Culturable Bacterial Diversity from the Basaltic Subsurface of the Young Volcanic Island of Surtsey, Iceland
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Site and Sample Collection
2.2. Media Preparation, Enrichment, and Strains Isolation
2.3. Identification of Isolates by 16S rRNA Gene Sequencing
2.4. Construction of Phylogenetic Trees
2.5. Bacterial Cultured Collection vs. 16S rRNA Amplicon Gene Sequencing
3. Results
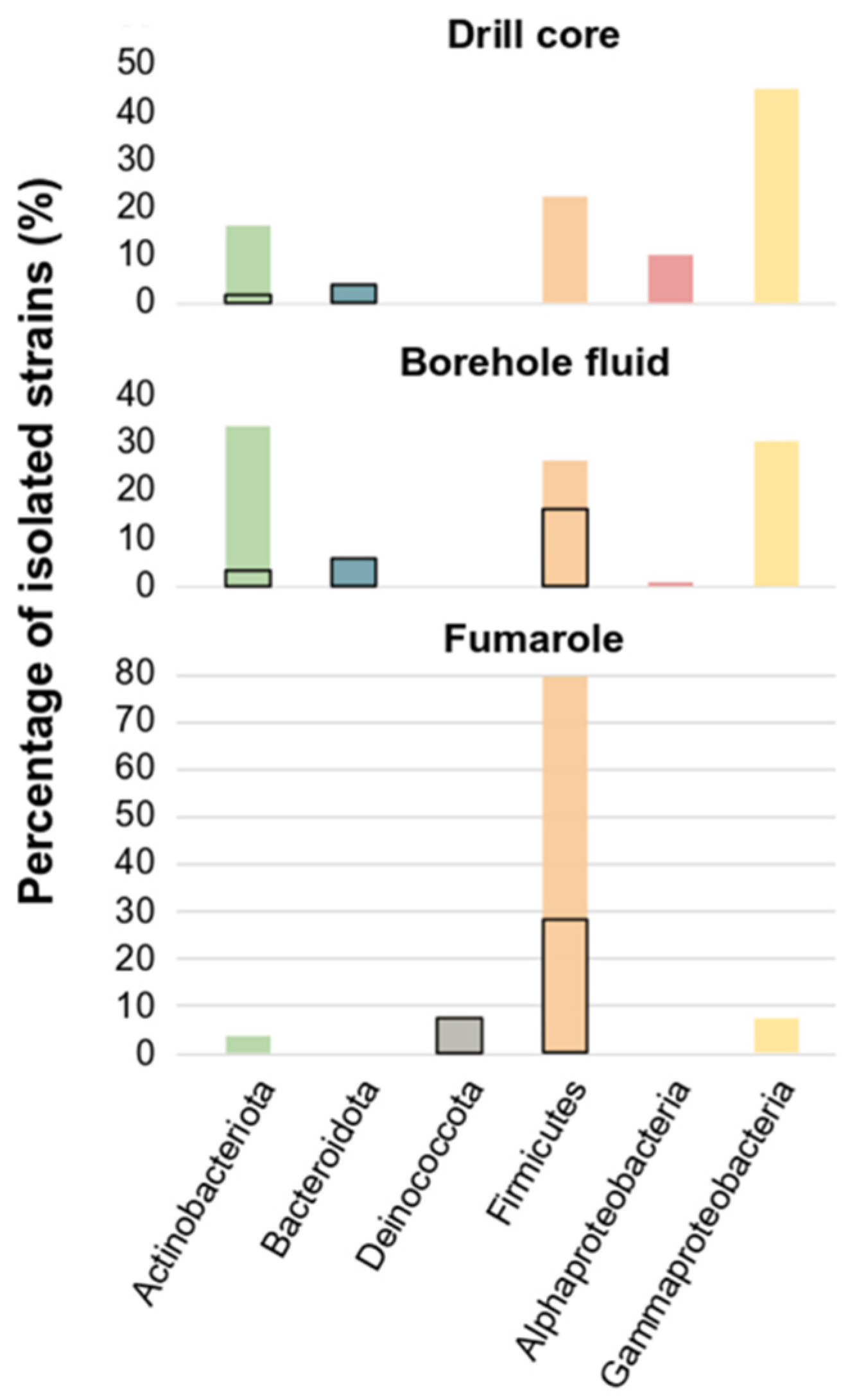
3.1. Cultivated Bacterial Diversity
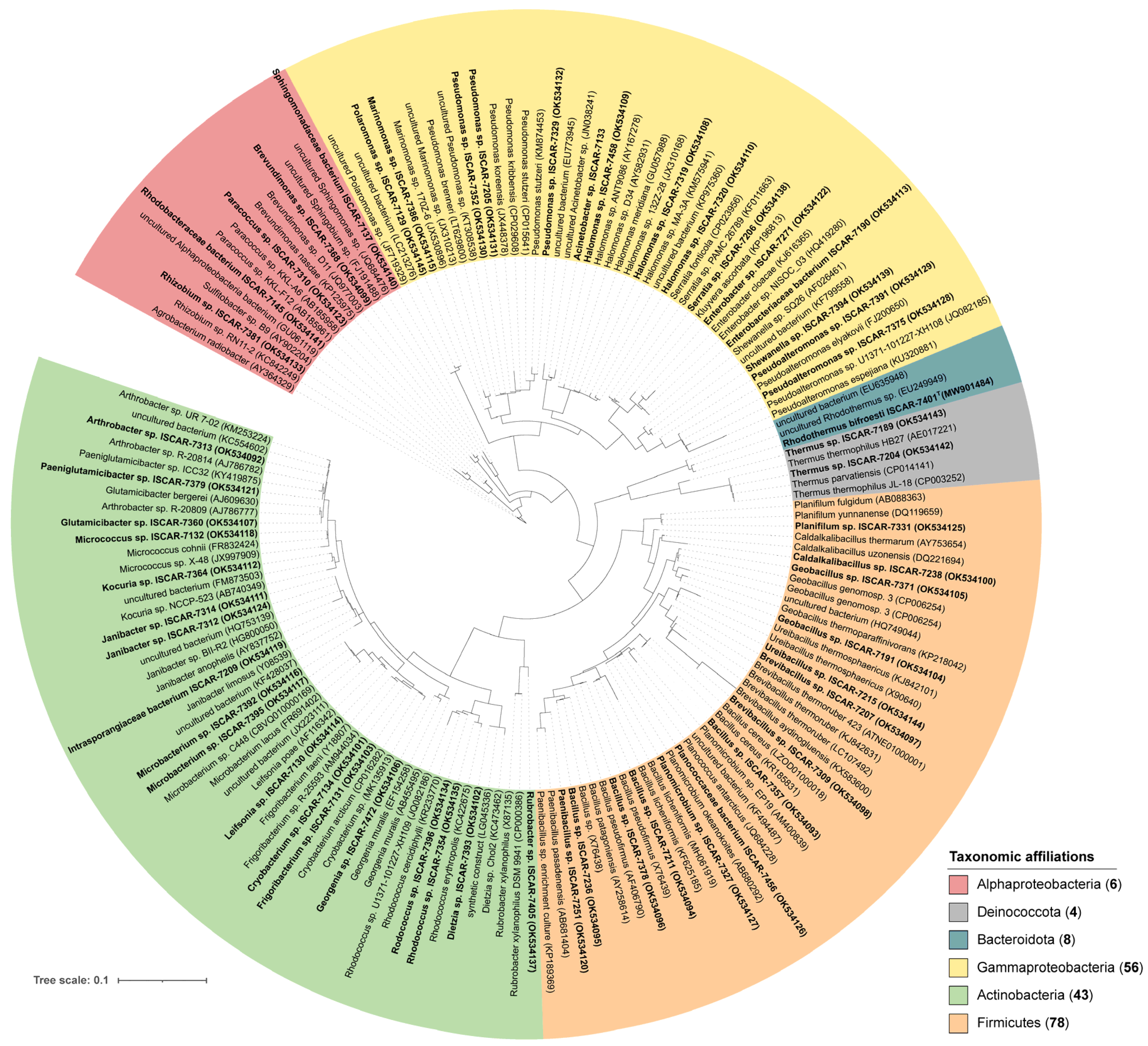
3.2. Phylogeny of the Isolates and Habitat of the Closest Relatives
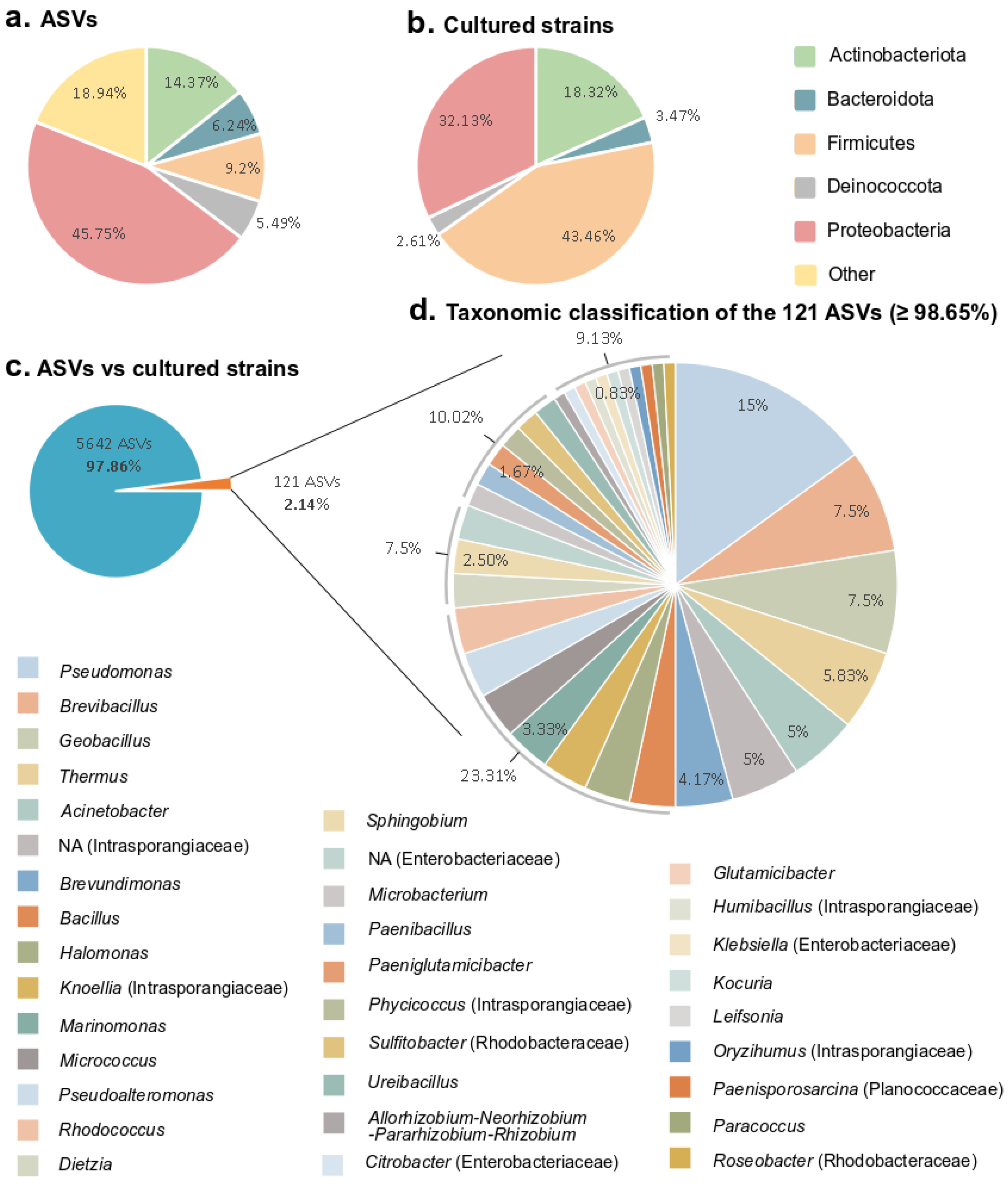
3.3. Comparison of the Bacterial Diversity Observed by Amplicon Sequencing from Environmental Samples and Cultured Diversity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Magnabosco, C.; Lin, L.H.; Dong, H.; Bomberg, M.; Ghiorse, W.; Stan-Lotter, H.; Pedersen, K.; Kieft, T.L.; van Heerden, E.; Onstott, T.C. The Biomass and Biodiversity of the Continental Subsurface. Nat. Geosci. 2018, 1, 707–717. [Google Scholar] [CrossRef]
- Huber, J.A.; Johnson, H.P.; Butterfield, D.A.; Baross, J.A. Microbial Life in Ridge Flank Crustal Fluids. Environ. Microbiol. 2006, 8, 88–99. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, S.; Inagaki, F.; Suzuki, Y.; Steinsbu, B.O.; Lever, M.A.; Takai, K.; Engelen, B.; Sako, Y.; Wheat, C.G.; Horikoshi, K. Microbial Community in Black Rust Exposed to Hot Ridge Flank Crustal Fluids. Appl. Environ. Microbiol. 2006, 72, 10. [Google Scholar] [CrossRef] [PubMed]
