, H.I. Mammalian pyrimidine biosynthesis: Fresh insights into an ancient pathway. J. Biol. Chem. 2017, 279, 33035–33038. [Google Scholar] [CrossRef]
- Wang, W.; Cui, J.; Ma, H.; Lu, W.; Huang, J. Targeting pyrimidine metabolism in the era of precision cancer medicine. Front. Oncol. 2021, 11, 684961. [Google Scholar] [CrossRef]
- Di Virgilio, F.; Adinolfi, E. Extracellular purines, purinergic receptors and tumor growth. Oncogene 2017, 36, 293–303. [Google Scholar] [CrossRef]
- Yin, J.; Ren, W.; Huang, X.; Deng, J.; Li, T.; Yin, Y. Potential mechanisms connecting purine metabolism and cancer therapy. Front. Immunol. 2018, 9, 1697. [Google Scholar] [CrossRef]
- Munger, J.; Bajad, S.U.; Coller, H.A.; Shenk, T.; Rabinowitz, J.D. Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog. 2006, 2, e132. [Google Scholar] [CrossRef] [PubMed]
- del Caño-Ochoa, F.; Moreno-Morcillo, M.; Ramón-Maiques, S. Cad, a multienzymatic protein at the head of de no pyrimidine biosynthesis. Subcell. Biochem. 2019, 93, 505–538. [Google Scholar] [CrossRef] [PubMed]
- Reis, R.A.G.; Calil, F.A.; Feliciano, P.R.; Pinheiro, M.P.; Nonato, M.C. The dihydroorotate dehydrogenases: Past and present. Arch. Biochem. Biophys. 2017, 632, 175–191. [Google Scholar] [CrossRef] [PubMed]
- Wittmann, J.G.; Heinrich, D.; Gasow, K.; Frey, A.; Diederichsen, U.; Rudolph, M.G. Structures of the human orotidine-5′-monophosphate decarboxylase support a covalent mechanism and provide a framework for drug design. Structure 2008, 16, 82–92. [Google Scholar] [CrossRef] [PubMed]
- Lane, A.N.; Fan, T.W.-M. Regulation of mammalian nucleotide metabolism and biosynthesis. Nucleic Acids Res. 2015, 43, 2466–2485. [Google Scholar] [CrossRef]
- Munier-Lehmann, H.; Vidalain, P.-O.; Tangy, F.; Janin, Y.L. On dihydroorotate dehydrogenases and their inhibitors and uses. J. Med. Chem. 2013, 56, 3148–3167. [Google Scholar] [CrossRef]
- Sanders, S.; Harisdangkul, V. Leflunomide for the treatment of rheumatoid arthritis and autoimmunity. Am. J. Med. Sci. 2002, 323, 190–193. [Google Scholar] [CrossRef]
- Oh, J.; O’Connor, P.W. An update of teriflunomide for treatment of multiple sclerosis. Ther. Clin. Risk Manag. 2013, 9, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Madak, J.T.; Bankhead, A.; Cuthbertson, C.R.; Showalter, H.D.; Neamati, N. Revisiting the role of dihydroorotate dehydrogenase as a therapeutic target for cancer. Pharmacol. Ther. 2019, 195, 111–131. [Google Scholar] [CrossRef]
- Zhou, Y.; Tao, L.; Zhou, X.; Zuo, Z.; Gong, J.; Liu, X.; Zhou, Y.; Liu, C.; Sang, N.; Liu, H.; et al. DHODH and cancer: Promising prospects to be explored. Cancer Metab. 2021, 9, 22. [Google Scholar] [CrossRef]
- Singh, A.; Maqbool, M.; Mobashir, M.; Hoda, N. Dihydroorotate dehydrogenase: A drug target for the development of antimalarials. Eur. J. Med. Chem. 2017, 125, 640–651. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, S.; Song, K.; Ye, J.; Li, W.; Zhong, Y.; Feng, Z.; Liang, S.; Cai, Z.; Xu, K. A broad antiviral strategy: Inhibitors of human DHODH pave the way for host-targeting antivirals against emerging and re-emerging viruses. Viruses 2022, 14, 928. [Google Scholar] [CrossRef]
- Lucas-Hourani, M.; Dauzonne, D.; Jorda, P.; Cousin, G.; Lupan, A.; Helynck, O.; Caignard, G.; Janvier, G.; Andre-Leroux, G.; Khiar, S.; et al. Inhibition of pyrimidine biosynthesis pathway suppresses viral growth through innate immunity. PLoS Pathog. 2013, 9, e1003678. [Google Scholar] [CrossRef] [PubMed]
- Cheung, N.N.; Lai, K.K.; Dai, J.; Kok, K.H.; Chen, H.; Chan, K.-H.; Yuen, K.-Y.; Kao, R.Y.T. Broad-spectrum inhibition of common respiratory RNA viruses by a pyrimidine synthesis inhibitor with involvement of the host antiviral response. J. Gen. Virol. 2017, 98, 946–954. [Google Scholar] [CrossRef] [PubMed]
- Luthra, P.; Naidoo, J.; Pietzsch, C.A.; De, S.; Khadka, S.; Anantpadma, M.; Williams, C.G.; Edwards, M.R.; Davey, R.A.; Bukreyev, A.; et al. Inhibiting pyrimidine biosynthesis impairs Ebola virus replication through depletion of nucleoside pools and activation of innate immune responses. Antivir. Res. 2018, 158, 288–302. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, W.; Xu, L.; Zhou, X.; Shokrollahi, E.; Felczak, K.; van der Laan, L.J.; Pankiewicz, K.W.; Sprengers, D.; Raat, N.J.; et al. Cross talk between nucleotide synthesis pathways with cellular immunity in constraining hepatitis E virus replication. Antimicrob. Agents Chemother. 2016, 60, 2834–2848. [Google Scholar] [CrossRef]
- Chung, D.-H.; Golden, J.E.; Adcock, R.S.; Schroeder, C.E.; Chu, Y.-K.; Sotsky, J.B.; Cramer, D.E.; Chilton, P.M.; Song, C.; Anantpadma, M.; et al. Discovery of a broad-spectrum antiviral compound that inhibits pyrimidine biosynthesis and establishes a type 1 interferon-independent antiviral state. Antimicrob. Agents Chemother. 2016, 60, 4552–4562. [Google Scholar] [CrossRef]
- Xiong, R.; Zhang, L.; Li, S.; Sun, Y.; Ding, M.; Wang, Y.; Zhao, Y.; Wu, Y.; Shang, W.; Jiang, X.; et al. Novel and potent inhibitors targeting DHODH are broad-spectrum antivirals against RNA viruses including newly-emerged coronavirus SARS-CoV-2. Protein Cell 2020, 11, 723–739. [Google Scholar] [CrossRef]
- Luban, J.; Sattler, R.A.; Mühlberger, E.; Graci, J.D.; Cao, L.; Weetall, M.; Trotta, C.; Colacino, J.M.; Bavari, S.; Strambio-De-Castillia, C.; et al. The DHODH inhibitor PTC299 arrests SARS-CoV-2 replication and suppresses induction of inflammatory cytokines. Virus Res. 2021, 292, 198246. [Google Scholar] [CrossRef]
- Balzarini, J.; Gago, F.; Kulik, W.; van Kuilenburg, A.B.P.; Karlsson, A.; Peterson, M.A.; Robins, M.J. Introduction of a fluorine atom at C3 of 3-deazauridine shifts its antimetabolic activity from inhibition of CTP synthetase to inhibition of orotidylate decarboxylase, an early event in the de novo pyrimidine nucleotide biosynthesis pathway. J. Biol. Chem. 2012, 287, 30444–30454. [Google Scholar] [CrossRef]
