Metagenome-Assembled Genome of Cyanocohniella sp. LLY from the Cyanosphere of Llayta, an Edible Andean Cyanobacterial Macrocolony
Abstract
:1. Introduction
2. Materials and Methods
3. Results
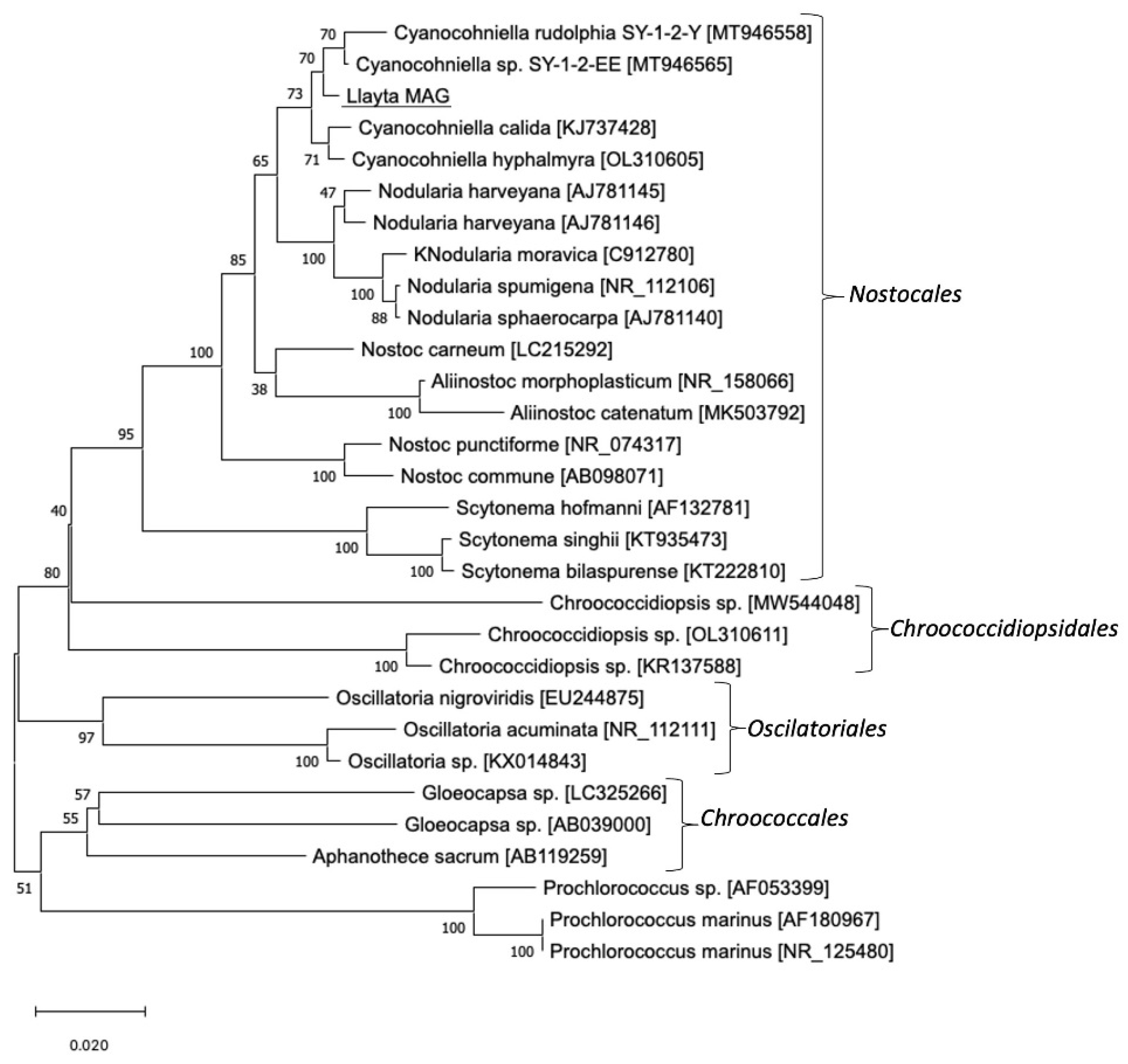
3.1. Genome Assembly and Phylogeny of Cyanocohniella sp. LLY
3.2. Functional Capabilities of Cyanocohniella sp. LLY
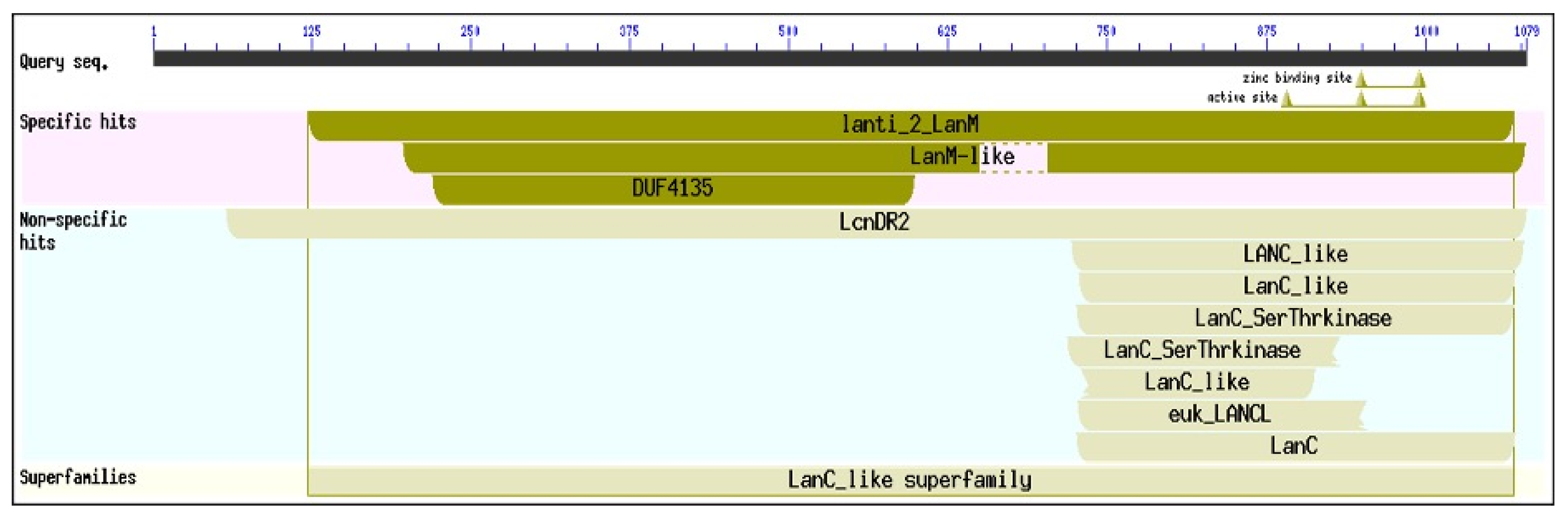
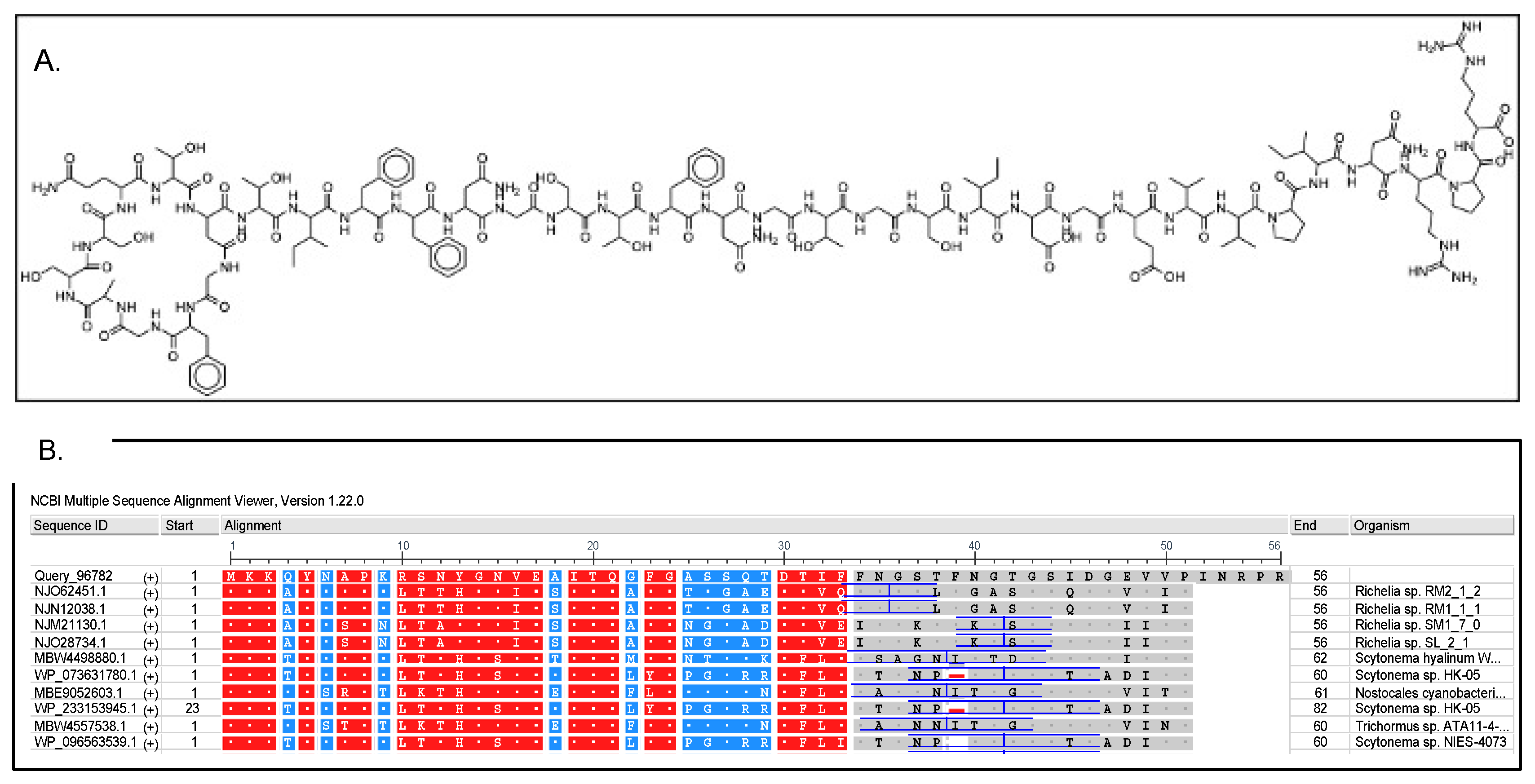
3.3. Secondary Metabolism of Cyanocohniella sp. LLY
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Aaronson, S. A role for algae as human food in antiquity. Food Foodways 1986, 1, 311–315. [Google Scholar] [CrossRef] [PubMed]
- Gao, K. Chinese studies on the edible blue-green alga, Nostoc flagelliforme: A review. J. Appl. Phycol. 1998, 10, 37–49. [Google Scholar] [CrossRef]
- Qiu, B.; Liu, J.; Liu, Z.; Liu, S. Distribution and ecology of the edible cyanobacterium Ge-Xian-Mi (Nostoc) in rice fields of Hefeng County in China. J. Appl. Phycol. 2002, 14, 423–429. [Google Scholar] [CrossRef]
