Development of a Novel Peptide Nucleic Acid Probe for the Detection of Legionella spp. in Water Samples
Abstract
1. Introduction
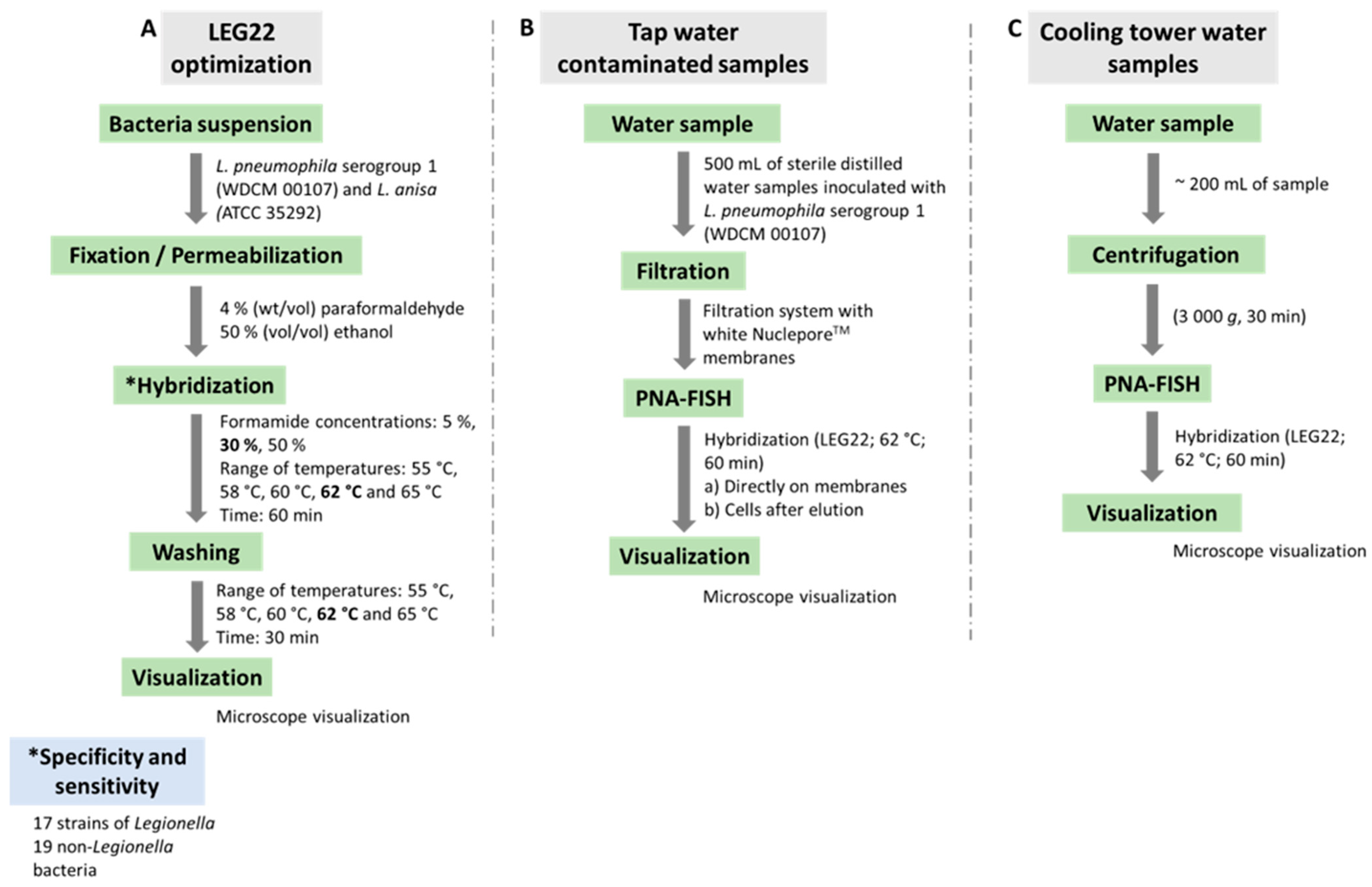
2. Materials and Methods
2.1. Strain and Growth Conditions
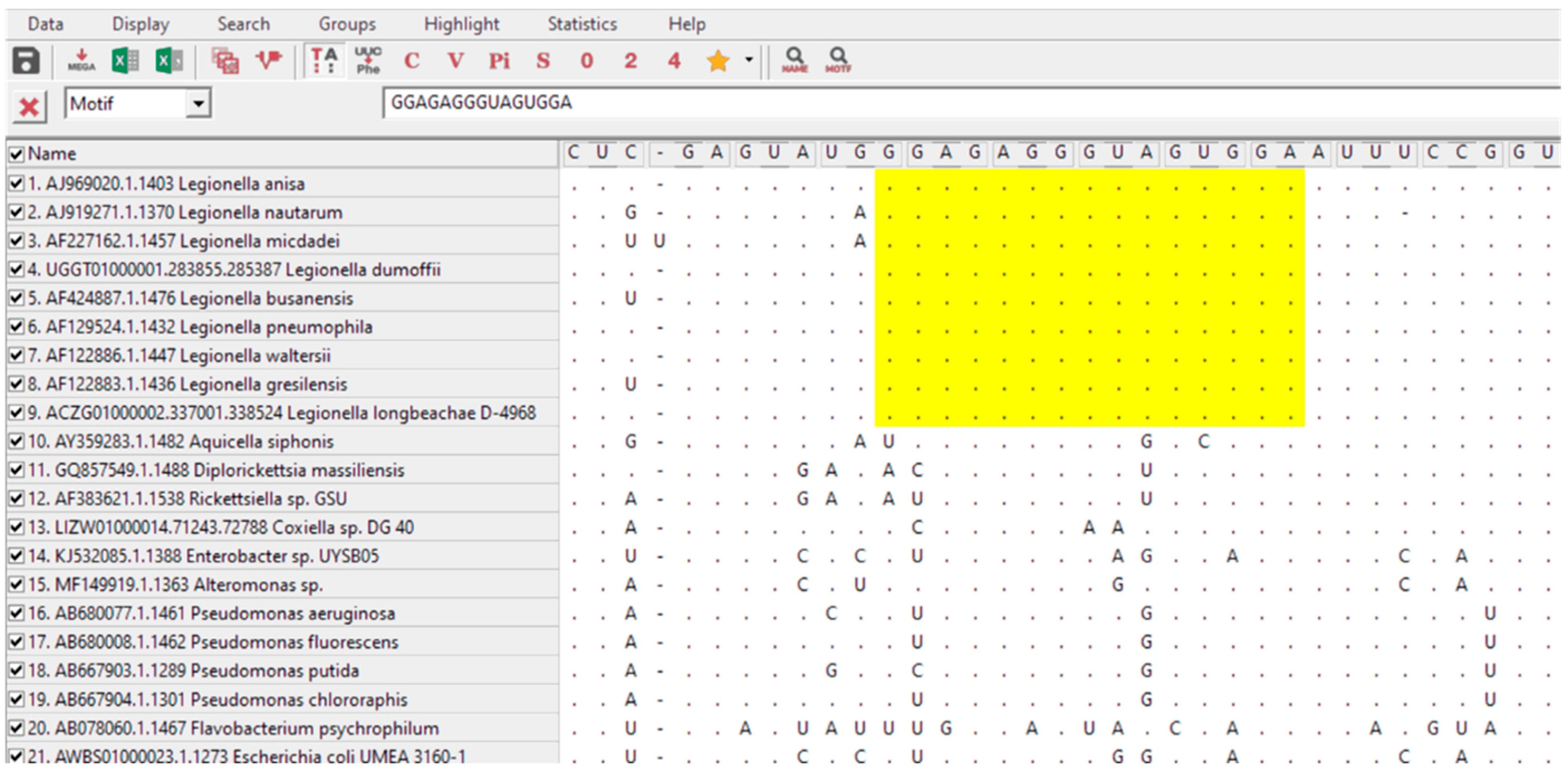
2.2. In Silico PNA Probe Design for the Specific Detection of Legionella
2.3. Hybridization Conditions
2.4. PNA-FISH Method Testing in Water Samples
2.5. Microscopy Visualization
2.6. Statistical Analysis
3. Results
3.1. Probe Design
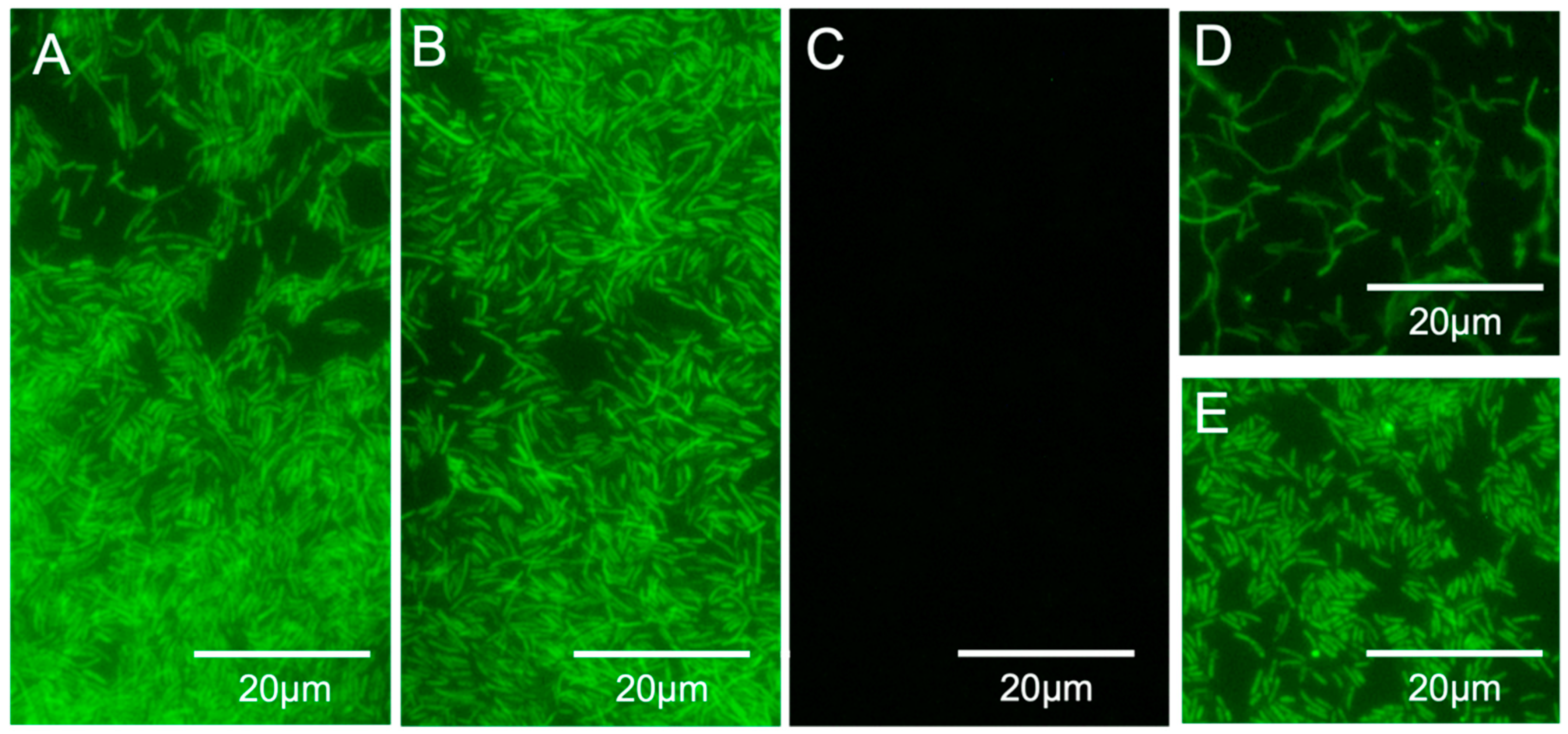
3.2. PNA-FISH Perfomance
3.3. Detection in Water Samples
4. Discussion
5. Conclusions
6. Patents
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chauhan, D.; Shames, S.R. Pathogenicity and Virulence of Legionella: Intracellular Replication and Host Response. Virulence 2021, 12, 1122–1144. [Google Scholar] [CrossRef]
- Hamilton, K.A.; Prussin, A.J.; Ahmed, W.; Haas, C.N. Outbreaks of Legionnaires’ Disease and Pontiac Fever 2006–2017. Curr. Environ. Health Rep. 2018, 5, 263–271. [Google Scholar] [CrossRef]
- Cross, K.E.; Mercante, J.W.; Benitez, A.J.; Brown, E.W.; Diaz, M.H.; Winchell, J.M. Simultaneous Detection of Legionella Species and L. Anisa, L. Bozemanii, L. Longbeachae and L. Micdadei Using Conserved Primers and Multiple Probes in a Multiplex Real-Time PCR Assay. Diagn. Microbiol. Infect. Dis. 2016, 85, 295–301. [Google Scholar] [CrossRef][Green Version]
- Ziltener, P.; Reinheckel, T.; Oxenius, A. Neutrophil and Alveolar Macrophage-Mediated Innate Immune Control of Legionella Pneumophila Lung Infection via TNF and ROS. PLoS Pathog. 2016, 12, e1005591. [Google Scholar] [CrossRef]
- Mercante, J.W.; Winchell, J.M. Current and Emerging Legionella Diagnostics for Laboratory and Outbreak Investigations. Clin. Microbiol. Rev. 2015, 28, 95–133. [Google Scholar] [CrossRef]