- Lee, K.; Kim, D.-E.; Jang, K.-S.; Kim, S.-J.; Cho, S.; Kim, C. Gemcitabine, a broad-spectrum antiviral drug, suppresses enterovirus infections through innate immunity induced by the inhibition of pyrimidine biosynthesis and nucleotide depletion. Oncotarget 2017, 8, 115315–115325. [Google Scholar] [CrossRef]
- Holmes, E.W.; Pehlke, D.M.; Kelley, W.N. Human IMP dehydrogenase: Kinetics and regulatory properties. Biochim. Biophys. Acta 1974, 364, 209–217. [Google Scholar] [CrossRef]
- Hedstrom, L. IMP dehydrogenase: Structure, mechanism and inhibition. Chem. Rev. 2009, 109, 2903–2928. [Google Scholar] [CrossRef]
- Carr, S.F.; Papp, E.; Wu, J.C.; Natsumeda, Y. Characterization of human type I and type II IMP dehydrogenase. J. Biol. Chem. 1993, 268, 27286–27290. [Google Scholar] [CrossRef]
- Naffouje, R.; Grover, P.; Yu, H.; Sendilnathan, A.; Wolfe, K.; Majd, N.; Smith, E.P.; Takeuchi, K.; Senda, T.; Kpfuji, S.; et al. Anti-tumor potential of IMP dehydrogenase inhibitors: A century-long story. Cancers 2019, 11, 1346. [Google Scholar] [CrossRef]
- Allison, A.C.; Eugui, E.M. Mycophenolate mofetil and its mechanisms of action. Immunopharmacology 2000, 47, 85–118. [Google Scholar] [CrossRef]
- Nair, V.; Shu, Q. Inosine monophosphate dehydrogenase as a probe in antiviral drug discovery. Antivir. Chem. Chemother. 2007, 18, 245–258. [Google Scholar] [CrossRef]
- Shipkova, M.; Armstrong, V.W.; Oellerich, M.; Wieland, E. Mycophenolate mofetil in organ transplantation: Focus on metabolism, safety and tolerability. Expert. Opin. Drug. Metab. Toxicol. 2005, 1, 505–526. [Google Scholar] [CrossRef]
- van Gelder, T.; Hesselink, D.A. Mycophenolate revisited. Transpl. Int. 2015, 28, 508–515. [Google Scholar] [CrossRef]
- Pan, Q.; Tilanus, H.W.; Metselaar, H.J.; Janssen, H.L.; van der Laan, L.J. Virus-drug interactions–molecular insight into immunosuppression and HCV. Nat. Rev. Gastroenterol. Hepatol. 2012, 9, 355–362. [Google Scholar] [CrossRef]
- Dang, W.; Yin, Y.; Wang, Y.; Wang, W.; Su, J.; Sprengers, D.; van der Laan, L.J.W.; Felczak, K.; Pankiewicz, K.W.; Chang, K.-O.; et al. Inhibition of calcineurin or IMP dehydrogenase exerts moderate to potent antiviral activity against Norovirus replication. Antimicrob. Agents Chemother. 2017, 61, e01095-17. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Jiang, H. Potential treatment of COVID-19 by inhibitors of human dihydroorotate dehydrogenase. Protein Cell 2020, 11, 699–702. [Google Scholar] [CrossRef]
- Davis, J.P.; Cain, G.A.; Pitts, W.J.; Magolda, R.L.; Copeland, R.A. The immunosuppressive metabolite of leflunomide is a potent inhibitor of human dihydroorotate dehydrogenase. Biochemistry 1996, 35, 1270–1273. [Google Scholar] [CrossRef]
- McLean, J.E.; Neidhardt, E.A.; Grossman, T.H.; Hedstrom, L. Multiple inhibitor analysis of the brequinar and leflunomide binding sites on human dihydroorotate dehydrogenase. Biochemistry 2001, 40, 2194–2200. [Google Scholar] [CrossRef]
- Knecht, W.; Bergjohann, U.; Gonski, S.; Kirschbaum, B.; Lóffler, M. Functional expression of a fragment of human dihydroorotate dehydrogenase by means of the baculovirus expression vector system, and kinetic investigation of the purified recombinant enzyme. Eur. J. Biochem. 1996, 240, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Fragoso, Y.D.; Brooks, J.B. Leflunomide and teriflunomide: Altering the metabolism of pyrimidines for the treatment of autoimmune diseases. Expert Rev. Clin. Pharmacol. 2015, 8, 315–320. [Google Scholar] [CrossRef] [PubMed]
- Waldman, W.J.; Knight, D.A.; Lurain, N.S.; Miller, D.M.; Sedmak, D.D.; Williams, J.W.; Chong, A.S. Novel mechanism of inhibition of cytomegalovirus by the experimental immunosuppressive agent leflunomide. Transplantation 1999, 68, 814–825. [Google Scholar] [CrossRef] [PubMed]
- Liacini, A.; Seamone, M.E.; Muruve, D.A.; Tibbles, L.A. Anti-BK virus mechanisms of sirolimus and leflunomide alone and in combination: Toward a new therapy for BK virus infection. Transplantation 2010, 90, 1450–1457. [Google Scholar] [CrossRef] [PubMed]
- Bilger, A.; Plowshay, J.; Ma, S.; Nawandar, D.; Barlow, E.A.; Romero-Masters, J.C.; Bristol, J.A.; Li, Z.; Tsai, M.H.; Delecluse, H.J.; et al. Leflunomide/teriflunomide inhibit Epstein-Barr virus (EBV)-induced lymphoproliferative disease and lytic viral replication. Oncotarget 2017, 8, 44266–44280. [Google Scholar] [CrossRef]
- Wang, E.; Jan, A.S.; Doan, V.P.; Ferguson, J.B.; Yeh, J.C. Leflunomide therapy for refractory cytomegalovirus infections in hematopoietic stem cell transplant recipients. J. Oncol. Pharm. Pract. 2019, 25, 1731–1737. [Google Scholar] [CrossRef]
- Sepúlveda, C.S.; García, C.C.; Damonte, E.B. Antiviral activity of A771726, the active metabolite of leflunomide, against Junín virus. J. Med. Virol. 2018, 90, 819–827. [Google Scholar] [CrossRef]
- Wang, J.; Sun, J.; Hu, J.; Wang, C.; Prinz, R.A.; Peng, D.; Liu, X.; Xu, X. A77 1726, the active metabolite of the anti-rheumatoid arthritis drug leflunomide, inhibits influenza A virus replication in vitro and in vivo by inhibiting the activity of Janus kinases. FASEB J. 2020, 34, 10132–10145. [Google Scholar] [CrossRef]
- Davis, I.C.; Lazarowski, E.R.; Hickman-Davis, J.M.; Fortenberry, J.A.; Chen, F.P.; Zhao, X.; Sorscher, E.; Graves, L.M.; Sullender, W.M.; Matalon, S. Leflunomide prevents alveolar fluid clearance inhibition by respiratory syncytial virus. Am. J. Respir. Crit. Care Med. 2006, 173, 673–682. [Google Scholar] [CrossRef]
- Davis, I.C.; Lazarowski, E.R.; Chen, F.P.; Hickman-Davis, J.M.; Sullender, W.M.; Matalon, S. Post-infection A77-1726 blocks pathophysiologic sequelae of respiratory syncytial virus infection. Am. J. Respir. Cell. Mol. Biol. 2007, 37, 379–386. [Google Scholar] [CrossRef]