- Lever, M.A.; Rouxel, O.; Alt, J.C.; Shimizu, N.; Ono, S.; Coggon, R.M.; Shanks, W.C.; Lapham, L.; Elvert, M.; Prieto-Mollar, X.; et al. Evidence for Microbial Carbon and Sulfur Cycling in Deeply Buried Ridge Flank Basalt. Science 2013, 339, 6125. [Google Scholar] [CrossRef]
- Jungbluth, S.P.; Lin, H.T.; Cowen, J.P.; Glazer, B.T.; Rappé, M.S. Phylogenetic Diversity of Microorganisms in Subseafloor Crustal Fluids from Holes 1025C and 1026B along the Juan de Fuca Ridge Flank. Front. Microbiol. 2014, 5, 119. [Google Scholar] [CrossRef][Green Version]
- Robador, A.; Jungbluth, S.P.; LaRowe, D.E.; Bowers, R.M.; Rappé, M.S.; Amend, J.P.; Cowen, J.P. Activity and Phylogenetic Diversity of Sulfate-Reducing Microorganisms in Low-Temperature Subsurface Fluids within the Upper Oceanic Crust. Front. Microbiol. 2015, 5, 748. [Google Scholar] [CrossRef]
- Baquiran, J.P.M.; Ramírez, G.A.; Haddad, A.G.; Toner, B.M.; Hulme, S.; Wheat, C.G.; Edwards, K.J.; Orcutt, B.N. Temperature and Redox Effect on Mineral Colonization in Juan de Fuca Ridge Flank Subsurface Crustal Fluids. Front. Microbiol. 2016, 7, 396. [Google Scholar] [CrossRef]
- Cowen, J.P.; Giovannoni, S.J.; Kenig, F.; Johnson, H.P.; Butterfield, D.; Rappé, M.S.; Hutnak, M.; Lam, P. Fluids from Aging Ocean Crust That Support Microbial Life. Science 2003, 299, 120–123. [Google Scholar] [CrossRef]
- Orcutt, B.N.; Bach, W.; Becker, K.; Fisher, A.T.; Hentscher, M.; Toner, B.M.; Wheat, C.G.; Edwards, K.J. Colonization of Subsurface Microbial Observatories Deployed in Young Ocean Crust. ISME J. 2011, 5, 692–703. [Google Scholar] [CrossRef]
- Jungbluth, S.P.; Grote, J.; Lin, H.T.; Cowen, J.P.; Rappé, M.S. Microbial Diversity within Basement Fluids of the Sediment-Buried Juan de Fuca Ridge Flank. ISME J. 2013, 7, 161–172. [Google Scholar] [CrossRef]