- Becker, E. Micro-algae as a source of protein. Biotechnol. Adv. 2007, 25, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Gantar, M.; Svirčev, Z. Microalgae and cyanobacteria: Food for thought. J. Phycol. 2008, 44, 260–268. [Google Scholar] [CrossRef] [PubMed]
- Becker, E.W. Microalgae for Human and Animal Nutrition. In Handbook of Microalgal Culture: Biotechnology and Applied Phycology; Richmond, A., Ed.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2013; pp. 461–503. [Google Scholar] [CrossRef]
- Wells, M.L.; Potin, P.; Craigie, J.S.; Raven, J.A.; Merchant, S.S.; Helliwell, K.E.; Smith, A.G.; Camire, M.E.; Brawley, S.H. Algae as nutritional and functional food sources: Revisiting our understanding. J. Appl. Phycol. 2017, 29, 949–982. [Google Scholar] [CrossRef]
- Nandagopal, P.; Steven, A.N.; Chan, L.-W.; Rahmat, Z.; Jamaluddin, H.; Noh, N.I.M. Bioactive Metabolites Produced by Cyanobacteria for Growth Adaptation and Their Pharmacological Properties. Biology 2021, 10, 1061. [Google Scholar] [CrossRef]
- Vargas, M.A.; Moreno, J.; Olivares, H.; Del Campo, J.A.; Rodriguez, H.; Rivas, J.; Guerrero, M.G. Biochemical composition and fatty acid content of filamentous nitrogen-fixing cyanobacteria. J. Phycol. 1998, 34, 812–817. [Google Scholar] [CrossRef]