- Hu, K.; Wang, M.; Zhao, Y.; Zhang, Y.; Wang, T.; Zheng, Z.; Li, X.; Zeng, S.; Zhao, D.; Li, H.; et al. A Small-scale medication of leflunomide as a treatment of COVID-19 in an open-label blank-controlled clinical trial. Virol. Sin. 2020, 35, 725–733. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhao, Y.; Hu, W.; Zhao, D.; Zhang, Y.; Wang, T.; Zheng, Z.; Li, X.; Zeng, S.; Liu, Z.; et al. Treatment of coronavirus disease 2019 patients with prolonged postsymptomatic viral shedding with leflunomide: A single-center randomized controlled clinical trial. Clin. Infect. Dis. 2021, 73, e4012–e4019. [Google Scholar] [CrossRef] [PubMed]
- Makowka, L.; Sher, L.S.; Cramer, D.V. The development of brequinar as an immunosuppressive drug for transplantation. Immunol. Rev. 1993, 136, 51–70. [Google Scholar] [CrossRef]
- Knecht, W.; Löffler, M. Species-related inhibition of human and rat dihydroorotate dehydrogenase by immunosuppressive isoxazol and cinchoninic acid derivatives. Biochem. Pharmacol. 1998, 56, 1259–1264. [Google Scholar] [CrossRef]
- Morales Vasquez, D.; Park, J.G.; Ávila-Pérez, G.; Nogales, A.; de la Torre, J.C.; Almazan, F.; Martinez-Sobrido, L. Identifcation of inhibitors of ZIKV replication. Viruses 2020, 12, 1041. [Google Scholar] [CrossRef] [PubMed]
- Qing, M.; Zou, G.; Wang, Q.Y.; Xu, H.Y.; Dong, H.; Yuan, Z.; Shi, P.Y. Characterization of dengue virus resistance to brequinar in cell culture. Antimicrob. Agents Chemother. 2010, 54, 3686–3695. [Google Scholar] [CrossRef] [PubMed]
- Evers, D.L.; Wanga, X.; Huonga, S.-M.; Andreonib, K.A.; Huang, E.-S. Inhibition of human cytomegalovirus signalling and replication by the immunosuppressant FK778. Antivir. Res. 2005, 65, 1–12. [Google Scholar] [CrossRef]
- Fu, H.; Zhang, Z.; Dai, Y.; Liu, S.; Fu, E. Brequinar inhibits enterovirus replication by targeting biosynthesis pathway of pyrimidines. Am. J. Transl. Res. 2020, 12, 8247–8255. [Google Scholar]
- Chen, S.; Ding, S.; Yin, Y.; Xu, L.; Li, P.; Peppelenbosch, M.P.; Pan, Q.; Wang, W. Suppression of pyrimidine biosynthesis by targeting DHODH enzyme robustly inhibits rotavirus replication. Antivir. Res. 2019, 167, 35–44. [Google Scholar] [CrossRef]
- Schultz, D.C.; Johnson, R.M.; Ayyanathan, K.; Miller, J.; Whig, K.; Kamalia, B.; Dittmar, M.; Weston, S.; Hammond, H.L.; Dillen, C.; et al. Pyrimidine inhibitors synergize with nucleoside analogues to block SARS-CoV-2. Nature 2022, 604, 134–140. [Google Scholar] [CrossRef]
- Diao, Y.Y.; Lu, W.Q.; Jin, H.T.; Zhu, J.S.; Han, L.; Xu, M.H.; Gao, R.; Shen, X.; Zhao, Z.J.; Liu, X.F.; et al. Discovery of diverse human dihydroorotate dehydrogenase inhibitors as immunosuppressive agents by structure-based virtual screening. J. Med. Chem. 2012, 55, 8341–8349. [Google Scholar] [CrossRef]
- Zhu, J.D.; Meng, W.; Wang, X.J.; Wang, H.C. Broad-spectrum antiviral agents. Front. Microbiol. 2015, 6, 517. [Google Scholar] [CrossRef]
- Cao, L.; Weetall, M.; Trotta, C.; Cintron, K.; Ma, J.; Kim, M.J.; Furia, B.; Romfo, C.; Graci, J.D.; Li, W.; et al. Targeting of hematologic malignancies with PTC299, a novel potent inhibitor of dihydroorotate dehydrogenase with favourable pharmaceutical properties. Mol. Cancer Ther. 2019, 18, 3–16. [Google Scholar] [CrossRef]
- Şimşek-Yavuz, S.; Komsuoğlu Çelikyurt, F.I. An update of anti-viral treatment of COVID-19. Turk. J. Med. Sci. 2021, 51, 3372–3390. [Google Scholar] [CrossRef]
- Muehler, A.; Kohlhof, H.; Groeppel, M.; Vitt, D. Safety, Tolerability and pharmacokinetics of vidofludimus calcium (IMU-838) after single and multiple ascending oral doses in healthy male subjects. Eur. J. Drug Metab. Pharmacokinet. 2020, 45, 557–573. [Google Scholar] [CrossRef]
- Muehler, A.; Peelen, E.; Kohlhof, H.; Gröppel, M.; Vitt, D. Vidofludimus calcium, a next generation DHODH inhibitor for the treatment of relapsing-remitting multiple sclerosis. Mult. Scler. Relat. Disord. 2020, 43, 102129. [Google Scholar] [CrossRef]
- Hahn, F.; Wangen, C.; Häge, S.; Peter, A.S.; Dobler, G.; Hurst, B.; Julander, J.; Fuchs, J.; Ruzsics, Z.; Überla, K.; et al. IMU-838, a developmental DHODH inhibitor in phase II for autoimmune disease, shows anti-SARS-CoV-2 and broad-spectrum antiviral efficacy in vitro. Viruses 2020, 12, 1394. [Google Scholar] [CrossRef]
- Hoffmann, H.H.; Kunz, A.; Simon, V.A.; Palese, P.; Shaw, M.L. Broad-spectrum antiviral that interferes with de novo pyrimidine biosynthesis. Proc. Natl. Acad. Sci. USA 2011, 108, 5777–5782. [Google Scholar] [CrossRef]
- Ortiz-Riaño, E.; Ngo, N.; Devito, S.; Eggink, D.; Munger, J.; Shaw, M.L.; de la Torre, J.C.; Martínez-Sobrido, L. Inhibition of arenavirus by A3, a pyrimidine biosynthesis inhibitor. J. Virol. 2014, 88, 878–889. [Google Scholar] [CrossRef]
- Christian, S.; Merz, C.; Evans, L.; Gradl, S.; Seidel, H.; Friberg, A.; Eheim, A.; Lejeune, P.; Brzezinka, K.; Zimmermann, K.; et al. The Novel Dihydroorotate Dehydrogenase (DHODH) Inhibitor BAY 2402234 triggers differentiation and is effective in the treatment of myeloid malignancies. Leukemia 2019, 33, 2403–2415. [Google Scholar] [CrossRef]
- Mathieu, C.; Touret, F.; Jacquemin, C.; Janin, Y.L.; Nougairède, A.; Brailly, M.; Mazelier, M.; Décimo, D.; Vasseur, V.; Hans, A.; et al. A bioluminescent 3CLPro activity assay to monitor SARS-CoV-2 replication and identify inhibitors. Viruses 2021, 13, 1814. [Google Scholar] [CrossRef]
- Sainas, S.; Pippione, A.C.; Lupino, E.; Giorgis, M.; Circosta, P.; Gaidano, V.; Goyal, P.; Bonanni, D.; Rolando, B.; Cignetti, A.; et al. Targeting myeloid differentiation using potent 2-hydroxypyrazol[1,5-a]pyridine scaffold-based human dihydroorotate dehydrogenase inhibitors. J. Med. Chem. 2018, 61, 6034–6055. [Google Scholar] [CrossRef]