- Jungbluth, S.P.; Bowers, R.M.; Lin, H.T.; Cowen, J.P.; Rappé, M.S. Novel Microbial Assemblages Inhabiting Crustal Fluids within Mid-Ocean Ridge Flank Subsurface Basalt. ISME J. 2016, 10, 2033–2047. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Rinke, C.; Chuvochina, M.; Chaumeil, P.-A.; Woodcroft, B.J.; Evans, P.N.; Hugenholtz, P.; Tyson, G.W. Recovery of Nearly 8,000 Metagenome-Assembled Genomes Substantially Expands the Tree of Life. Nat. Microbiol. 2017, 2, 1533–1542. [Google Scholar] [CrossRef] [PubMed]
- Lynch, M.D.J.; Neufeld, J.D. Ecology and Exploration of the Rare Biosphere. Nat. Rev. Microbiol. 2015, 13, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Hug, L.A.; Baker, B.J.; Anantharaman, K.; Brown, C.T.; Probst, A.J.; Castelle, C.J.; Butterfield, C.N.; Hernsdorf, A.W.; Amano, Y.; Ise, K.; et al. A New View of the Tree of Life. Nat. Microbiol. 2016, 1, 16048. [Google Scholar] [CrossRef]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A Communal Catalogue Reveals Earth’s Multiscale Microbial Diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef]
- Nunoura, T.; Chikaraishi, Y.; Izaki, R.; Suwa, T.; Sato, T.; Harada, T.; Mori, K.; Kato, Y.; Miyazaki, M.; Shimamura, S.; et al. A Primordial and Reversible TCA Cycle in a Facultatively Chemolithoautotrophic Thermophile. Science 2018, 359, 559–563. [Google Scholar] [CrossRef]
- Amann, R.I.; Ludwig, W.; Schleifer, K.H. Phylogenetic Identification and in Situ Detection of Individual Microbial Cells without Cultivation. Microbiol. Rev. 1995, 59, 143–169. [Google Scholar] [CrossRef]