- Galetović, A.; Araya, J.; Gómez-Silva, B. Composición bioquímica y toxicidad de colonias comestibles de la cianobacteria andina Nostoc sp. Llayta. Rev. Chil. Nutr. 2017, 44, 360–370. [Google Scholar] [CrossRef] [Green Version]
- Guedes, A.C.; Amaro, H.M.; Malcata, F.X. Microalgae as Sources of Carotenoids. Mar. Drugs 2011, 9, 625–644. [Google Scholar] [CrossRef] [PubMed]
- Galetović, A.; Seura, F.; Gallardo, V.; Graves, R.; Cortés, J.; Valdivia, C.; Núñez, J.; Tapia, C.; Neira, I.; Sanzana, S.; et al. Use of Phycobiliproteins from Atacama Cyanobacteria as Food Colorants in a Dairy Beverage Prototype. Foods 2020, 9, 244. [Google Scholar] [CrossRef] [Green Version]
- Bull, A.T.; Asenjo, J.A.; Goodfellow, M.; Gómez-Silva, B. The Atacama Desert: Technical Resources and the Growing Importance of Novel Microbial Diversity. Annu. Rev. Microbiol. 2016, 70, 215–234. [Google Scholar] [CrossRef] [Green Version]
- Gómez-Silva, B. Lithobiontic life: Atacama rocks are well and alive. Antoine Van Leeuwenhoek 2018, 111, 1333–1345. [Google Scholar] [CrossRef] [PubMed]