- Luganini, A.; Sibille, G.; Mognetti, B.; Sainas, S.; Pippione, A.C.; Giorgis, M.; Boschi, D.; Lolli, M.L.; Gribaudo, G. Effective deploying of a novel DHODH inhibitor against herpes simplex type 1 and type 2 replication. Antivir. Res. 2021, 189, 105057. [Google Scholar] [CrossRef]
- Calistri, A.; Luganini, A.; Mognetti, B.; Elder, E.; Sibille, G.; Conciatori, V.; Del Vecchio, C.; Sainas, S.; Boschi, D.; Montserrat, N.; et al. The new generation hDHODH inhibitor MEDS433 hinders the in vitro replication of SARS-CoV-2 and other human coronaviruses. Microorganisms 2021, 9, 1731. [Google Scholar] [CrossRef]
- Yang, Y.; Cao, L.; Gao, H.; Wu, Y.; Wang, Y.; Fang, F.; Lan, T.; Lou, Z.; Rao, Y. Discovery, optimization, and target Identification of novel potent broad-spectrum antiviral inhibitors. J. Med. Chem. 2019, 62, 4056–4073. [Google Scholar] [CrossRef]
- Gong, M.; Yang, Y.; Huang, Y.; Gan, T.; Wu, Y.; Gao, H.; Li, Q.; Nie, J.; Huang, W.; Wang, Y.; et al. Novel quinolone derivatives targeting human dihydroorotate dehydrogenase suppress Ebola virus infection in vitro. Antivir. Res. 2021, 194, 105161. [Google Scholar] [CrossRef]
- Yang, C.F.; Gopula, B.; Liang, J.J.; Li, J.K.; Chen, S.Y.; Lee, Y.L.; Chen, C.S.; Lin, Y.L. Novel AR-12 derivatives, P12-23 and P12-34, inhibit flavivirus replication by blocking host de novo pyrimidine biosynthesis. Emerg. Microbes Infect. 2018, 7, 187. [Google Scholar] [CrossRef]
- Pankiewicz, K.W.; Goldstein, B.M. Chapter 1 Inosine monophosphate dehydrogenase and its inhibitors: An overview. In Inosine Monophosphate Dehydrogenase; ACS Symposium Series; American Chemical Society: Washington, DC, USA, 2003. [Google Scholar] [CrossRef]
- Buey, R.; Ledesma-Amaro, R.; Velázquez-Campoy, A.; Balsera, M.; Chagoyen, M.; de Pereda, J.M.; Revuelta, J.L. Guanine nucleotide binding to the Bateman domain mediates the allosteric inhibition of eukaryotic IMP dehydrogenases. Nat. Commun. 2015, 6, 8923. [Google Scholar] [CrossRef]
- Neyts, S.J.; Andrei, G.; De Clercq, E. The novel immunosuppressive agent mycophenolate mofetil markedly potentiates the antiherpesvirus activities of acyclovir, ganciclovir, and penciclovir in vitro and in vivo. Antimicrob. Agents Chemother. 1998, 42, 216–222. [Google Scholar] [CrossRef]
- De Clercq, E. Trends in the development of new antiviral agents for the chemotherapy of infections caused by herpes viruses and retroviruses. Rev. Med. Virol. 1995, 5, 149–164. [Google Scholar] [CrossRef]
- Sidwell, R.W.; Huffman, J.H.; Khare, G.P.; Allen, L.B.; Witkowski, J.T.; Robins, R.K. Broad-spectrum antiviral activity of Virazole: 1-β-D-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science 1972, 177, 705–706. [Google Scholar] [CrossRef]
- Graci, J.D.; Cameron, C.E. Mechanisms of action of ribavirin against distinct viruses. Rev. Med. Virol. 2006, 16, 37–48. [Google Scholar] [CrossRef]
- Nyström, K.; Wanrooij, P.H.; Waldenström, J.; Adamek, L.; Brunet, S.; Said, J.; Nilsson, S.; Wind-Rotolo, M.; Hellstrand, K.; Norder, H.; et al. Inosine triphosphate pyrophosphatase dephosphorylates ribavirin triphosphate and reduced enzymatic activity potentiates mutagenesis in hepatitis C virus. J. Virol. 2018, 92, e01087-18. [Google Scholar] [CrossRef]
- Hultgren, C.; Milich, D.R.; Weiland, O.; Sallberg, M. The antiviral compound ribavirin modulates the T helper (Th) 1/Th2 subset balance in hepatitis B and C virus-specific immune responses. J. Gen. Virol. 1998, 79, 2381–2391. [Google Scholar] [CrossRef]
- Tam, R.C.; Lau, Y.Y.N.; Hong, Z. Mechanisms of action of ribavirin in antiviral therapies. Antivir. Chem. Chemother. 2002, 12, 261–272. [Google Scholar] [CrossRef]
- Yan, Y.; Svitkin, Y.; Lee, J.M.; Bisaillon, M.; Pelletier, J. Ribavirin is not a functional mimic of the 7-methyl guanosine mRNA cap. RNA 2005, 11, 1238–1244. [Google Scholar] [CrossRef]
- Vo, N.V.; Young, K.C.; Lai, M.M. Mutagenic and inhibitory effects of ribavirin on hepatitis C virus RNA polymerase. Biochemistry 2003, 42, 10462–10471. [Google Scholar] [CrossRef]
- Hadj Hassine, I.; Ben M’hadheb, M.; Menéndez-Arias, L. Lethal mutagenesis of RNA viruses and approved drugs with antiviral mutagenic activity. Viruses 2022, 14, 841. [Google Scholar] [CrossRef]
- Cameron, C.E.; Castro, C. The mechanism of action of ribavirin: Lethal mutagenesis of RNA virus genomes mediated by the viral RNA-dependent RNA polymerase. Curr. Opin. Infect. Dis. 2001, 14, 757–764. [Google Scholar] [CrossRef]
- Nyström, K.; Waldenström, J.; Tang, K.; Lagging, M. Ribavirin: Pharmacology, multiple modes of action and possible future perspectives. Future Virol. 2019, 14, 153–160. [Google Scholar] [CrossRef]
- Cooper, A.C.; Banasiak, N.C.; Allen, P.J. Management and prevention strategies for respiratory syncytial virus (RSV) bronchiolitis in infants and young children: A review of evidence based practice interventions. Pediatr. Nurs. 2003, 29, 452–456. [Google Scholar] [PubMed]
- Pawlotsky, J.-M. Mechanisms of antiviral treatment efficacy and failure in chronic hepatitis C. Antivir. Res. 2003, 59, 1–11. [Google Scholar] [CrossRef]
- Huggins, J.W. Prospects for treatment of viral hemorrhagic fevers with ribavirin, a broad-spectrum antiviral drug. Rev. Infect. Dis. 1989, 4, S750–S761. [Google Scholar] [CrossRef] [PubMed]
- Hayden, F.G.; Shindo, N. Influenza virus polymerase inhibitors in clinical development. Curr. Opin. Infect. Dis. 2019, 32, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Chaturaka, R.; Herath, T.; Wickramarachchi, U.; Fernando, D.; Rajapakse, S. Treatment of chikungunya-associated joint pain: A systematic review of controlled clinical trials. Trans. R. Soc. Trop. Med. Hyg. 2022, 6, trac045. [Google Scholar] [CrossRef]