- Hugenholtz, P.; Goebel, B.M.; Pace, N.R. Impact of Culture-Independent Studies on the Emerging Phylogenetic View of Bacterial Diversity. J. Bacteriol. 1998, 180, 4765–4774. [Google Scholar] [CrossRef]
- Rappé, M.S.; Giovannoni, S.J. The Uncultured Microbial Majority. Annu. Rev. Microbiol. 2003, 57, 369–394. [Google Scholar] [CrossRef]
- Steen, A.D.; Crits-Christoph, A.; Carini, P.; DeAngelis, K.M.; Fierer, N.; Lloyd, K.G.; Cameron Thrash, J. High Proportions of Bacteria and Archaea across Most Biomes Remain Uncultured. ISME J. 2019, 13, 3126–3130. [Google Scholar] [CrossRef]
- Thorarinsson, S.; Þórarinsson, S. The Surtsey Eruption: Course of Events and the Development of the New Island. Surtsey Res. Prog. Rep. 1965, 1, 51–55. [Google Scholar]
- Jakobsson, S.P.; Moore, J.G. The Surtsey Research Drilling Project of 1979. Surtsey Res. Prog. Rep. 1982, 9, 76–93. [Google Scholar]
- Jakobsson, S.P.; Thors, K.; Vésteinsson, Á.T.; Ásbjörnsdóttir, L. Some Aspects of the Seafloor Morphology at Surtsey Volcano: The New Multibeam Bathymetric Survey of 2007, Surtsey Research. Surtsey Res. Prog. Rep. 2009, 12, 9–20. [Google Scholar]
- Jakobsson, S.P.; Moore, J.G. Hydrothermal Minerals and Alteration Rates at Surtsey Volcano, Iceland. Geol. Soc. Am. Bull. 1986, 97, 648–659. [Google Scholar] [CrossRef]
- Takai, K.; Nakamura, K.; Toki, T.; Tsunogai, U.; Miyazaki, M.; Miyazaki, J.; Hirayama, H.; Nakagawa, S.; Nunoura, T.; Horikoshi, K. Cell Proliferation at 122 °C and Isotopically Heavy CH4 Production by a Hyperthermophilic Methanogen under High-Pressure Cultivation. Proc. Natl. Acad. Sci. USA 2008, 105, 10949–10954. [Google Scholar] [CrossRef] [PubMed]
