Raman Microscopic Analysis of Dry-Surface Biofilms on Clinically Relevant Materials
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Growth
2.2. Preparation of the Dry Surface Biofilms
2.3. Sample Preparation for Raman Measurements and Spectral Recording
2.4. Data Preprocessing and Mapping Representation
2.5. Predictive Model for the Identification of Microorganisms
3. Results and Discussion
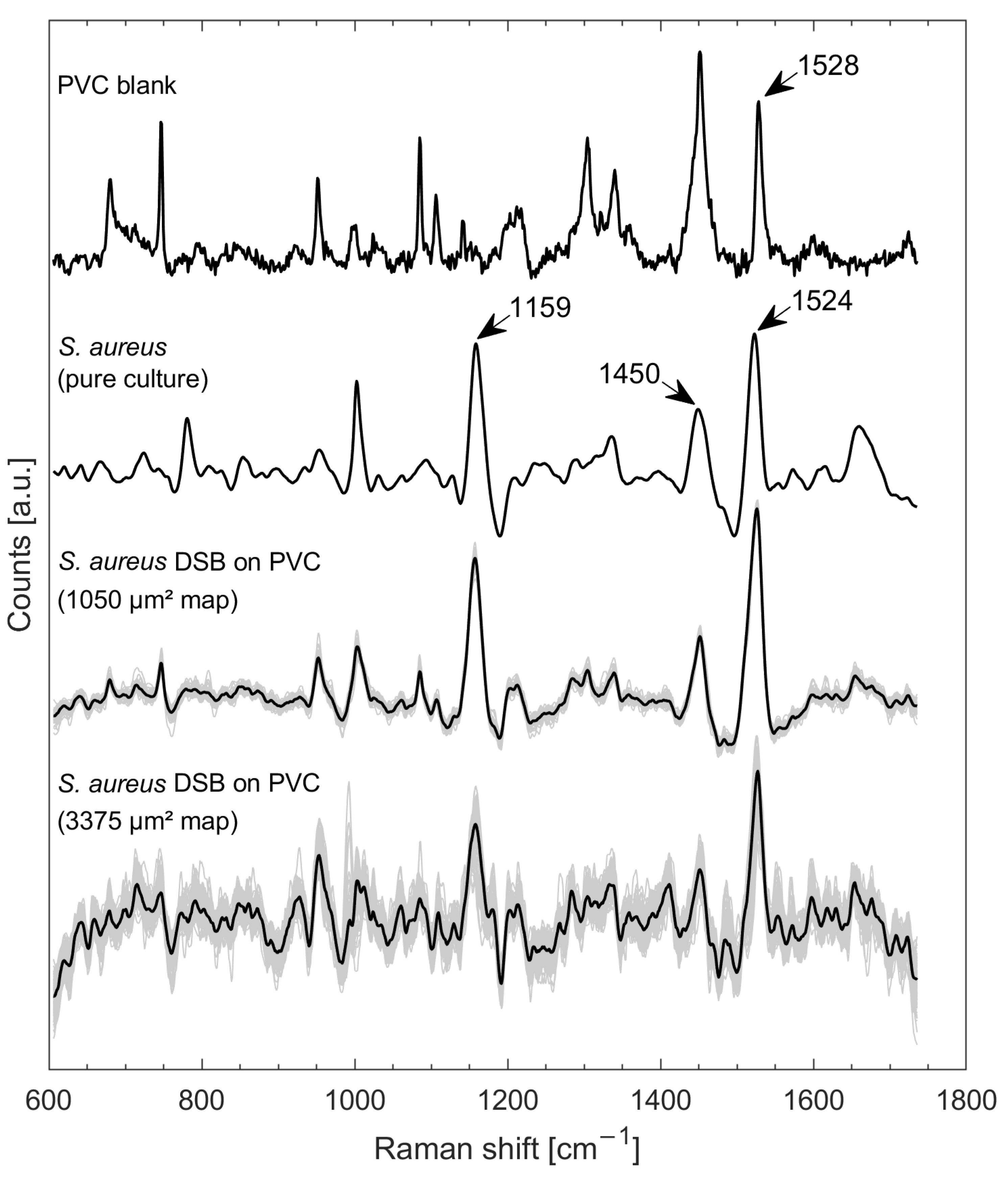
3.1. Raman Mapping of Monospecies DSBs on a PVC Blank
3.2. Raman Measurements of Single Cells in DSBs
3.3. Raman Mapping of dual-species DSBs on Steel Surfaces
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the Natural environment to infectious diseases. Nat. Rev. Microbiol. 2004, 2, 95–108. [Google Scholar] [CrossRef] [PubMed]
- Nkemngong, C.A.; Voorn, M.G.; Li, X.; Teska, P.J.; Oliver, H.F. A rapid model for developing dry surface biofilms of Staphylococcus aureus and Pseudomonas aeruginosa for in vitro disinfectant efficacy testing. Antimicrob. Resist. Infect. Control 2020, 9, 134. [Google Scholar] [CrossRef] [PubMed]
- Vickery, K.; Deva, A.; Jacombs, A.; Allan, J.; Valente, P.; Gosbell, I.B. Presence of biofilm containing viable multiresistant organisms despite terminal cleaning on clinical surfaces in an intensive care unit. J Hosp. Infect. 2012, 80, 52–55. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Johani, K.; Gosbell, I.B.; Jacombs AS, W.; Almatroudi, A.; Whiteley, G.S.; Deva, A.K.; Vickery, K. Intensive care unit environmental surfaces are contaminated by multidrug-resistant bacteria in biofilms: Combined results of conventional culture, pyrosequencing, scanning electron microscopy, and confocal laser microscopy. J Hosp. Infect. 2015, 91, 35–44. [Google Scholar] [CrossRef]
- Ledwoch, K.; Dancer, S.J.; Otter, J.A.; Kerr, K.; Roposte, D.; Rushton, L.; Weiser, R.; Mahenthiralingam, E.; Muir, D.D.; Maillard, J.-Y. Beware biofilm! Dry biofilms containing bacterial pathogens on multiple healthcare surfaces; a multi-centre study. J. Hosp. Infect. 2018, 100, e47–e56. [Google Scholar] [CrossRef]
- Almatroudi, A.; Gosbell, I.B.; Hu, H.; Jensen, S.O.; Espedido, B.A.; Tahir, S.; Glasbey, T.O.; Legge, P.; Whiteley, G.; Deva, A.; et al. Staphylococcus aureus dry-surface biofilms are not killed by sodium hypochlorite: Implications for infection control. J. Hosp. Infect. 2016, 93, 263–270. [Google Scholar] [CrossRef]
- Almatroudi, A.; Tahir, S.; Hu, H.; Chowdhury, D.; Gosbell, I.B.; Jensen, S.O.; Whiteley, G.S.; Deva, A.K.; Glasbey, T.; Vickery, K. Staphylococcus aureus dry-surface biofilms are more resistant to heat treatment than traditional hydrated biofilms. J. Hosp. Infect. 2018, 98, 161–167. [Google Scholar] [CrossRef]