- Ramírez-Olivencia, G.; Estébanez, M.; Membrillo, F.J.; Ybarra, M.D.C. Use of ribavirin in viruses other than hepatitis C. A review of the evidence. Enferm. Infect. Microbiol. Clin. 2019, 37, 602–608. [Google Scholar] [CrossRef]
- Salam, A.P.; Cheng, V.; Edwards, T.; Olliaro, P.; Sterne, J.; Horby, P. Time to reconsider the role of ribavirin in Lassa fever. PLoS Negl. Trop. Dis. 2021, 15, e0009522. [Google Scholar] [CrossRef]
- Ying, C.; Colonno, R.; De Clercq, E.; Neyts, J. Ribavirin and mycophenolic acid markedly potentiate the anti-hepatitis B virus activity of entecavir. Antivir. Res. 2007, 73, 192–196. [Google Scholar] [CrossRef]
- Balzarini, J.; Lee, C.K.; Herdewijn, P.; De Clercq, E. Mechanism of the potentiating effect of ribavirin on the activity of 2′,3′-dideoxyinosine against human immunodeficiency virus. J. Biol. Chem. 1991, 266, 21509–21514. [Google Scholar] [CrossRef]
- Shigeta, S.; Mori, S.; Baba, M.; Ito, M.; Honzumi, K.; Nakamura, K.; Oshitani, H.; Numazaki, Y.; Matsuda, A.; Obara, T.; et al. 5-ethynyl-1-β-d-ribofuranosylimidazole-4-carboxamide, and 6′-(R)-6′-C-methylneplanocin A against several ortho- and paramyxoviruses. Antimicrob. Agents Chemother. 1992, 36, 435–439. [Google Scholar] [CrossRef]
- Pancheva, S.N. Potentiating effect of ribavirin on the anti-herpes activity of acyclovir. Antivir. Res. 1991, 16, 151–161. [Google Scholar] [CrossRef]
- Vogt, M.W.; Hartshorn, K.L.; Furman, P.A.; Chou, T.C.; Fyfe, J.A.; Coleman, L.A.; Crumpacker, C.; Schooley, R.T.; Hirsch, M.S. Ribavirin antagonizes the effect of azidothymidine on HIV replication. Science 1987, 235, 376–379. [Google Scholar] [CrossRef] [PubMed]
- Baba, M.; Pauwels, R.; Balzarini, J.; Herdewijn, P.; DeClercq, E.; Desmyter, J. Ribavirin antagonizes inhibitory effects of pyrimidine 2′,3′-dideoxynucleosides but enhances inhibitory effects of purine 2′,3′-dideoxynucleosides on replication of human immunodeficiency virus in vitro. Antimicrob. Agents Chemother. 1987, 31, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Mangia, A.; Foster, G.R.; Berg, C.P.; Curescu, M.; Ledinghen, V.; Habersetzer, F.; Manolakopoulos, S.; Negri, E.; Papatheodoridis, G.; Ahlers, S.; et al. Efficacy and safety profile of boceprevir- or telaprevir-based triple therapy or dual peginterferon alfa-2a or alfa-2b plus ribavirin therapy in chronic hepatitis C: The real-world PegBase observational study. Ann. Gastroenterol. 2017, 30, 327–343. [Google Scholar] [CrossRef] [PubMed]
- Hager, P.W.; Collart, F.R.; Huberman, E.; Mitchell, B.S. Recombinant human inosine monophosphate dehydrogenase type I and type II proteins. Purification and characterization of inhibitor binding. Biochem. Pharmacol. 1995, 49, 1323–1329. [Google Scholar] [CrossRef]
- Hosoya, M.; Shigeta, S.; Ishii, T.; Suzuki, H.; De Clercq, E. Comparative inhibitory effects of various nucleoside and nonnucleoside analogues on replication of influenza virus types A and B in vitro and in ovo. J. Infect. Dis. 1993, 168, 641646. [Google Scholar] [CrossRef]
- Shigeta, S. Recent progress in antiviral chemotherapy for respiratory syncytial virus infections. Expert Opin. Investig. Drugs 2000, 9, 221–235. [Google Scholar] [CrossRef]
- Yokota, S. Mizoribine: Mode of action and effects in clinical use. Pediatr. Int. 2002, 44, 196–198. [Google Scholar] [CrossRef]
- Yoshimura, N.; Ushigome, H.; Akioka, K.; Nobori, S.; Suzuki, T.; Sakai, K.; Okamoto, M. The beneficial effect of high-dose mizoribine combined with cyclosporine, basiliximab, and corticosteroids on CMV infection in renal transplant recipients. Clin. Exp. Nephrol. 2013, 17, 127–133. [Google Scholar] [CrossRef]
- Jishi, L.; Zhang, K.; Ji, Y.; Zhang, W.; Li, W.; Yong, Q.; Wang, J.; Sun, J.; Zhang, H. Safety and efficacy of mizoribine treatment in nephrotic syndrome complicated with hepatitis B virus infection: A clinical study. Ren. Fail. 2016, 38, 723–727. [Google Scholar] [CrossRef]
- Itoh, H.; Komatsuda, A.; Wakui, H.; Miura, A.B.; Tashima, Y. Mammalian HSP60 is a major target for an immunosuppressant mizoribine. J. Biol. Chem. 1999, 274, 35147–35151. [Google Scholar] [CrossRef] [PubMed]
- Saijo, M.; Morikawa, S.; Fukushi, S.; Mizutani, T.; Hasegawa, H.; Nagata, N.; Iwata, N.; Kurane, I. Inhibitory effect of mizoribine and ribavirin on the replication of severe acute respiratory syndrome (SARS)-associated coronavirus. Antivir. Res. 2005, 66, 159–163. [Google Scholar] [CrossRef]
- De Clercq, E.; Cools, M.; Balzarini, J.; Snoeck, R.; Andrei, G.; Hosoya, M.; Shigeta, S.; Ueda, T.; Minakawa, N.; Matsuda, A. Antiviral activities of 5-ethynyl-1-β-D-ribofuranosylimidazole-4- carboxamide and related compounds. Antimicrob. Agents Chemother. 1991, 35, 679–684. [Google Scholar] [CrossRef] [PubMed]
- Leyssen, P.; De Clercq, E.; Neyts, J. The anti-yellow fever virus activity of ribavirin is independent of error-prone replication. Mol. Pharmacol. 2006, 69, 1461–1467. [Google Scholar] [CrossRef] [PubMed]
- Ölschläger, S.; Neyts, J.; Günther, S. Depletion of GTP pool is not the predominant mechanism by which ribavirin exerts its antiviral effect on Lassa virus. Antivir. Res. 2011, 91, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Papov, V.V.; Minakawa, N.; Matsuda, A.; Biemann, K.; Hedstrom, L. Inactivation of inosine 5’-monophosphate dehydrogenase by the antiviral agent 5-ethynyl-1-β-d-ribofuranosylimidazole-4-carboxamide 5′-monophosphate. Biochemistry 1996, 35, 95–101. [Google Scholar] [CrossRef]
- Barnard, D.L.; Day, C.W.; Bailey, K.; Heiner, M.; Montgomery, R.; Lauridsen, L.; Winslow, S.; Hoopes, J.; Li, J.K.; Lee, J.; et al. Enhancement of the infectivity of SARS-CoV in BALB/c mice by IMP dehydrogenase inhibitors, including ribavirin. Antivir. Res. 2006, 71, 53–63. [Google Scholar] [CrossRef]