- Ólafsson, M.; Jakobsson, S.P. Chemical Composition of Hydrothermal Water and Water-Rock Interactions on Surtsey Volcanic Island: A Preliminary Report. Surtsey Res. 2009, 12, 29–38. [Google Scholar]
- Marteinsson, V.; Klonowski, A.; Reynisson, E.; Vannier, P.; Sigurdsson, B.D.; Ólafsson, M. Microbial Colonization in Diverse Surface Soil Types in Surtsey and Diversity Analysis of Its Subsurface Microbiota. Biogeosciences 2015, 12, 1191–1203. [Google Scholar] [CrossRef]
- Jackson, M.D.; Couper, S.; Stan, C.V.; Ivarsson, M.; Czabaj, M.W.; Tamura, N.; Parkinson, D.; Miyagi, L.M.; Moore, J.G. Authigenic Mineral Texture in Submarine 1979 Basalt Drill Core, Surtsey Volcano, Iceland. Geochem. Geophys. Geosyst. 2019, 20, 3751–3773. [Google Scholar] [CrossRef]
- Jackson, M.D.; Gudmundsson, M.T.; Bach, W.; Cappelletti, P.; Coleman, N.J.; Ivarsson, M.; Jónasson, K.; Jørgensen, S.L.; Marteinsson, V.; McPhie, J.; et al. Time-Lapse Characterization of Hydrothermal Seawater and Microbial Interactions with Basaltic Tephra at Surtsey Volcano. Sci. Drill. 2015, 20, 51–58. [Google Scholar] [CrossRef]
- Jackson, M.D.; Gudmundsson, M.T.; Weisenberger, T.B.; Rhodes, J.M.; Stefánsson, A.; Kleine, B.I.; Lippert, P.C.; Marquardt, J.M.; Reynolds, H.I.; Kück, J.; et al. SUSTAIN Drilling at Surtsey Volcano, Iceland, Tracks Hydrothermal and Microbiological Interactions in Basalt 50 Years after Eruption. Sci. Drill. 2019, 25, 35–46. [Google Scholar] [CrossRef]
