Differences in the Propensity of Different Antimicrobial Resistance Determinants to Be Disseminated via Transformation in Campylobacter jejuni and Campylobacter coli
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Growth Conditions
2.2. DNA Isolation from Donor Strains
2.3. Transformation and Determination of Transformation and Mutation Frequencies
2.4. Minimum Inhibitory Concentration (MIC) Determinations
2.5. Confirmation of Resistance Determinants
2.6. Bioinformatics Analysis of Gentamicin Resistance Regions in Donor and Recipient Genomes
2.7. Statistical Analysis
3. Results and Discussion
3.1. Differences in Transformation Frequencies of C. jejuni SN:CM for Different Resistance Markers
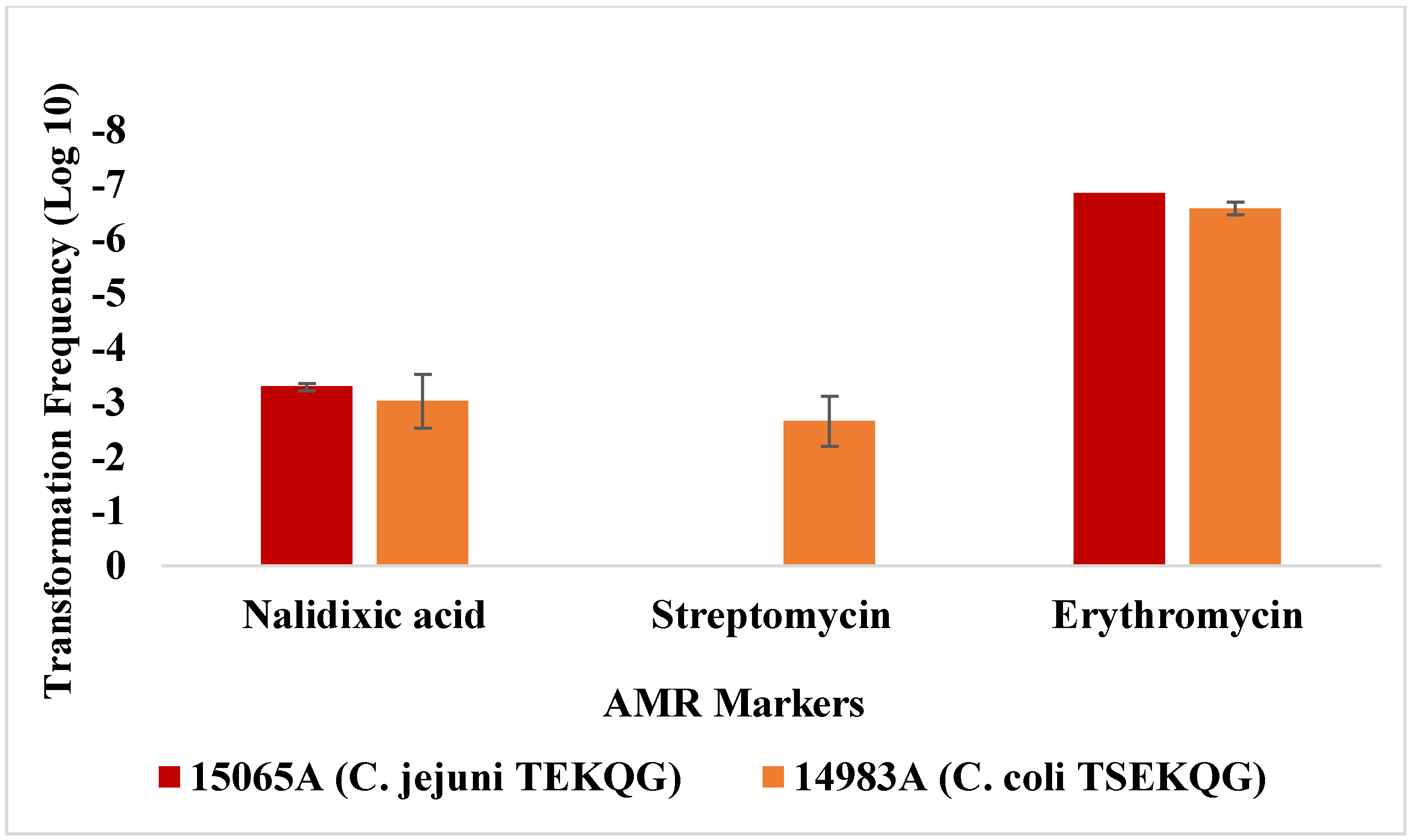
3.2. Contemporary Industry-Relevant Campylobacter Strains Exhibit Variable Outcomes for Transformation to Gentamicin Resistance
3.3. Transformation Frequency of C. jejuni SN:CM to Erythromycin Resistance Is Markedly Lower Than Observed with Contemporary Campylobacter Strains from Poultry
4. Conclusions and Further Research
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scallan, E.; Hoekstra, R.M.; Angulo, F.J.; Tauxe, R.V.; Widdowson, M.A.; Roy, S.L.; Jones, J.L.; Griffin, P.M. Foodborne illness acquired in the United States-Major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. [Google Scholar] [CrossRef] [PubMed]
- The Interagency Food Safety Analytics Collaboration (IFSAC). Foodborne Illness Source Attribution Estimates for 2017 for Salmonella, Escherichia coli O157, Listeria monocytogenes, and Campylobacter Using Multi-Year Outbreak Surveillance Data, United States. 2019. Available online: https://www.cdc.gov/foodsafety/ifsac/pdf/P19-2017-report-TriAgency-508.pdf (accessed on 7 June 2022).