- Ostrowski, T.; Zeidler, J. Synthesis of 5-ethynyl-1-β-D-ribofuranosyl-1H-[1,2,3]triazole-4-carboxylic acid amide (isosteric to EICAR) and its derivatives. Nucleic Acids Symp. Ser. 2008, 52, 585–586. [Google Scholar] [CrossRef]
- Okano, Y.; Saito-Tarashima, N.; Kurosawa, M.; Iwabu, A.; Ota, M.; Watanabe, T.; Kato, F.; Hishiki, T.; Fujimuro, M.; Minakawa, N. Synthesis and biological evaluation of novel imidazole nucleosides as potential anti-dengue virus agents. Bioorg. Med. Chem. 2019, 27, 2181–2186. [Google Scholar] [CrossRef]
- Gutowski, G.E.; Sweeney, M.J.; De Long, D.C.; Hamill, R.L.; Gerzon, K.; Dyke, R.W. Biochemistry and biological effects of the pyrazofurins (pyrazomycins): Initial clinical trial. Ann. N. Y. Acad. Sci. 1975, 255, 544–551. [Google Scholar] [CrossRef]
- Cooney, D.A.; Jayaram, H.N.; Gebeyehu, G.; Betts, C.R.; Kelley, J.A.; Marquez, V.E.; Johns, D.G. The conversion of tiazofurin to an analogue of NAD with potent IMP dehydrogenase-inhibitory properties. Biochem. Pharmacol. 1982, 31, 2133–2136. [Google Scholar] [CrossRef]
- Srivastava, P.C.; Pickering, M.V.; Allen, L.B.; Streeter, D.G.; Campbell, M.T.; Witkowski, J.T.; Sidwell, R.W.; Robins, R.K. Synthesis and antiviral activity of certain thiazole C-nucleosides. J. Med. Chem. 1977, 20, 256–262. [Google Scholar] [CrossRef] [PubMed]
- Descamps, J.; De Clercq, E. Broad-spectrum antiviral activity of pyrazofurin (pyrazomycin). In Current Chemotherapy, Proceedings of the Tenth International Congress of Chemotherap, Zürich, Switzerland, 18–23 September 1977; Siegenthaler, W., Lüthy, R., Eds.; American Society for Microbiology: Washington, DC, USA, 1978; pp. 354–357. [Google Scholar]
- Kirsi, J.J.; North, J.A.; Mc Kernan, P.A.; Murray, B.K.; Canonico, P.G.; Huggins, J.W.; Srivastava, P.C.; Robins, R.K. Broad-spectrum antiviral activity of 2-β-d-ribofuranosylselenazole-4-carboxamide, a new antiviral agent. Antimicrob. Agents Chemother. 1983, 24, 353–361. [Google Scholar] [CrossRef] [PubMed]
- De Clercq, E. C-nucleosides to be revisited. J. Med. Chem. 2016, 59, 2301–2311. [Google Scholar] [CrossRef]
- Warren, T.K.; Wells, J.; Panchal, R.G.; Stuthman, K.S.; Garza, N.L.; van Tongeren, S.A.; Dong, L.; Retterer, C.J.; Eaton, B.P.; Pegoraro, G.; et al. Protection against filovirus diseases by a novel broad-spectrum nucleoside analogue BCX4430. Nature 2014, 508, 402–405. [Google Scholar] [CrossRef]
- Cho, A.; Zhang, L.; Xu, J.; Lee, R.; Butler, T.; Metobo, S.; Aktoudianakis, V.; Lew, W.; Ye, H.; Clarke, M.; et al. Discovery of the first C-nucleoside HCV polymerase inhibitor (GS-6620) with demonstrated antiviral response in HCV infected patients. J. Med. Chem. 2014, 57, 1812–1825. [Google Scholar] [CrossRef]
- Ransom, J.T. Mechanism of action of mycophenolate mofetil. Ther. Drug. Monit. 1995, 17, 681–684. [Google Scholar] [CrossRef]
- Kunio, A.; Suzuki, S.; Tamura, G.; Arima, K. Antiviral activity of mycophenolic acid. Studies on antiviral and antitumor antibiotics. J. Antibiot. 1968, 21, 649–652. [Google Scholar] [CrossRef][Green Version]
- Williams, R.H.; Lively, D.H.; De Long, D.C.; Cline, J.C.; Sweeny, M.J. Mycophenolic acid: Antiviral and antitumor properties. J. Antibiot. 1968, 21, 463–464. [Google Scholar] [CrossRef]
- Cline, J.C.; Nelson, J.; Gerzon, K.; Williams, R.H.; Delong, D.C. In vitro antiviral activity of mycophenolic acid and its reversal by guanine-type compounds. Appl. Microbiol. 1969, 18, 14–20. [Google Scholar] [CrossRef]
- Hart, B.J.; Dyall, J.; Postnikova, E.; Zhou, H.; Kindrachuk, J.; Johnson, R.F.; Olinger, G.G.; Frieman, M.B.; Holbrook, M.R.; Jahrling, P.B.; et al. Interferon-β and mycophenolic acid are potent inhibitors of Middle East respiratory syndrome coronavirus in cell-based assays. J. Gen. Virol. 2014, 95, 571–577. [Google Scholar] [CrossRef] [PubMed]
- To, K.K.W.; Mok, K.Y.; Chan, A.S.F.; Cheung, N.N.; Wang, P.; Lui, Y.M.; Chan, J.F.W.; Chen, H.; Chan, K.H.; Kao, R.Y.T.; et al. Mycophenolic acid, an immunomodulator, has potent and broad-spectrum in vitro antiviral activity against pandemic, seasonal and avian influenza viruses affecting humans. J. Gen. Virol. 2016, 97, 1807–1817. [Google Scholar] [CrossRef] [PubMed]
- Shen, L.; Niu, J.; Wang, C.; Huang, B.; Wang, W.; Zhu, N.; Deng, Y.; Wang, H.; Ye, F.; Cen, S.; et al. High-throughput screening and identification of potent broad-spectrum inhibitors of coronaviruses. J. Virol. 2019, 93, e00023-19. [Google Scholar] [CrossRef] [PubMed]
- Kato, F.; Matsuyama, S.; Kawase, M.; Hishiki, T.; Kato, H.; Takeda, M. Antiviral activities of mycophenolic acid and IMD-0354 against SARS-CoV-2. Microbiol. Immunol. 2020, 64, 635–639. [Google Scholar] [CrossRef] [PubMed]
- Barrows, N.; Campos, R.K.; Powell, S.T.; Routh, A.; Bradrick, S.S.; Garcia-Blanco, M.A. A screen of FDA-approved drugs for inhibitors of Zika virus infection. Cell Host Microbe 2016, 20, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Yin, Y.; Wang, Y.; Dang, W.; Xu, L.; Su, J.; Zhou, X.; Wang, W.; Felczak, K.; van der Laan, L.J.W.; Pankiewicz, K.W.; et al. Mycophenolic acid potently inhibits rotavirus infection with a high barrier to resistance development. Antivir. Res. 2016, 133, 41–49. [Google Scholar] [CrossRef]
- Uematsu, J.; Sakai-Sugino, K.; Kihira-Nakanishi, S.; Yamamoto, H.; Hirai, K.; Kawano, M.; Nishio, M.; Tsurudome, M.; O’Brien, M.; Komada, H. Inhibitions of human parainfluenza virus type 2 replication by ribavirin and mycophenolate mofetil are restored by guanosine and S-(4-nitrobenzyl)-6-thioinosine. Drug Discov. Ther. 2019, 13, 314–321. [Google Scholar] [CrossRef]
- Sampey, G.; Iordanskiy, S.; Pleet, M.L.; De Marino, C.; Romerio, F.; Mahieux, R.; Kashanchi, F. Identification of modulators of HIV-1 proviral transcription from a library of FDA-approved pharmaceuticals. Viruses 2020, 23, 1067. [Google Scholar] [CrossRef]