- Weisenberger, T.B.; Gudmundsson, M.T.; Jackson, M.D.; Gorny, C.F.; Türke, A.; Kleine, B.I.; Marshall, B.; Jørgensen, S.L.; Marteinsson, V.T.; Stefánsson, A.; et al. Operational Report for the 2017 Surtsey Underwater Volcanic System for Thermophiles, Alteration Processes and INnovative Concretes (SUSTAIN) Drilling Project at Surtsey Volcano, Iceland; GFZ German Research Centre for Geosciences: Potsdam, Germany, 2019; 240p. [Google Scholar] [CrossRef]
- Schipper, C.I.; Le Voyer, M.; Moussallam, Y.; White, J.D.L.; Thordarson, T.; Kimura, J.I.; Chang, Q. Degassing and Magma Mixing during the Eruption of Surtsey Volcano (Iceland, 1963–1967): The Signatures of a Dynamic and Discrete Rift Propagation Event. Bull. Volcanol. 2016, 78, 33. [Google Scholar] [CrossRef]
- Kleine, B.I.; Stefánsson, A.; Kjartansdóttir, R.; Prause, S.; Weisenberger, T.B.; Reynolds, H.I.; Sveinbjörnsdóttir, Á.E.; Jackson, M.D.; Gudmundsson, M.T. The Surtsey Volcano Geothermal System: An Analogue for Seawater-Oceanic Crust Interaction with Implications for the Elemental Budget of the Oceanic Crust. Chem. Geol. 2020, 550, 119702. [Google Scholar] [CrossRef]
- McPhie, J.; White, J.D.L.; Gorny, C.; Jackson, M.D.; Gudmundsson, M.T.; Couper, S. Lithofacies from the 1963-1967 Surtsey Eruption in SUSTAIN Drill Cores SE-2a, SE-2b and SE-03. Surtsey Res. 2020, 14, 85–90. [Google Scholar] [CrossRef]
- Moore, J.C.; Jackson, M.D. Observations on the Structure of Surtsey. Surtsey Res. 2020, 14, 33–45. [Google Scholar] [CrossRef]
- Prause, S.; Weisenberger, T.B.; Cappelletti, P.; Grimaldi, C.; Rispoli, C.; Jónasson, K.; Jackson, M.D.; Gudmundsson, M.T. Alteration Progress within the Surtsey Hydrothermal System, SW Iceland—A Time-Lapse Petrographic Study of Cores Drilled in 1979 and 2017. J. Volcanol. Geotherm. Res. 2020, 392, 106754. [Google Scholar] [CrossRef]