- International Organization for Standardization. ISO 11731:2017 Water Quality—Enumeration of Legionella; International Organization for Standardization: Geneva, Switzerland, 2017; Volume 2017. [Google Scholar]
- Sciuto, E.L.; Laganà, P.; Filice, S.; Scalese, S.; Libertino, S.; Corso, D.; Faro, G.; Coniglio, M.A. Environmental Management of Legionella in Domestic Water Systems: Consolidated and Innovative Approaches for Disinfection Methods and Risk Assessment. Microorganisms 2021, 9, 577. [Google Scholar] [CrossRef]
- ISO. ISO/TS 12869 Water Quality—Detection and Quantification of Legionella Spp. and/or Legionella Pneumophila by Concentration and Genic Amplification by Quantitative Polymerase Chain Reaction (QPCR); ISO: Geneva, Switzerland, 2019. [Google Scholar]
- European Working Group for Legionella Infections (EWGLI). European Technical Guidelines for the Prevention, Control and Investigation, of Infections Caused by Legionella Species; European Centre for Disease Prevention and Control: Stockholm, Sweden, 2017; p. 126.
- Young, C.; Smith, D.; Wafer, T.; Crook, B. Rapid Testing and Interventions to Control Legionella Proliferation Following a Legionnaires’ Disease Outbreak Associated with Cooling Towers. Microorganisms 2021, 9, 615. [Google Scholar] [CrossRef]
- Walker, J.T.; McDermott, P.J. Confirming the Presence of Legionella Pneumophila in Your Water System: A Review of Current Legionella Testing Methods. J. AOAC Int. 2021, 104, 1135–1147. [Google Scholar] [CrossRef]
- Azevedo, A.S.; Sousa, I.M.; Fernandes, R.M.; Azevedo, N.F.; Almeida, C. Optimizing Locked Nucleic Acid/2′-O-Methyl-RNA Fluorescence In Situ Hybridization (LNA/2′OMe-FISH) Procedure for Bacterial Detection. PLoS ONE 2019, 14, e0217689. [Google Scholar] [CrossRef]