- Houston, J. Variability of precipitation in the Atacama Desert: Its causes and hydrological impact. Int. J. Clim. 2006, 26, 2181–2198. [Google Scholar] [CrossRef]
- Rivera, M.; Galetović, A.; Licuime, R.; Gómez-Silva, B. A Microethnographic and Ethnobotanical Approach to Llayta Consumption Among Andes Feeding Practices. Foods 2018, 7, 202. [Google Scholar] [CrossRef] [Green Version]
- Villagrán, C.; Castro, V.; Sánchez, G.; Hinojosa, F.; Latorre, C. The Altiplano tradition: Ethnobotanic study at the Iquique Andes, I. Region, Chile. Chungara 1999, 31, 81–186. [Google Scholar]
- Pardo, O.; Pizarro, J.L. Chile: Pre-Hispanic Edible Plants; Ediciones Parina: Arica, Chile, 2003. [Google Scholar]
- Gómez-Silva, B.; Mendizabal, I.; Tapia, I.; Olivares, H. Microalgas del norte de Chile. IV. Composición química de Nostoc commune Llaita. Rev. Invest. Cient. Tecnol. Cs. 1994, 3, 19–25. Available online: http://bioquimica.uantof.cl/files/Publicaciones/1994%20Rev%20Invest%20%20Nostoc%20Llaita25.pdf (accessed on 11 October 2021).