- Tang, H.; Liu, Y.; Ren, R.; Liu, Y.; He, Y.; Qi, Z.; Peng, H.; Zhao, P. Identification of clinical candidates against West Nile virus by activity screening in vitro and effect evaluation in vivo. J. Med. Virol. 2022. [Google Scholar] [CrossRef]
- Park, J.-G.; Ávila-Pérez, G.; Nogales, A.; Blanco-Lobo, P.; de la Torre, J.C.; Martínez-Sobrido, L. Identification and characterization of novel compounds with broad-spectrum antiviral activity against influenza A and B viruses. J Virol. 2020, 94, e02149-19. [Google Scholar] [CrossRef]
- Wu, J.; Liu, Q.; Xie, H.; Chen, R.; Huang, W.; Liang, C.; Xiao, X.; Yu, Y.; Wang, Y. Screening and evaluation of potential inhibitors against vaccinia virus from 767 approved drugs. J. Med. Virol. 2019, 91, 2016–2024. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, X.; Debing, Y.; Chen, K.; van der Laan, L.J.W.; Neyts, J.; Janssen, H.L.; Metselaar, H.J.; Peppelenbosch, M.P.; Pan, Q. Calcineurin inhibitors stimulate and mycophenolic acid inhibits replication of hepatitis E virus. Gastroenterology 2014, 146, 1775–1783. [Google Scholar] [CrossRef] [PubMed]
- Takebe, N.; Cheng, X.; Bauer, K.; Tricot, G.; Karp, J.; Ross, D. Induction of apoptosis in multiple myeloma (MM) cell lines using mycophenolate mofetil (Cellcept). Blood 2001, 98, 312B. [Google Scholar] [CrossRef]
- Fang, S.; Su, J.; Liang, B.; Li, X.; Li, Y.; Jiang, J.; Huang, J.; Zhou, B.; Ning, C.; Li, J.; et al. Suppression of autophagy by mycophenolic acid contributes to inhibition of HCV replication in human hepatoma cells. Sci. Rep. 2017, 7, 44039. [Google Scholar] [CrossRef] [PubMed]
- Manchala, N.R.; Dungdung, R.; Trivedi, P.; Unniyampurath, U.; Pilankatta, R. Mycophenolic acid (MPA) modulates host cellular autophagy progression in sub genomic dengue virus-2 replicon cells. Microb. Pathog. 2019, 137, 103762. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.-W.; Cheng, S.-C.; Chen, W.-Y.; Lin, M.-H.; Chuang, S.-J.; Cheng, I.-H.; Sun, C.-Y.; Chou, C.-Y. Thiopurine analogs and mycophenolic acid synergistically inhibit the papain-like protease of Middle East respiratory syndrome coronavirus. Antivir. Res. 2015, 115, 9–16. [Google Scholar] [CrossRef]
- Hirunsatitpron, P.; Hanprasertpong, N.; Noppakun, K.; Pruksakorn, D.; Teekachunhatean, S.; Koonrungsesomboon, N. Mycophenolic acid and cancer risk in solid organ transplant recipients: Systematic review and meta-analysis. Br. J. Clin. Pharmacol. 2022, 88, 476–489. [Google Scholar] [CrossRef] [PubMed]
- Allison, A.C. Mechanisms of action of mycophenolate mofetil. Lupus 2005, 14, 2–8. [Google Scholar] [CrossRef]
- Borroto-Esoda, K.; Myrick, F.; Feng, J.; Jeffrey, J.; Furman, P. In vitro combination of amdoxovir and the inosine monophosphate dehydrogenase inhibitors mycophenolic acid and ribavirin demonstrates potent activity against wild-type and drug-resistant variants of human immunodeficiency virus type 1. Antimicrob. Agents Chemother. 2004, 48, 4387–4394. [Google Scholar] [CrossRef]
- Pan, Q.; de Ruiter, P.E.; Metselaar, H.J.; Kwekkeboom, J.; de Jonge, J.; Tilanus, H.W.; Janssen, H.L.A.; van der Laan, L.J.W. Mycophenolic acid augments interferon-stimulated gene expression and inhibits hepatitis C Virus infection in vitro and in vivo. Hepatology 2012, 55, 1673–1683. [Google Scholar] [CrossRef]
- Ying, C.; De Clercq, E.; Neyts, J. Ribavirin and mycophenolic acid potentiate the activity of guanine- and diaminopurine-based nucleoside analogues against hepatitis B virus. Antivir. Res. 2000, 48, 117–124. [Google Scholar] [CrossRef]
- Sebastian, L.; Desai, A.; Yogeeswari, P.; Sriram, D.; Madhusudana, S.N.; Ravi, V. Combination of N-methylisatin-β-thiosemicarbazone derivative (SCH16) with ribavirin and mycophenolic acid potentiates the antiviral activity of SCH16 against Japanese encephalitis virus in vitro. Lett. Appl. Microbiol. 2012, 55, 234–239. [Google Scholar] [CrossRef] [PubMed]
- Ruiter, P.; Gadjradj, Y.; de Knegt, R.J.; Metselaar, H.J.; Ijzermans, J.N.; van der Laan, L.J. Interaction of immunosuppressants with HCV antivirals daclatasvir and asunaprevir: Combined effects with mycophenolic acid. World J. Transpl. 2018, 8, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Minjae, K.; Hwang, S.; Ahn, C.-S.; Moon, D.-B.; Ha, T.-Y.; Song, G.-W.; Jung, D.-H.; Park, G.-C.; Kim, K.-H.; Namgoong, J.-M.; et al. Twenty-year longitudinal follow-up after liver transplantation: A single-center experience with 251 consecutive patients. Korean J. Transplant. 2022, 36, 45–53. [Google Scholar] [CrossRef]
- Favi, E.; Signorini, L.; Villani, S.; Dolci, M.; Ticozzi, R.; Basile, G.; Ferrante, P.; Ferraresso, M.; Delbue, S. In vitro study evaluating the effect of different immunosuppressive agents on human polyomavirus BK replication. Transplant. Proc. 2022, in press. [CrossRef]
- Sintchak, M.D.; Nimmesgern, E. The structure of inosine 5X-monophosphate dehydrogenase and the design of novel inhibitors. Immunopharmacology 2000, 47, 163–184. [Google Scholar] [CrossRef]
- Markland, W.; Mc Quaid, T.J.; Jain, J.; Kwong, A.D. Broad-spectrum antiviral activity of the IMP dehydrogenase inhibitor VX-497: A comparison with ribavirin and demonstration of antiviral additivity with alpha interferon. Antimicrob. Agents Chemother. 2000, 44, 859–866. [Google Scholar] [CrossRef] [PubMed]
- Rustgi, V.K.; Lee, W.M.; Lawitz, E.; Gordon, S.C.; Afdhal, N.; Poordad, F.; Bonkovsky, H.L.; Bengtsson, L.; Chandorkar, G.; Harding, M.; et al. Merimepodib triple combination study group. Merimepodib, pegylated interferon, and ribavirin in genotype 1 chronic hepatitis C pegylated interferon and ribavirin nonresponders. Hepatology 2009, 50, 1719–1726. [Google Scholar] [CrossRef]