- Cerqueira, L.; Moura, S.; Almeida, C.; Vieira, M.J.; Azevedo, N.F. Establishment of a New PNA-FISH Method for Aspergillus Fumigatus Identification: First Insights for Future Use in Pulmonary Samples. Microorganisms 2020, 8, 1950. [Google Scholar] [CrossRef]
- Kuo, J.-T.; Chang, L.-L.; Yen, C.-Y.; Tsai, T.-H.; Chang, Y.-C.; Huang, Y.-T.; Chung, Y.-C. Development of Fluorescence In Situ Hybridization as a Rapid, Accurate Method for Detecting Coliforms in Water Samples. Biosensors 2021, 11, 8. [Google Scholar] [CrossRef]
- Manz, W.; Amann, R.; Szewzyk, R.; Szewzyk, U.; Stenström, T.A.; Hutzler, P.; Schleifer, K.H. In Situ Identification of Legionellaceae Using 16S RRNA-Targeted Oligonucleotide Probes and Confocal Laser Scanning Microscopy. Microbiology 1995, 141, 29–39. [Google Scholar] [CrossRef]
- Rocha, R.; Santos, R.S.; Madureira, P.; Almeida, C.; Azevedo, N.F. Optimization of Peptide Nucleic Acid Fluorescence In Situ Hybridization (PNA-FISH) for the Detection of Bacteria: The Effect of PH, Dextran Sulfate and Probe Concentration. J. Biotechnol. 2016, 226, 1–7. [Google Scholar] [CrossRef]
- Emerson, J.B.; Adams, R.I.; Román, C.M.B.; Brooks, B.; Coil, D.A.; Dahlhausen, K.; Ganz, H.H.; Hartmann, E.M.; Hsu, T.; Justice, N.B.; et al. Schrödinger’s Microbes: Tools for Distinguishing the Living from the Dead in Microbial Ecosystems. Microbiome 2017, 5, 86. [Google Scholar] [CrossRef]
- Cerqueira, L.; Fernandes, R.M.; Ferreira, R.M.; Oleastro, M.; Carneiro, F.; Brandão, C.; Pimentel-Nunes, P.; Dinis-Ribeiro, M.; Figueiredo, C.; Keevil, C.W.; et al. Validation of a Fluorescence In Situ Hybridization Method Using Peptide Nucleic Acid Probes for Detection of Helicobacter Pylori Clarithromycin Resistance in Gastric Biopsy Specimens. J. Clin. Microbiol. 2013, 51, 1887–1893. [Google Scholar] [CrossRef]