- Silva, J.; Leite, D.; Fernandes, M.; Mena, C.; Gibbs, P.A.; Teixeira, P. Campylobacter spp. as a foodborne pathogen: A review. Front. Microbiol. 2011, 2, 200. [Google Scholar] [CrossRef] [PubMed]
- Tack, D.M.; Ray, L.; Griffin, P.M.; Cieslak, P.R.; Dunn, J.; Rissman, T.; Jervis, R.; Lathrop, S.; Muse, A.; Duwell, M.; et al. Preliminary incidence and trends of infections with pathogens transmitted commonly through food—Foodborne diseases active surveillance network, 10 U.S. sites, 2016–2019. MMWR Morb. Mortal. Wkly. Rep. 2019, 69, 509–514. [Google Scholar] [CrossRef] [PubMed]
- Kaakoush, N.O.; Castaño-Rodríguez, N.; Mitchell, H.M.; Man, S.M. Global epidemiology of Campylobacter infection. Clin. Microbiol. Rev. 2015, 28, 687–720. [Google Scholar] [CrossRef]
- Man, S.M. The clinical importance of emerging Campylobacter species. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 669–685. [Google Scholar] [CrossRef]
- Nachamkin, I.; Allos, B.M.; Ho, T. Campylobacter species and Guillain-Barré syndrome. Clin. Microbiol. Rev. 1998, 11, 555–567. [Google Scholar] [CrossRef]
- Scallan Walter, E.J.; Crim, S.M.; Bruce, B.B.; Griffin, P.M. Incidence of Campylobacter-associated Guillain-Barré Syndrome estimated from health insurance data. Foodborne Pathog. Dis. 2020, 17, 23–28. [Google Scholar] [CrossRef]
- Facciolà, A.; Riso, R.; Avventuroso, E.; Visalli, G.; Delia, S.A.; Laganà, P. Campylobacter: From microbiology to prevention. J. Prev. Med. Hyg. 2017, 58, E79–E92. [Google Scholar] [CrossRef]
- Friedman, C.R.; Neimann, J.; Wegener, H.C.; Tauxe, R.V. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations. In Campylobacter; ASM International: Washington, DC, USA, 2000; pp. 121–138. [Google Scholar]
- Scallan, E.; Hoekstra, R.M.; Mahon, B.E.; Jones, T.F.; Griffin, P.M. An assessment of the human health impact of seven leading foodborne pathogens in the United States using disability adjusted life years. Epidemiol. Infect. 2015, 143, 2795–2804. [Google Scholar] [CrossRef]
- Centers for Disease Control and Prevention (CDC). Antibiotic resistance Threats in the United States. 2019. Available online: https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf (accessed on 27 April 2022).
- Bolinger, H.; Kathariou, S. The current state of macrolide resistance in Campylobacter spp.: Trends and impacts of resistance mechanisms. Appl. Environ. Microbiol. 2017, 83, e00416-17. [Google Scholar] [CrossRef]
- Sproston, E.L.; Wimalarathna, H.M.L.; Sheppard, S.K. Trends in fluoroquinolone resistance in Campylobacter. Microb. Genom. 2018, 4, e000198. [Google Scholar] [CrossRef]
- The National Antimicrobial Resistance Monitoring System (NARMS). The National Antimicrobial Resistance Monitoring System Integrated Report: Enteric Bacteria. 2014. Available online: https://fda.report/media/101511/2014-NARMS-Report.pdf (accessed on 27 April 2022).