- Tong, X.; Smith, J.; Bukreyeva, N.; Koma, T.; Manning, J.T.; Kalkeri, R.; Kwong, A.D.; Paessler, S. Merimepodib, an IMPDH inhibitor, suppresses replication of Zika virus and other emerging viral pathogens. Antivir. Res. 2018, 149, 34–40. [Google Scholar] [CrossRef]
- Cholewiński, G.; Iwaszkiewicz-Grześ, D.; Prejs, M.; Głowacka, A.; Dzierzbicka, K. Synthesis of the inosine 5′-monophosphate dehydrogenase (IMPDH) inhibitors. J. Enzyme Inhib. Med. Chem. 2015, 30, 550–563. [Google Scholar] [CrossRef] [PubMed]
- Shah, C.P.; Kharkar, P.S. Inosine 5′-monophosphate dehydrogenase inhibitors as antimicrobial agents: Recent progress and future perspectives. Future Med. Chem. 2015, 7, 1415–1429. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.-J.; Sun, W.-F.; Zhong, Z.-J.; Gao, R.-M.; Yi, H.; Li, Y.-H.; Peng, Z.-G.; Li, Z.-R. Synthesis and broad-spectrum antiviral activity of some novel benzo-heterocyclic amine compounds. Molecules 2014, 19, 925–939. [Google Scholar] [CrossRef]
- Hu, J.; Ma, L.; Wang, H.; Yan, H.; Zhang, D.; Li, Z.; Jiang, J.; Li, Y. A novel benzo-heterocyclic amine derivative N30 inhibits influenza virus replication by depression of Inosine-5′-monophospate dehydrogenase activity. Virol. J. 2017, 14, 55. [Google Scholar] [CrossRef]
- Kumarasamy, D.; Roy, B.G.; Rocha-Pereira, J.; Neyts, J.; Nanjappan, S.; Maity, S.; Mookerjee, M.; Naesens, L. Synthesis and in vitro antiviral evaluation of 4-substituted 3,4-dihydropyrimidinones. Bioorg. Med. Chem. Lett. 2017, 27, 139–142. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, J.M.; Harding, M.W.; Genna, T.; Bol, D.K. A phase I dose-ranging study of the pharmacokinetics, pharmacodynamics, safety, and tolerability of AVN944, an IMPDH Inhibitor, in healthy male volunteers. J. Clin. Pharmacol. 2009, 49, 30–38. [Google Scholar] [CrossRef]
- Dunhama, E.C.; Leskeb, A.; Shiffletta, K.; Watta, A.; Feldmanna, H.; Hoenena, T.; Grosetha, A. Lifecycle modelling systems support inosine monophosphate dehydrogenase (IMPDH) as a pro-viral factor and antiviral target for New World arenaviruses. Antivir. Res. 2018, 157, 140–150. [Google Scholar] [CrossRef]
- Vanderlinden, E.; Marchand, A.; Van Berwaer, R.; Van Dam, W.; Arzel, P.; Klaassen, H.; Persoons, L.; Chaltin, P.; Naesens, L. A broad influenza virus inhibitor acting via IMP dehydrogenase and in synergism with ribavirin. Antivir. Res. 2021, 196, 105208. [Google Scholar] [CrossRef]
- Juvale, K.; Shaik, A.; Kirubakaran, S. Inhibitors of inosine 5′-monophosphate dehydrogenase as emerging new generation antimicrobial agents. Med. Chem. Commun. 2019, 10, 1290. [Google Scholar] [CrossRef]

| DHODH Inhibitor | Viral Model | Efficacy [EC50] | Ref. | DHODH Inhibitor | Viral Model | Efficacy [EC50] | Ref. | |
|---|---|---|---|---|---|---|---|---|
| Leflunomide | BKV | 40 µg/mL | [54] | Vidofludimus calcium (IMU-838) | SARS-CoV-2 | 3.2–7.6 µM | [77] | |
| EBV | 10 µg/mL | [55] | HCMV | 7.4 µM | [77] | |||
| SARS-CoV-2 | 41.49–48.98 µM | [34] | HCV | 4.5 µM | [77] | |||
| HIV-1 | 2.1 µM | [77] | ||||||
| Teriflunomide (A77 1726) | HCMV | 40–60 µM | [53] | A3 | IAV | 170 nM | [78] | |
| JUNV | 16–45 µM | [57] | HIV-1 | 205 nM | [78] | |||
| IAV | 2.73–35.02 µM | [58] | HCV | <2 µM | [78] | |||
| ZIKV | 17.72 µM | [34] | AdV-5 | <2 µM | [78] | |||
| EBOV | 3.41 µM | [34] | LCMV | 82 nM | [79] | |||
| SARS-CoV-2 | 6.0–26.06 µM | [34] | JUNV | <1 µM | [79] | |||
| Brequinar | WNV | 3 µM | [66] | FA-613 | IAV | 3.8 µM | [30] | |
| YFV | 3 µM | [66] | IBV | 0.2 µM | [30] | |||
| DENV | 78 nM | [66] | EA71 | 8.6 µM | [30] | |||
| ZIKV | 0.3–1.51 µM | [20] | RSV | 10.1 µM | [30] | |||
| EBOV | 0.1 µM | [34] | MERS-CoV | >30 µM | [30] | |||
| HCMV | 17 nM | [67] | SARS-CoV-1 | >30 µM | [30] | |||
| IAV | 0.24 µM | [34] | Rhinovirus AV | 9.7 µM | [30] | |||
| EV71 | 82.40 nM | [68] | MEDS433 | HSV-1 | 78–116 nM | [83] | ||
| EV70 | 29.26 nM | [68] | HSV-2 | 61–95 nM | [83] | |||
| CVB3 | 35.14 nM | [68] | HCoV-OC43 | 12 nM | [84] | |||
| RV | 49.17 nM | [69] | HCoV-229E | 22 nM | [84] | |||
| SARS-CoV-2 | 0.06–0.794 µM | [34,70] | SARS-CoV-2 | 63–76 nM | [84] | |||
| S312/S416 | IAV | 0.013–13.7 µM | [34] | |||||
| ZIKV | 0.019–1.24 µM | [34] | RYL-634 | HCV | 1.56 µM | [85] | ||
| EBOV | 0.018–11.39 µM | [34] | DENV | 7 nM | [85] | |||
| SARS-CoV-2 | 0.014–1.59 µM | [34] | ZIKV | 20 nM | [85] | |||
| Emvododstat (PTC299) | SARS-CoV-2 | 2.0–31.6 nM | [35] | EV71 | 4 nM | [85] | ||
| EBOV | 9.1 nM | [35] | HIV | 13 nM | [85] | |||
| PV | 0.57 nM | [35] | IAV | <1 µM | [85] | |||
| HCV | 36 nM | [35] | AR-12 derivatives | DENV | 62.2–68 nM | [87] | ||
| RVFV | 13 nM | [35] | HIV | 118.6–130.3 nM | [87] | |||
| BAY2402234 | SARS-CoV-2 | 5–11nM | [70] | IAV | 53.2–56.1 nM | [87] | ||
| GSK983/SW835 | EBOV | 0.009–1 µM | [31] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sepúlveda, C.S.; García, C.C.; Damonte, E.B. Inhibitors of Nucleotide Biosynthesis as Candidates for a Wide Spectrum of Antiviral Chemotherapy. Microorganisms 2022, 10, 1631. https://doi.org/10.3390/microorganisms10081631
Sepúlveda CS, García CC, Damonte EB. Inhibitors of Nucleotide Biosynthesis as Candidates for a Wide Spectrum of Antiviral Chemotherapy. Microorganisms. 2022; 10(8):1631. https://doi.org/10.3390/microorganisms10081631
Chicago/Turabian StyleSepúlveda, Claudia Soledad, Cybele Carina García, and Elsa Beatriz Damonte. 2022. "Inhibitors of Nucleotide Biosynthesis as Candidates for a Wide Spectrum of Antiviral Chemotherapy" Microorganisms 10, no. 8: 1631. https://doi.org/10.3390/microorganisms10081631
APA StyleSepúlveda, C. S., García, C. C., & Damonte, E. B. (2022). Inhibitors of Nucleotide Biosynthesis as Candidates for a Wide Spectrum of Antiviral Chemotherapy. Microorganisms, 10(8), 1631. https://doi.org/10.3390/microorganisms10081631