- Cerqueira, L.; Azevedo, N.F.; Almeida, C.; Jardim, T.; Keevil, C.W.; Vieira, M.J. DNA Mimics for the Rapid Identification of Microorganisms by Fluorescence In Situ Hybridization (FISH). Int. J. Mol. Sci. 2008, 9, 1944–1960. [Google Scholar] [CrossRef]
- Stender, H.; Fiandaca, M.; Hyldig-Nielsen, J.J.; Coull, J. PNA for Rapid Microbiology. J. Microbiol. Methods 2002, 48, 1–17. [Google Scholar] [CrossRef]
- Chan, J.H.; And, S.L.; Wong, W.F. Antisense Oligonucleotides: From Design To Therapeutic Application. Clin. Exp. Pharmacol. Physiol. 2006, 33, 480–481. [Google Scholar] [CrossRef]
- Schoch, K.M.; Miller, T.M. Antisense Oligonucleotides: Translation from Mouse Models to Human Neurodegenerative Diseases Kathleen. Physiol. Behav. 2017, 176, 139–148. [Google Scholar] [CrossRef]
- Cerqueira, L.; Fernandes, R.M.; Ferreira, R.M.; Carneiro, F.; Dinis-Ribeiro, M.; Figueiredo, C.; Keevil, C.W.; Azevedo, N.F.; Vieira, M.J. PNA-FISH as a New Diagnostic Method for the Determination of Clarithromycin Resistance of Helicobacter Pylori. BMC Microbiol. 2011, 11, 101. [Google Scholar] [CrossRef]
- Rocha, R.; Sousa, J.M.; Cerqueira, L.; Vieira, M.J.; Almeida, C.; Azevedo, N.F. Development and Application of Peptide Nucleic Acid Fluorescence In Situ Hybridization for the Specific Detection of Listeria Monocytogenes. Food Microbiol. 2019, 80, 1–8. [Google Scholar] [CrossRef]
- Lehtola, M.J.; Torvinen, E.; Miettinen, I.T.; Keevil, C.W. Fluorescence In Situ Hybridization Using Peptide Nucleic Acid Probes for Rapid Detection of Mycobacterium Avium Subsp. Avium and Mycobacterium Avium Subsp. Paratuberculosis in Potable-Water Biofilms. Appl. Environ. Microbiol. 2006, 72, 848–853. [Google Scholar] [CrossRef][Green Version]