- Taylor, D. Genetics of Campylobacter and Heliobacter. Annu. Rev. Microbiol. 1992, 46, 35–64. [Google Scholar] [CrossRef]
- Wang, Y.; Taylor, D.E. Natural transformation in Campylobacter species. J. Bacteriol. 1990, 172, 949–955. [Google Scholar] [CrossRef]
- Kim, J.S.; Kim, J.W.; Kathariou, S. Differential effects of temperature on natural transformation to erythromycin and nalidixic acid resistance in Campylobacter coli. Appl. Environ. Microbiol. 2008, 74, 6121–6125. [Google Scholar] [CrossRef]
- Kim, J.S.; Carver, D.K.; Kathariou, S. Natural transformation-mediated transfer of erythromycin resistance in Campylobacter coli strains from turkeys and swine. Appl. Environ. Microbiol. 2006, 72, 1316–1321. [Google Scholar] [CrossRef]
- Vegge, C.S.; Brøndsted, L.; Ligowska-Marzeta, M.; Ingmer, H. Natural transformation of Campylobacter jejuni occurs beyond limits of growth. PLoS ONE 2012, 7, e45467. [Google Scholar] [CrossRef]
- Bae, J.; Oh, E.; Jeon, B. Enhanced transmission of antibiotic resistance in Campylobacter jejuni biofilms by natural transformation. Antimicrob. Agents Chemother. 2014, 58, 7573–7575. [Google Scholar] [CrossRef]
- Wilson, D.L.; Bell, J.A.; Young, V.B.; Wilder, S.R.; Mansfield, L.S.; Linz, J.E. Variation of the natural transformation frequency of Campylobacter jejuni in liquid shake culture. Microbiology 2003, 149, 3603–3615. [Google Scholar] [CrossRef][Green Version]