- Bertonio, L. Vocabulario de la Lengua Aymara; Colección Biblioteca Nacional de Chile: Impreso en la Compañía de Jesús, Perú, 1612; Electronic Document; Available online: http://www.memoriachilena.cl/602/w3-article-8656.html (accessed on 11 October 2021).
- Lagerheim, M.G. La Yuyucha. La Nuova Notarisia 1892, 3, 1376–1377. Available online: https://ia601307.us.archive.org/23/items/lanotarisiacomme7293levi/lanotarisiacomme7293levi.pdf (accessed on 11 October 2021).
- Mannheim, B. Poetic form in Guaman Poma’s Wariqsa Arawi. Amerindian. Am. Indian Q. 1986, 11, 41–67. Available online: https://sedyl.cnrs.fr/amerindia/articles/pdf/A_11_03.pdf (accessed on 11 October 2021).
- Galetović, A.; Azevedo, J.; Castelo-Branco, R.; Oliveira, F.; Gómez-Silva, B.; Vasconcelos, V. Absence of Cyanotoxins in Llayta, Edible Nostocaceae Colonies from the Andes Highlands. Toxins 2020, 12, 382. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Guo, M. Healthy efficacy of Nostoc commune Vaucher. Oncotarget 2018, 9, 14669–14679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niccolai, A.; Bigagli, E.; Biondi, N.; Rodolfi, L.; Cinci, L.; Luceri, C.; Tredici, M.R. In vitro toxicity of microalgal and cyanobacterial strains of interest as food source. J. Appl. Phycol. 2017, 29, 199–209. [Google Scholar] [CrossRef]
- Dodds, W.; Gudder, D.A.; Mollenhauer, D. The ecology of Nostoc. J. Phycol. 1995, 31, 2–18. [Google Scholar] [CrossRef]
- Secker, N.H.; Chua, J.P.S.; Laurie, R.E.; McNoe, L.; Guy, P.; Orlovich, D.A.; Summerfield, T.C. Characterization of the cyanobacteria and associated bacterial community from an ephemeral wetland in New Zealand. J. Phycol. 2016, 52, 761–773. [Google Scholar] [CrossRef] [PubMed]
- Aguilar, P.; Dorador, C.; Vila, I.; Sommaruga, R. Bacterial Communities Associated with Spherical Nostoc Macrocolonies. Front. Microbiol. 2019, 10, 483. [Google Scholar] [CrossRef] [PubMed]
- Satjarak, A.; Graham, L.E.; Piotrowski, M.J.; Trest, M.T.; Wilcox, L.W.; Cook, M.E.; Knack, J.J.; Arancibia-Avila, P. Shotgun metagenomics and microscopy indicate diverse cyanophytes, other bacteria, and microeukaryotes in the epimicrobiota of a northern Chilean wetland Nostoc (cyanobacteria). J. Phycol. 2021, 57, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Arnon, D.I.; McSwain, B.D.; Tsujimoto, H.Y.; Wada, K. Photochemical activity and components of membrane preparations from blue-green algae. I. Coexistence of two photosystems in relation to chlorophyll a and removal of phycocyanin. Biochim. Biophys. Acta 1974, 357, 231–245. [Google Scholar] [CrossRef]
- Li, D.; Luo, R.; Liu, C.-M.; Leung, C.-M.; Ting, H.-F.; Sadakane, K.; Yamashita, H.; Lam, T.-W. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 2016, 102, 3–11. [Google Scholar] [CrossRef]
- Wattam, A.R.; Abraham, D.; Dalay, O.; Disz, T.L.; Driscoll, T.; Gabbard, J.L.; Gillespie, J.J.; Gough, R.; Hix, D.; Kenyon, R.; et al. PATRIC, the bacterial bioinformatics database and analysis resource. Nucleic Acids Res. 2014, 42, D581–D591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 75. [Google Scholar] [CrossRef] [Green Version]