- Lehtola, M.J.; Loades, C.J.; Keevil, C.W. Advantages of Peptide Nucleic Acid Oligonucleotides for Sensitive Site Directed 16S RRNA Fluorescence In Situ Hybridization (FISH) Detection of Campylobacter Jejuni, Campylobacter Coli and Campylobacter Lari. J. Microbiol. Methods 2005, 62, 211–219. [Google Scholar] [CrossRef]
- Bragança, S.M.; Azevedo, N.F.; Simöes, L.C.; Keevil, C.W.; Vieira, M.J. Use of Fluorescent In Situ Hybridisation for the Visualisation of Helicobacter Pylori in Real Drinking Water Biofilms. Water Sci. Technol. 2007, 55, 387–393. [Google Scholar] [CrossRef]
- Hall, L.; Le Febre, K.M.; Deml, S.M.; Wohlfiel, S.L.; Wengenack, N.L. Evaluation of the Yeast Traffic Light PNA FISH Probes for Identification of Candida Species from Positive Blood Cultures. J. Clin. Microbiol. 2012, 50, 1446–1448. [Google Scholar] [CrossRef] [PubMed]
- Santos, R.S.; Lima, C.C.; Carvalho, D.; Meireles, F.; Guimarães, N.; Azevedo, N.F. Response Surface Methodology to Optimize Peptide Nucleic Acid Fluorescence In Situ Hybridization (PNA-FISH) in Saccharomyces Cerevisiae. LWT Food Sci. Technol. 2017, 80, 27–31. [Google Scholar] [CrossRef]
- Shinozaki, M.; Okubo, Y.; Sasai, D.; Nakayama, H.; Murayama, S.Y.; Ide, T.; Wakayama, M.; Hiruta, N.; Shibuya, K. Identification of Fusarium Species in Formalin-Fixed and Paraffin-Embedded Sections by In Situ Hybridization Using Peptide Nucleic Acid Probes. J. Clin. Microbiol. 2011, 49, 808–813. [Google Scholar] [CrossRef]
- Teertstra, W.R.; Lugones, L.G.; Wösten, H.A.B. In Situ Hybridisation in Filamentous Fungi Using Peptide Nucleic Acid Probes. Fungal Genet. Biol. 2004, 41, 1099–1103. [Google Scholar] [CrossRef]