- Taylor, D.E.; Eaton, M.; Yan, W.; Chang, N. Genome maps of Campylobacter jejuni and Campylobacter coli. J. Bacteriol. 1992, 174, 2332–2337. [Google Scholar] [CrossRef][Green Version]
- Karlyshev, A.V.; Henderson, J.; Ketley, J.M.; Wren, B.W. An improved physical and genetic map of Campylobacter jejuni NCTC 11168 (UA580). Microbiology 1998, 144, 503–508. [Google Scholar] [CrossRef][Green Version]
- Weingarten, R.A.; Grimes, J.L.; Olson, J.W. Role of Campylobacter jejuni respiratory oxidases and reductases in host colonization. Appl. Environ. Microbiol. 2008, 74, 1367–1375. [Google Scholar] [CrossRef]
- Crespo, M.D.; Altermann, E.; Olson, J.; Miller, W.G.; Chandrashekhar, K.; Kathariou, S. Novel plasmid conferring kanamycin and tetracycline resistance in the turkey-derived Campylobacter jejuni strain 11601MD. Plasmid 2016, 86, 32–37. [Google Scholar] [CrossRef]
- Miller, W.G.; Huynh, S.; Parker, C.T.; Niedermeyer, J.A.; Kathariou, S. Complete genome sequences of multidrug-resistant Campylobacter jejuni strain 14980A (turkey feces) and Campylobacter coli strain 14983A (housefly from a turkey farm), harboring a novel gentamicin resistance mobile element. Genome Announc. 2016, 4, 16–17. [Google Scholar] [CrossRef]
- Guernier-Cambert, V.; Trachsel, J.; Maki, J.; Qi, J.; Sylte, M.J.; Hanafy, Z.; Kathariou, S.; Looft, T. Natural horizontal gene transfer of antimicrobial resistance genes in Campylobacter spp. from turkeys and swine. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef] [PubMed]