- Skinnider, M.A.; Merwin, N.J.; Johnston, C.W.; Magarvey, N.A. PRISM 3: Expanded prediction of natural product chemical structures from microbial genomes. Nucleic Acids Res. 2017, 45, W49–W54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blin, K.; Shaw, S.; Kloosterman, A.M.; Charlop-Powers, Z.; van Wezel, G.P.; Medema, M.H.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef] [PubMed]
- Cordero, R.R.; Damiani, A.; Jorquera, J.; Sepúlveda, E.; Caballero, M.; Fernandez, S.; Feron, S.; Llanillo, P.J.; Carrasco, J.; Laroze, D.; et al. Ultraviolet radiation in the Atacama Desert. Antonie Van Leeuwenhoek 2018, 111, 1301–1313. [Google Scholar] [CrossRef] [PubMed]
- Garneau, J.E.; Dupuis, M.-È.; Villion, M.; Romero, D.A.; Barrangou, R.; Boyaval, P.; Fremaux, C.; Horvath, P.; Magadáán, A.H.; Moineau, S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 2010, 468, 67–71. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, N.; Wolf, Y.I.; Makarova, K.S.; Koonin, E.V. Nature and Intensity of Selection Pressure on CRISPR-Associated Genes. J. Bacteriol. 2012, 194, 1216–1225. [Google Scholar] [CrossRef] [Green Version]
- Marraffini, L.A. CRISPR-Cas immunity in prokaryotes. Nature 2015, 526, 55–61. [Google Scholar] [CrossRef]
- Grissa, I.; Vergnaud, G.; Pourcel, C. The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinform. 2007, 8, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Shmakov, S.A.; Sitnik, V.; Makarova, K.S.; Wolf, Y.; Severinov, K.V.; Koonin, E.V. The CRISPR Spacer Space Is Dominated by Sequences from Species-Specific Mobilomes. mBio 2017, 8, e01397-17. [Google Scholar] [CrossRef] [Green Version]
- Shao, Y.; Cocozaki, A.I.; Ramia, N.F.; Terns, R.M.; Terns, M.P.; Li, H. Structure of the Cmr2-Cmr3 Subcomplex of the Cmr RNA Silencing Complex. Structure 2013, 21, 376–384. [Google Scholar] [CrossRef] [Green Version]
- Jungblut, A.D.; Raymond, F.; Dion, M.B.; Moineau, S.; Mohit, V.; Nguyen, G.Q.; Déraspe, M.; Francovic-Fontaine, C.; Lovejoy, C.; Culley, A.I.; et al. Genomic diversity and CRISPR-Cas systems in the cyanobacterium Nostoc in the High Arctic. Environ. Microbiol. 2021, 23, 2955–2968. [Google Scholar] [CrossRef]
- Albarracin, V.H.; Kurth, D.; Ordoñez, O.F.; Belfiore, C.; Luccini, E.; Salum, G.; Piacentini, R.D.; Farias, M.E. High-Up: A Remote Reservoir of Microbial Extremophiles in Central Andean Wetlands. Front. Microbiol. 2015, 6, 1404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vilo, C.; Galetovic, A.; Araya, J.E.; Gómez-Silva, B.; Dong, Q. Draft Genome Sequence of a Bacillus Bacterium from the Atacama Desert Wetlands Metagenome. Genome Announc. 2015, 3, e00955-15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaštovský, J.; Gómez, E.B.; Hladil, J.; Johansen, J.R. Cyanocohniella calida gen. et sp. nov. (Cyanobacteria: Aphanizomenonaceae) a new cyanobacterium from the thermal springs from Karlovy Vary, Czech Republic. Phytotaxa 2014, 181, 279. [Google Scholar] [CrossRef]