- Wilks, S.A.; Keevil, C.W. Targeting Species-Specific Low-Affinity 16S RRNA Binding Sites by Using Peptide Nucleic Acids for Detection of Legionellae in Biofilms. Appl. Environ. Microbiol. 2006, 72, 5453–5462. [Google Scholar] [CrossRef]
- Simões, L.C.; Simões, M.; Oliveira, R.; Vieira, M.J. Potential of the Adhesion of Bacteria Isolated from Drinking Water to Materials. J. Basic Microbiol. 2007, 47, 174–183. [Google Scholar] [CrossRef]
- Teixeira, H.; Sousa, A.L.; Azevedo, A.S. Bioinformatic Tools and Guidelines for the Design of Fluorescence In Situ Hybridization Probes. In Fluorescence In Situ Hybridization (FISH) for Microbial Cells—Methods and Concepts; Humana: New York, NY, USA, 2021; Volume 2246, pp. 35–50. [Google Scholar]
- Almeida, C.; Azevedo, N.F.; Fernandes, R.M.; Keevil, C.W.; Vieira, M.J. Fluorescence In Situ Hybridization Method Using a Peptide Nucleic Acid Probe for Identification of Salmonella Spp. in a Broad Spectrum of Samples. Appl. Environ. Microbiol. 2010, 76, 4476–4485. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, R.; Azevedo, A.S.; Mendes, L. Application of Nucleic Acid Mimics in Fluorescence In Situ Hybridization. In Fluorescence In Situ Hybridization (FISH) for Microbial Cells—Methods and Concepts; Humana: New York, NY, USA, 2021; Volume 2246, pp. 69–86. [Google Scholar]
- Ameida, D.Q.; Silva, T.; Rodrigues, V.; Ladeira, R.; Sousa, F.; Capucho, R.; Duarte, G. Surto de Doença Dos Legionários Na Costa Norte de Portugal Durante a Pandemia Da Covid-19. Acta Med. Port. 2021, 34, 2021. [Google Scholar] [CrossRef]
- European Centre for Disease Prevention and Control. ELDSNET Operating Procedures for the Surveillance of Travel-Associated Legionnaires’ Disease in the EU/EEA; ECDC: Stockholm, Sweden, 2017; Volume 22.