- Crespo, M.D.; Olson, J.W.; Altermann, E.; Siletzky, R.M.; Kathariou, S. Chromosomal tet(O)-harboring regions in Campylobacter coli isolates from turkeys and swine. Appl. Environ. Microbiol. 2012, 78, 8488–8491. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Mukherjee, S.; Chen, Y.; Li, C.; Young, S.; Warren, M.; Abbott, J.; Friedman, S.; Kabera, C.; Karlsson, M.; et al. Novel gentamicin resistance genes from Campylobacter isolated from humans and retail meats in the U.S.A. J. Antimicrob. Chemother. 2015, 70, 1314–1321. [Google Scholar] [CrossRef] [PubMed]
- Carver, T.J.; Rutherford, K.M.; Berriman, M.; Rajandream, M.A.; Barrell, B.G.; Parkhill, J. ACT: The Artemis comparison tool. Bioinformatics 2005, 21, 3422–3423. [Google Scholar] [CrossRef]
- Zhao, S.; Mukherjee, S.; Li, C.; Jones, S.B.; Young, S.; Mcdermott, P.F. Cloning and expression of novel aminoglycoside phosphotransferase genes from Campylobacter and their role in the resistance to six aminoglycosides. Antimicrob. Agents Chemother. 2017, 62, e01682-17. [Google Scholar] [CrossRef]
- Taylor, D.E.; De Grandis, S.A.; Karmali, M.A.; Fleming, P.C. Transmissible plasmids from Campylobacter jejuni. Antimicrob. Agents Chemother. 1981, 19, 831–835. [Google Scholar] [CrossRef]
- Lefébure, T.; Bitar, P.D.P.; Suzuki, H.; Stanhope, M.J. Evolutionary dynamics of complete Campylobacter pan-genomes and the bacterial species concept. Genome Biol. Evol. 2010, 2, 646–655. [Google Scholar] [CrossRef]