- Jung, P.; Mikhailyuk, T.; Emrich, D.; Baumann, K.; Dultz, S.; Büdel, B. Shifting Boundaries: Ecological and Geographical Range extension Based on Three New Species in the Cyanobacterial Genera Cyanocohniella, Oculatella, and, Aliterella. J. Phycol. 2020, 56, 1216–1231. [Google Scholar] [CrossRef] [PubMed]
- Panou, M.; Gkelis, S. Unravelling unknown cyanobacteria diversity linked with HCN production. Mol. Phylogenet. Evol. 2022, 166, 107322. [Google Scholar] [CrossRef] [PubMed]
- Sommer, V.; Mikhailyuk, T.; Glaser, K.; Karsten, U. Uncovering Unique Green Algae and Cyanobacteria Isolated from Biocrusts in Highly Saline Potash Tailing Pile Habitats, Using an Integrative Approach. Microorganisms 2020, 8, 1667. [Google Scholar] [CrossRef]
- Jung, P.; Sommer, V.; Karsten, U.; Lakatos, M. Salty Twins: Salt-Tolerance of Terrestrial Cyanocohniella Strains (Cyanobacteria) and Description of C. rudolphia sp. nov. Point towards a Marine Origin of the Genus and Terrestrial Long Distance Dispersal Patterns. Microorganisms 2022, 10, 968. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Pichel, F.; Castenholz, R.W. Occurrence of UV-Absorbing, Mycosporine-Like Compounds among Cyanobacterial Isolates and an Estimate of Their Screening Capacity. Appl. Environ. Microbiol. 1993, 59, 163–169. [Google Scholar] [CrossRef] [Green Version]
- Tietz, J.I.; Schwalen, C.J.; Patel, P.S.; Maxson, T.; Blair, P.M.; Tai, H.-C.; Zakai, U.I.; Mitchell, D.A. A new genome-mining tool redefines the lasso peptide biosynthetic landscape. Nat. Chem. Biol. 2017, 13, 470–478. [Google Scholar] [CrossRef]
- Martin-Gómez, H.; Tulla-Puche, J. Lasso peptides: Chemical approaches and structural elucidation. Org. Biomol. Chem. 2018, 16, 5065–5080. [Google Scholar] [CrossRef]
- Mevaere, J.; Goulard, C.; Schneider, O.; Sekurova, O.N.; Ma, H.; Zirah, S.; Afonso, C.; Rebuffat, S.; Zotchev, S.B.; Li, Y. An orthogonal system for heterologous expression of actinobacterial lasso peptides in Streptomyces hosts. Sci. Rep. 2018, 8, 8232. [Google Scholar] [CrossRef]
- Fei, X.; Yin, X.; Zhang, L.; Zabriskie, T.M. Roles of VioG and VioQ in the Incorporation and Modification of the Capreomycidine Residue in the Peptide Antibiotic Viomycin. J. Nat. Prod. 2007, 70, 618–622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ziemert, N.; Ishida, K.; Weiz, A.; Hertweck, C.; Dittmann, E. Exploiting the Natural Diversity of Microviridin Gene Clusters for Discovery of Novel Tricyclic Depsipeptides. Appl. Environ. Microbiol. 2010, 76, 3568–3574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sorek, R.; Lawrence, C.M.; Wiedenheft, B. CRISPR-Mediated Adaptive Immune Systems in Bacteria and Archaea. Annu. Rev. Biochem. 2013, 82, 237–266. [Google Scholar] [CrossRef] [Green Version]
- Komárek, J.; Kaštovský, J.; Mareš, J.; Johansen, J.R. Taxonomic classification of cyanoprokaryotes (cyanobacterial genera) 2014, using a polyphasic approach. Preslia 2014, 86, 295–335. [Google Scholar]
- Gómez-Silva, B.; Batista-García, R.A. The Atacama Desert: A Biodiversity Hotspot and Not Just a Mineral-Rich Region. Front. Microbiol. 2022, 13, 812842. [Google Scholar] [CrossRef]