- George, F.; Shivaji, T.; Pinto, C.S.; Serra, L.A.O.; Valente, J.; Albuquerque, M.J.; Vicêncio, P.C.O.; San-Bento, A.; Diegues, P.; Nogueira, P.J.; et al. A Large Outbreak of Legionnaires’ Disease in an Industrial Town in Portugal. Rev. Port. Saúde Pública 2016, 34, 199–208. [Google Scholar] [CrossRef][Green Version]
- Frickmann, H.; Zautner, A.E.; Moter, A.; Kikhney, J.; Hagen, R.M.; Stender, H.; Poppert, S. Fluorescence In Situ Hybridization (FISH) in the Microbiological Diagnostic Routine Laboratory: A Review. Crit. Rev. Microbiol. 2017, 43, 263–293. [Google Scholar] [CrossRef] [PubMed]
- Moter, A.; Göbel, U.B. Fluorescence In Situ Hybridization (FISH) for Direct Visualization of Microorganisms. J. Microbiol. Methods 2000, 41, 85–112. [Google Scholar] [CrossRef]
- Almeida, C.; Cerqueira, L.; Azevedo, N.F.; Vieira, M.J. Detection of Salmonella Enterica Serovar Enteritidis Using Real Time PCR, Immunocapture Assay, PNA FISH and Standard Culture Methods in Different Types of Food Samples. Int. J. Food Microbiol. 2013, 161, 16–22. [Google Scholar] [CrossRef]
- Amann, R.; Fuchs, B.M. Single-Cell Identification in Microbial Communities by Improved Fluorescence In Situ Hybridization Techniques. Nat. Rev. Microbiol. 2008, 6, 339–348. [Google Scholar] [CrossRef]
- Yilmaz, L.Ş.; Ökten, H.E.; Noguera, D.R. Making All Parts of the 16S RRNA of Escherichia Coli Accessible In Situ to Single DNA Oligonucleotides. Appl. Environ. Microbiol. 2006, 72, 733–744. [Google Scholar] [CrossRef]
- Dar, D.; Dar, N.; Cai, L.; Newman, D.K. In Situ Single-Cell Activities of Microbial Populations Revealed by Spatial Transcriptomics. bioRxiv 2021. [Google Scholar] [CrossRef]
- Almeida, C.; Azevedo, N.F. An Introduction to Fluorescence In Situ Hybridization in Microorganisms. In Fluorescence In Situ Hybridization (FISH) for Microbial Cells—Methods and Concepts; Humana: New York, NY, USA, 2021; Volume 2246, pp. 1–15. [Google Scholar]
- Farrell, R.E. RNA Methodologies A Laboratory Guide for Isolation and Characterization; Academic Press: New York, NY, USA, 2010. [Google Scholar]
- Santos, R.S.; Guimarães, N.; Madureira, P.; Azevedo, N.F. Optimization of a Peptide Nucleic Acid Fluorescence In Situ Hybridization (PNA-FISH) Method for the Detection of Bacteria and Disclosure of a Formamide Effect. J. Biotechnol. 2014, 187, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Almeida, C.; Azevedo, N.F.; Iversen, C.; Fanning, S.; Keevil, C.W.; Vieira, M.J. Development and Application of a Novel Peptide Nucleic Acid Probe for the Specific Detection of Cronobacter Genomospecies (Enterobacter Sakazakii) in Powdered Infant Formula. Appl. Environ. Microbiol. 2009, 75, 2925–2930. [Google Scholar] [CrossRef] [PubMed]
- McEgan, R.; Rodrigues, C.A.P.; Sbodio, A.; Suslow, T.V.; Goodridge, L.D.; Danyluk, M.D. Detection of Salmonella Spp. from Large Volumes of Water by Modified Moore Swabs and Tangential Flow Filtration. Lett. Appl. Microbiol. 2013, 56, 88–94. [Google Scholar] [CrossRef] [PubMed]

| Microorganisms | Origin | PNA-FISH Outcome |
|---|---|---|
| L. anisa * 1 | ATCC 35292 | + |
| L. anisa 144 1 | Treated Drinking Water | + |
| L. bozemanii1 | External Quality Assessment | + |
| L. feeleii 106 1 | Thermal Water | + |
| L. feeleii 187 1 | Cooling Tower Water | + |
| L. feeleii 242 1 | Irrigation Water | + |
| L. feeleii 263 1 | Treated Drinking Water | + |
| L. geestiana1 | Cooling Tower Water | + |