- Pascoe, B.; Williams, L.K.; Calland, J.K.; Meric, G.; Hitchings, M.D.; Dyer, M.; Ryder, J.; Shaw, S.; Lopes, B.S.; Chintoan-Uta, C.; et al. Domestication of Campylobacter jejuni NCTC 11168. Microb. Genom. 2019, 5, 000279. [Google Scholar] [CrossRef]
- Zhang, X.; Zhou, Q.; Tang, M.; Pu, J.; Zhang, J.; Lu, J.; Zhang, Y.; Gao, Y. Aminoglycoside resistance and possible mechanisms in Campylobacter spp. isolated from chicken and swine in Jiangsu, China. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef]
- Qin, S.; Wang, Y.; Zhang, Q.; Chen, X.; Shen, Z.; Deng, F.; Wu, C.; Shen, J. Identification of a novel genomic island conferring resistance to multiple aminoglycoside antibiotics in Campylobacter coli. Antimicrob. Agents Chemother. 2012, 56, 5332–5339. [Google Scholar] [CrossRef]
- Yao, H.; Liu, D.; Wang, Y.; Zhang, Q.; Shen, Z. High prevalence and predominance of the aph(2′)-if gene conferring aminoglycoside resistance in Campylobacter. Antimicrob. Agents Chemother. 2017, 61, 112–117. [Google Scholar] [CrossRef]
- Payot, S.; Bolla, J.-M.; Corcoran, D.; Fanning, S.; Mégraud, F.; Zhang, Q. Mechanisms of fluoroquinolone and macrolide resistance in Campylobacter spp. Microbes Infect. 2006, 8, 1967–1971. [Google Scholar] [CrossRef]