- Ledwoch, K.; Magoga, M.; Williams, D.; Fabbri, S.; Walsh, J.; Maillard, J.-Y. Is a reduction in viability enough to determine biofilm susceptibility to a biocide? Infect. Control Hosp. Epidemiol. 2021, 42, 1486–1492. [Google Scholar] [CrossRef]
- Maquelin, K.; Kirschner, C.; Choo-Smith, L.-P.; van den Braak, N.; Endtz, H.P.; Naumann, D.; Puppels, G.J. Identification of medically relevant microorganisms by vibrational spectroscopy. J. Microbiol. Methods 2002, 51, 255–271. [Google Scholar] [CrossRef]
- Yang, H.; Irudayaraj, J. Rapid detection of foodborne microorganisms on food surface using Fourier transform Raman spectroscopy. J. Mol. Struct. 2003, 646, 35–43. [Google Scholar] [CrossRef]
- Stöckel, S.; Kirchhoff, J.; Neugebauer, U.; Rösch, P.; Popp, J. The application of Raman spectroscopy for the detection and identification of microorganisms. J. Raman Spectrosc. 2016, 47, 89–109. [Google Scholar] [CrossRef]
- Rösch, P.; Harz, M.; Krause, M.; Popp, J. Fast and reliable identification of microorganisms by means of Raman spectroscopy. In Biophotonics 2007: Optics in Life Science; Paper 6633_45; Optical Society of America: Washington, DC, USA, 2007. [Google Scholar] [CrossRef]
- Di Martino, P. Extracellular polymeric substances, a key element in understanding biofilm phenotype. AIMS Microbiol. 2018, 4, 274–288. [Google Scholar] [CrossRef]
- Ivleva, N.P.; Wagner, M.; Horn, H.; Niessner, R.; Haisch, C. In Situ Surface-Enhanced Raman Scattering Analysis of Biofilm. Anal. Chem. 2008, 80, 8538–8544. [Google Scholar] [CrossRef] [PubMed]
- Ivleva, N.P.; Wagner, M.; Horn, H.; Niessner, R.; Haisch, C. Towards a nondestructive chemical characterization of biofilm matrix by Raman microscopy. Anal. Bioanal. Chem. 2009, 393, 197–206. [Google Scholar] [CrossRef]
- Beier, B.D.; Quivey, R.G.; Berger, A.J. Raman microspectroscopy for species identification and mapping within bacterial biofilms. AMB Express 2012, 2, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ledwoch, K.; Said, J.; Norville, P.; Maillard, J.Y. Artificial dry surface biofilm models for testing the efficacy of cleaning and disinfection. Lett. Appl. Microbiol. 2019, 68, 329–336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tewes, T.J.; Kerst, M.; Platte, F.; Bockmühl, D.P. Raman Microscopic Identification of Microorganisms on Metal Surfaces via Support Vector Machines. Microorganisms 2022, 10, 556. [Google Scholar] [CrossRef]
- Kumar, B.N.V.; Kampe, B.; Rösch, P.; Popp, J. Characterization of carotenoids in soil bacteria and investigation of their photodegradation by UVA radiation via resonance Raman spectroscopy. Analyst 2015, 140, 4584–4593. [Google Scholar] [CrossRef]
- Strola, S.A.; Baritaux, J.-C.; Schultz, E.; Simon, A.C.; Allier, C.; Espagnon IJary, D.; Dintent, J.-M. Single bacteria identification by Raman spectroscopy. J. Biomed. Opt. 2014, 19, 111610. [Google Scholar] [CrossRef]