| L. gormanii1 | Thermal Water | + |
| L. jamestowniensis1 | External Quality Assessment | + |
| L. jordanis1 | External Quality Assessment | + |
| L. longbeachae1 | External Quality Assessment | + |
| L. micdadei 94 1 | External Quality Assessment | + |
| L. micdadei 99 1 | Untreated Drinking Water | + |
| L. oakridgensis1 | Thermal Water | + |
| L. pneumophila (serogroup 1) * 1 | WDCM 00107 | + |
| L. pneumophila (serogroup 2–15) 1 | Treated Drinking Water | + |
| Acinetobacter calcoaceticus2 | Treated Drinking Water | − |
| Aerococcus viridans1 | External Quality Assessment | − |
| Aeromonas hydrofila1 | Environment | − |
| Burkholderia cepacian2 | Treated Drinking Water | − |
| Enterobacter cloacae1 | Environment | − |
| Enterobacter aerogenes1 | External Quality Assessment | − |
| Enterococos faecalis1 | External Quality Assessment | − |
| Escherichia vulneris1 | Environment | − |
| Escherichia coli1 | External Quality Assessment | − |
| Klebsiella oxytoca1 | External Quality Assessment | − |
| Leifsonia aquatica1 | External Quality Assessment | − |
| Micrococcus luteus1 | External Quality Assessment | − |
| Pseudomonas fluorescens1 | Environment | − |
| Pseudomonas aeruginosa1 | Environment | − |
| Salmonella Wentworth1 | External Quality Assessment | − |
| Serratia liquefaciens1 | Environment | − |
| Staphylococcus warnerii1 | Environment | − |
| Staphylococcus aureus1 | External Quality Assessment | − |
| Stenotrophomonas maltophilia2 | Treated Drinking Water | − |
| Probe | Target Gene | Sequence (5′–3′) | Length (bp) | GC% | Specificity a% | Sensitivity a% | Position | Ref. |
|---|---|---|---|---|---|---|---|---|
| PLEG200 | 16S rRNA | GACGCAGGCTAATCT | 15 | 53.3 | 100.0 | 76.4 | 226–240 | [32] |
| LEG22 | 16S rRNA | TCCACTACCCTCTCC | 15 | 60.0 | 99.9 | 96.9 | 634–648 | This work |
| Artificially Inoculated Tap Water | ||
|---|---|---|
| CFU/L | PNA-FISH Outcome | Traditional Culture |
| 105 | + | + |
| 104 | + | + |
| 103 | + | + |
| 102 | + | + |
| 101 | + | + |
| 100 | − | + |
| 10−1 | − | + |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nácher-Vázquez, M.; Barbosa, A.; Armelim, I.; Azevedo, A.S.; Almeida, G.N.; Pizarro, C.; Azevedo, N.F.; Almeida, C.; Cerqueira, L. Development of a Novel Peptide Nucleic Acid Probe for the Detection of Legionella spp. in Water Samples. Microorganisms 2022, 10, 1409. https://doi.org/10.3390/microorganisms10071409
Nácher-Vázquez M, Barbosa A, Armelim I, Azevedo AS, Almeida GN, Pizarro C, Azevedo NF, Almeida C, Cerqueira L. Development of a Novel Peptide Nucleic Acid Probe for the Detection of Legionella spp. in Water Samples. Microorganisms. 2022; 10(7):1409. https://doi.org/10.3390/microorganisms10071409
Chicago/Turabian StyleNácher-Vázquez, Montserrat, Ana Barbosa, Inês Armelim, Andreia Sofia Azevedo, Gonçalo Nieto Almeida, Cristina Pizarro, Nuno Filipe Azevedo, Carina Almeida, and Laura Cerqueira. 2022. "Development of a Novel Peptide Nucleic Acid Probe for the Detection of Legionella spp. in Water Samples" Microorganisms 10, no. 7: 1409. https://doi.org/10.3390/microorganisms10071409
APA StyleNácher-Vázquez, M., Barbosa, A., Armelim, I., Azevedo, A. S., Almeida, G. N., Pizarro, C., Azevedo, N. F., Almeida, C., & Cerqueira, L. (2022). Development of a Novel Peptide Nucleic Acid Probe for the Detection of Legionella spp. in Water Samples. Microorganisms, 10(7), 1409. https://doi.org/10.3390/microorganisms10071409