- Luangtongkum, T.; Jeon, B.; Han, J.; Plummer, P.; Logue, C.M.; Qijing, Z. Antibiotic resistance in Campylobacter: Emergence, transmission and persistence. Futur. Microbiol. 2009, 4, 189–200. [Google Scholar] [CrossRef]
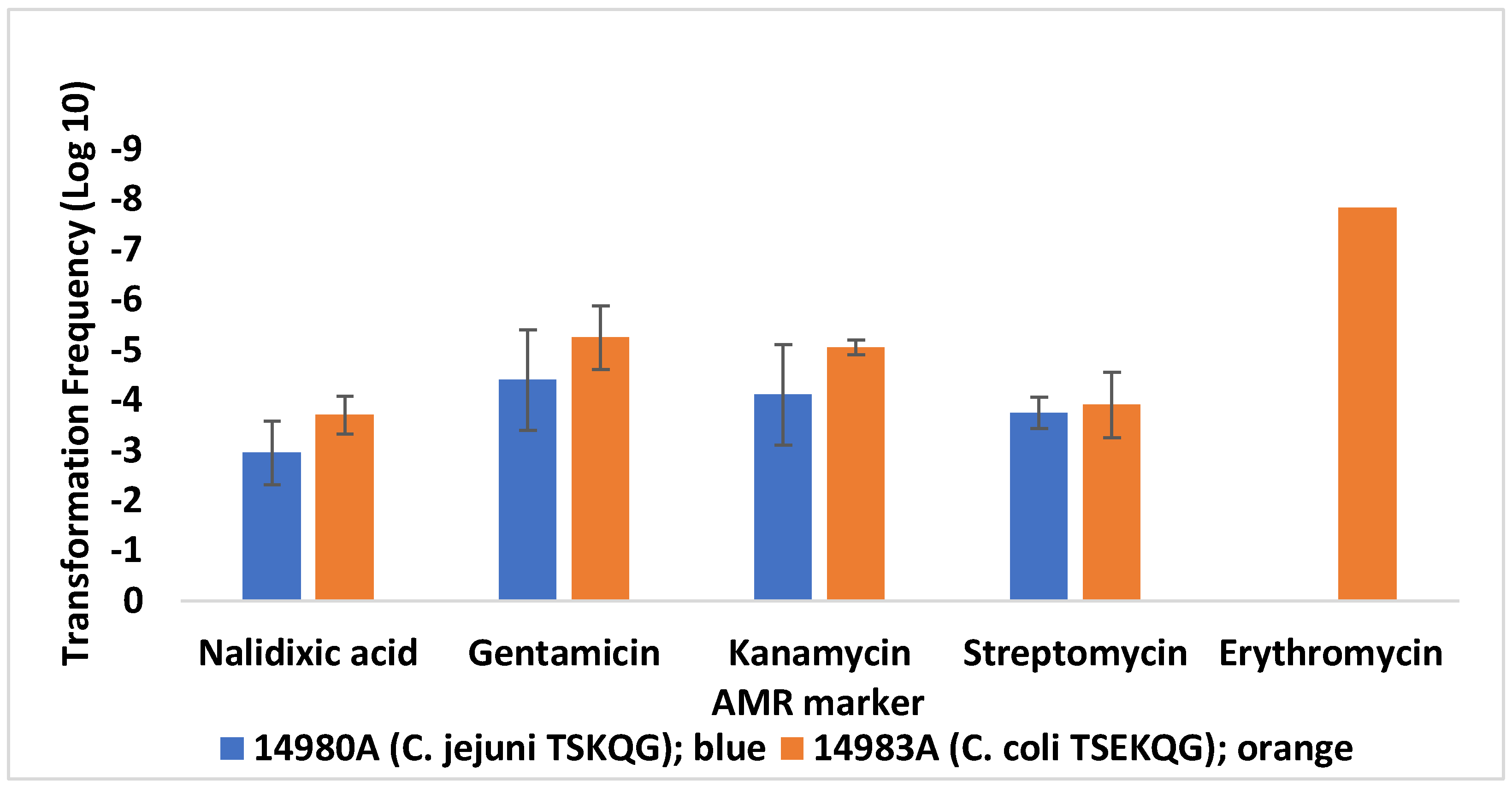
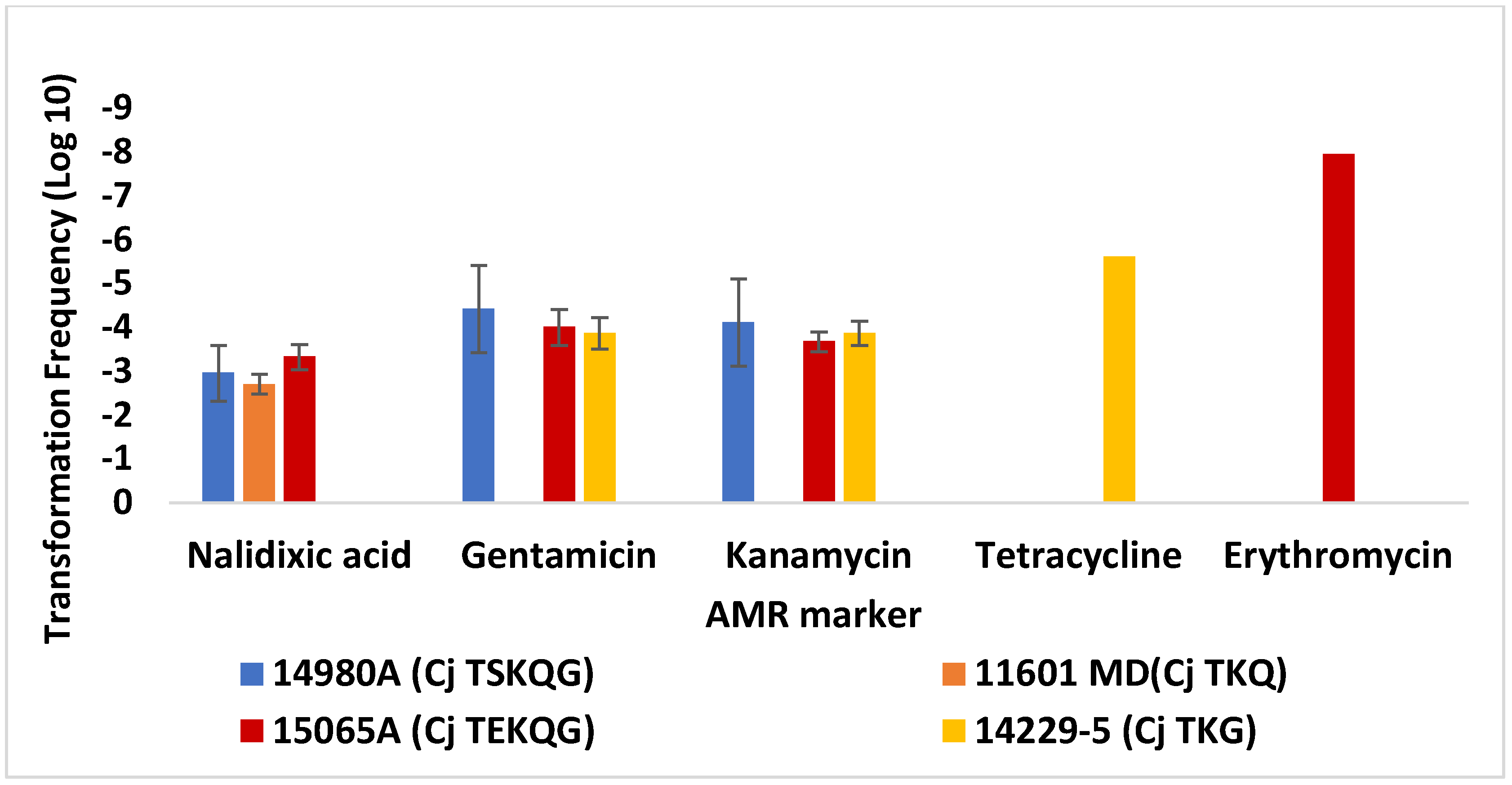
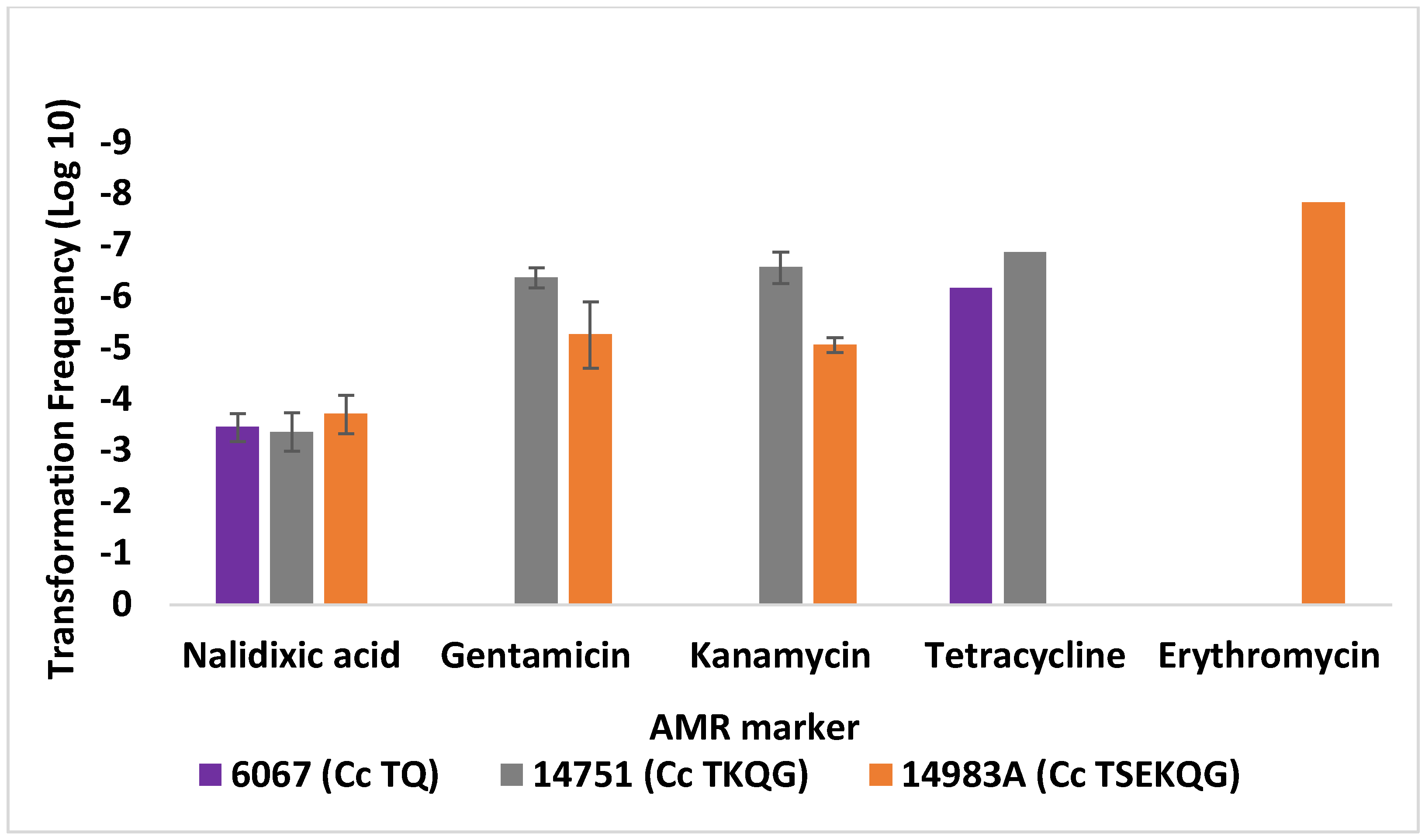

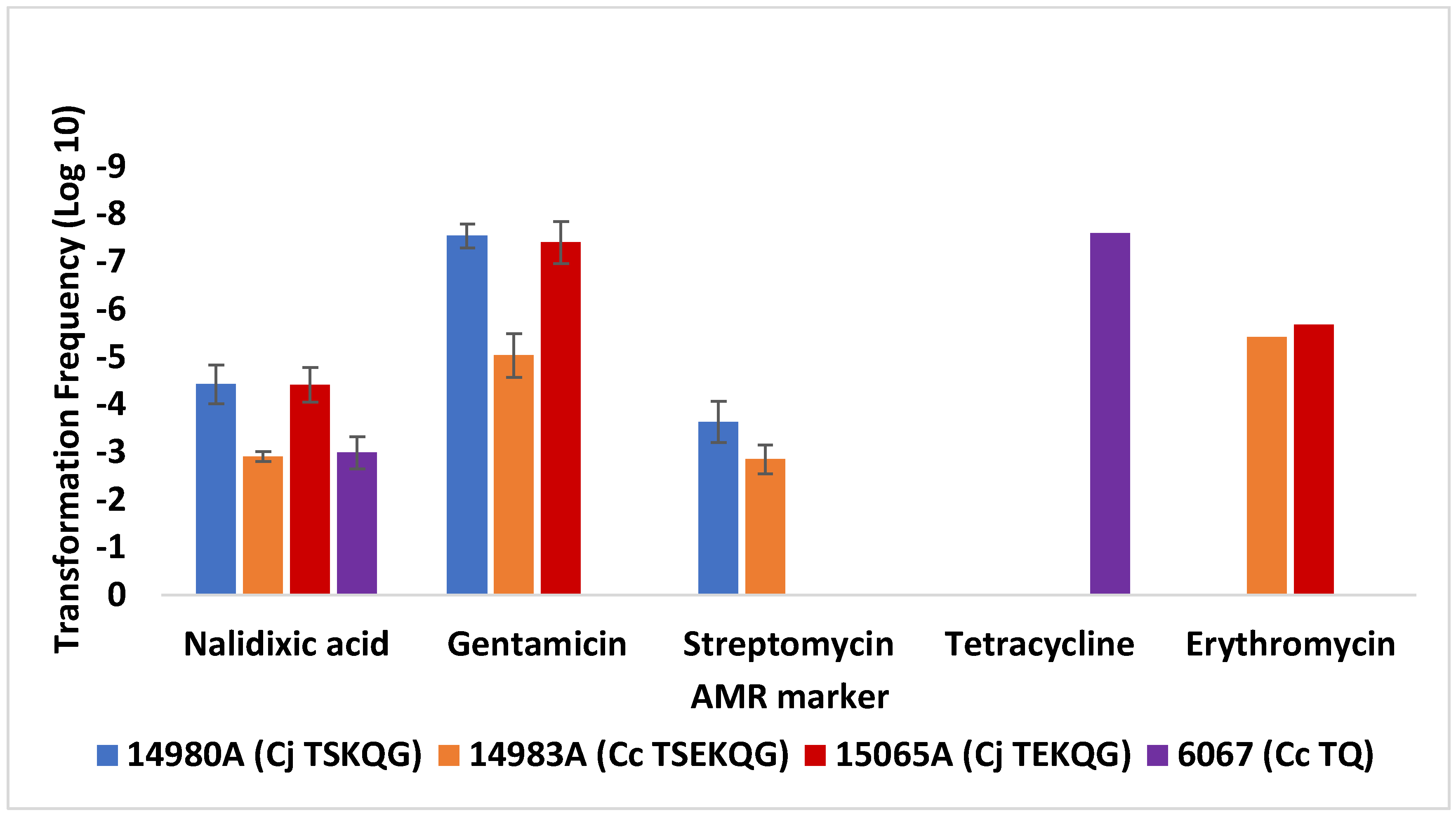
| Strain 1 | Source [Reference] 2 | AMR Profile 3 | Use | Genome Accession 4 |
|---|---|---|---|---|
| C. jejuni | ||||
| SN:CM | NCTC 11168 derivative, strain Cj0264c::Cm [25] | Cm | Recipient | AL111168 |
| 11601MD | Jejunum, turkey, 2006 [26] | TKQ | Donor | LKCR00000000 |
| 14980A | Feces, turkey, 2014 [27] | TSKQG | Donor | CP017029 |
| 14229-5 | Feces, turkey, 2013 [28] | TKG | Donor | https://www.ebi.ac.uk/biosamples/samples/SAMEA7848382 (accessed on 17 May 2022) |
| 15065A | Cecum, turkey, 2014 | TEKQG | Donor | CP092017 |
| FSIS 11810577 | Young chicken, 2018 | Pan-sensitive | Recipient | AACOOF000000000 |
| C. coli | ||||
| 6067 | Water, turkey house, 2003 [29] | TQ | Donor | LKCQ00000000 |
| 14983A | Housefly (Musca domestica) from turkey farm, 2014 [27] | TSEKQG | Donor | CP017025 |
| 14751 | Cecum, turkey, 2013 | TKQG | Donor | N/A |
| FSIS 1710488 | Young chicken, 2017 | Pan-sensitive | Recipient | AACKVZ000000000 |