| ORF | Description Based on Subsystem Annotations | Accession | Identity (%) |
|---|---|---|---|
| Orf 3147 | Demethyl 4-deoxygadusol synthase MysA | WP_069074324.1 | 93 |
| Orf 3148 | O-methyltransferase MysB | BBC27542.1 | 85 |
| Orf 3149 | ATP-grasp ligase forming mycosporine-glycine, MysC | BBC27543.1 | 82 |
| Orf 4592 | Scytonemin biosynthesis protein ScyA | WP_206262883.1 | 83 |
| Orf 4593 | Tryptophan dehydrogenase ScyB | WP_086764771.1 | 84 |
| ORF | Description Based on Subsystem Annotations | Accession | Identity (%) |
|---|---|---|---|
| Orf 2275 | CRISPR-associated RAMP Cmr2 | WP_179048547.1 | 92 |
| Orf 2276 | CRISPR-associated RAMP Cmr3 | WP_102220820.1 | 95 |
| Orf 2277 | CRISPR-associated RAMP Cmr4 | WP_179048549.1 | 94 |
| Orf 2278 | CRISPR-associated RAMP Cmr5 | WP_102220822.1 | 95 |
| Orf 2279 | CRISPR-associated RAMP Cmr6 | WP_179048551.1 | 78 |
| Orf 2288 | CRISPR-associated protein Cas1 | WP_218653184.1 | 94 |
| Orf 2289 | CRISPR-associated protein Cas2 | WP_179048556.1 | 94 |
| Orf 3613 | CRISPR-associated negative autoregulator Cas7/Cst2 | WP_194144297.1 | 94 |
| Orf 3614 | CRISPR-associated protein Cas5 | MBW4428053.1 | 96 |
| Orf 5125 | CRISPR-associated protein Cas2 | WP_190898341.1 | 95 |
| Orf 5126 | CRISPR-associated protein Cas1 | MBN3899475.1 | 93 |
| Orf 5227 | CRISPR-associated RecB family exonuclease Cas4 | MBW4675378.1 | 91 |
| Orf 5128 | CRISPR-associated endoribonuclease Cas6 | WP_096682797.1 | 94 |
| Orf 5137 | CRISPR-associated helicase Cas3 | WP_179075640.1 | 97 |
| Orf 5452 | CRISPR-associated RAMP Cmr2 | MBD2365142.1 | 92 |
| Orf 5453 | CRISPR-associated RAMP Cmr3 | WP_190709980.1 | 95 |
| Orf 5454 | CRISPR-associated RAMP Cmr4 | WP_190709978.1 | 93 |
| Orf 5455 | CRISPR-associated RAMP Cmr5 | WP_190709976.1 | 95 |
| Orf 5456 | CRISPR-associated RAMP Cmr6 | WP_190709975.1 | 91 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vilo, C.; Dong, Q.; Galetovic, A.; Gómez-Silva, B. Metagenome-Assembled Genome of Cyanocohniella sp. LLY from the Cyanosphere of Llayta, an Edible Andean Cyanobacterial Macrocolony. Microorganisms 2022, 10, 1517. https://doi.org/10.3390/microorganisms10081517
Vilo C, Dong Q, Galetovic A, Gómez-Silva B. Metagenome-Assembled Genome of Cyanocohniella sp. LLY from the Cyanosphere of Llayta, an Edible Andean Cyanobacterial Macrocolony. Microorganisms. 2022; 10(8):1517. https://doi.org/10.3390/microorganisms10081517
Chicago/Turabian StyleVilo, Claudia, Qunfeng Dong, Alexandra Galetovic, and Benito Gómez-Silva. 2022. "Metagenome-Assembled Genome of Cyanocohniella sp. LLY from the Cyanosphere of Llayta, an Edible Andean Cyanobacterial Macrocolony" Microorganisms 10, no. 8: 1517. https://doi.org/10.3390/microorganisms10081517
APA StyleVilo, C., Dong, Q., Galetovic, A., & Gómez-Silva, B. (2022). Metagenome-Assembled Genome of Cyanocohniella sp. LLY from the Cyanosphere of Llayta, an Edible Andean Cyanobacterial Macrocolony. Microorganisms, 10(8), 1517. https://doi.org/10.3390/microorganisms10081517