- Schuster, K.C.; Reese, I.; Urlaub, E.; Gapes, J.R.; Lendl, B. Multidimensional Information on the Chemical Composition of Single Bacterial Cells by Confocal Raman Microspectroscopy. Anal. Chem. 2000, 72, 5529–5534. [Google Scholar] [CrossRef]
- De Gelder, J.; De Gussem, K.; Vandenabeele, P.; Moens, L. Reference database of Raman spectra of biological molecules. J. Raman Spectrosc. 2007, 38, 1133–1147. [Google Scholar] [CrossRef]
- Huang, S.; Chen, D.; Pelczar, P.; Vepachedu, V.; Setlow, P.; Li, Y. Levels of Ca2+-Dipicolinic Acid in Individual Bacillus Spores Determined Using Microfluidic Raman Tweezers. J. Bacteriol. 2007, 189, 4681–4687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mlynáriková, K.; Samek, O.; Bernatová, S.; Růžička, F.; Ježek, J.; Hároniková, A.; Šiler, M.; Zemánek, P.; Holá, V. Influence of culture media on microbial fingerprints using raman spectroscopy. Sensors 2015, 15, 29635–29647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harz, M.; Rösch, P.; Peschke, K.D.; Ronneberger, O.; Burkhardt, H.; Popp, J. Micro-Raman spectroscopic identification of bacterial cells of the genus Staphylococcus and dependence on their cultivation conditions. Analyst 2005, 130, 1543–1550. [Google Scholar] [CrossRef] [PubMed]
- Hutsebaut, D.; Maquelin, K.; De Vos, P.; Vandenabeele, P.; Moens, L.; Puppels, G.J. Effect of Culture Conditions on the Achievable Taxonomic Resolution of Raman Spectroscopy Disclosed by Three Bacillus Species. Anal. Chem. 2004, 76, 6274–6281. [Google Scholar] [CrossRef]
- Liu, X.-Y.; Guo, S.; Bocklitz, T.; Rösch, P.; Popp, J.; Yu, H.-Q. Nondestructive 3D imaging and quantification of hydrated biofilm matrix by confocal Raman microscopy coupled with non-negative matrix factorization. Water Res. 2022, 210, 117973. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tewes, T.J.; Centeleghe, I.; Maillard, J.-Y.; Platte, F.; Bockmühl, D.P. Raman Microscopic Analysis of Dry-Surface Biofilms on Clinically Relevant Materials. Microorganisms 2022, 10, 1369. https://doi.org/10.3390/microorganisms10071369
Tewes TJ, Centeleghe I, Maillard J-Y, Platte F, Bockmühl DP. Raman Microscopic Analysis of Dry-Surface Biofilms on Clinically Relevant Materials. Microorganisms. 2022; 10(7):1369. https://doi.org/10.3390/microorganisms10071369
Chicago/Turabian StyleTewes, Thomas J., Isabella Centeleghe, Jean-Yves Maillard, Frank Platte, and Dirk P. Bockmühl. 2022. "Raman Microscopic Analysis of Dry-Surface Biofilms on Clinically Relevant Materials" Microorganisms 10, no. 7: 1369. https://doi.org/10.3390/microorganisms10071369
APA StyleTewes, T. J., Centeleghe, I., Maillard, J.-Y., Platte, F., & Bockmühl, D. P. (2022). Raman Microscopic Analysis of Dry-Surface Biofilms on Clinically Relevant Materials. Microorganisms, 10(7), 1369. https://doi.org/10.3390/microorganisms10071369