| CVM N43850 | Chicken breast, 2013 | TKG | Recipient | Biosample SAMN03344552; Assembly GCA_001417185.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hanafy, Z.; Osborne, J.A.; Miller, W.G.; Parker, C.T.; Olson, J.W.; Jackson, J.H., III; Kathariou, S. Differences in the Propensity of Different Antimicrobial Resistance Determinants to Be Disseminated via Transformation in Campylobacter jejuni and Campylobacter coli. Microorganisms 2022, 10, 1194. https://doi.org/10.3390/microorganisms10061194
Hanafy Z, Osborne JA, Miller WG, Parker CT, Olson JW, Jackson JH III, Kathariou S. Differences in the Propensity of Different Antimicrobial Resistance Determinants to Be Disseminated via Transformation in Campylobacter jejuni and Campylobacter coli. Microorganisms. 2022; 10(6):1194. https://doi.org/10.3390/microorganisms10061194
Chicago/Turabian StyleHanafy, Zahra, Jason A. Osborne, William G. Miller, Craig T. Parker, Jonathan W. Olson, James H. Jackson, III, and Sophia Kathariou. 2022. "Differences in the Propensity of Different Antimicrobial Resistance Determinants to Be Disseminated via Transformation in Campylobacter jejuni and Campylobacter coli" Microorganisms 10, no. 6: 1194. https://doi.org/10.3390/microorganisms10061194
APA StyleHanafy, Z., Osborne, J. A., Miller, W. G., Parker, C. T., Olson, J. W., Jackson, J. H., III, & Kathariou, S. (2022). Differences in the Propensity of Different Antimicrobial Resistance Determinants to Be Disseminated via Transformation in Campylobacter jejuni and Campylobacter coli. Microorganisms, 10(6), 1194. https://doi.org/10.3390/microorganisms10061194