- Bergsten, P.; Vannier, P.; Klonowski, A.M.; Knobloch, S.; Gudmundsson, M.T.; Jackson, M.D.; Marteinsson, V.T. Basalt-Hosted Microbial Communities in the Subsurface of the Young Volcanic Island of Surtsey, Iceland. Front. Microbiol. 2021, 12, 2789. [Google Scholar] [CrossRef]
- Wolin, E.A.; Wolin, M.J.; Wolfe, R.S. Formation of methane by bacterial extracts. J. Biol. Chem. 1963, 238, 2882–2886. [Google Scholar] [CrossRef]
- Hjorleifsdottir, S.; Skirnisdottir, S.; Hreggvidsson, G.O.; Holst, O.; Kristjansson, J.K. Species Composition of Cultivated and Noncultivated Bacteria from Short Filaments in an Icelandic Hot Spring at 88 °C. Microb. Ecol. 2001, 42, 117–125. [Google Scholar] [CrossRef]
- Button, D.K.; Schut, F.; Quang, P.; Martin, R.; Robertson, B.R. Viability and Isolation of Marine Bacteria by Dilution Culture: Theory, Procedures, and Initial Results. Appl. Environ. Microbiol. 1993, 559, 881–891. [Google Scholar] [CrossRef]
- Walsh, P.S.; Metzger, D.A.; Higuchi, R. Chelex® 100 as a Medium for Simple Extraction of DNA for PCR-Based Typing from Forensic Material. BioTechniques 1991, 10, 506–513. [Google Scholar] [CrossRef]
- Skirnisdottir, S.; Hreggvidsson, G.O.; Hjörleifsdottir, S.; Marteinsson, V.T.; Petursdottir, S.K.; Holst, O.; Kristjansson, J.K. Influence of Sulfide and Temperature on Species Composition and Community Structure of Hot Spring Microbial Mats. Appl. Environ. Microbiol. 2000, 66, 2835–2841. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A Fast, Scalable and User-Friendly Tool for Maximum Likelihood Phylogenetic Inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Miller, M.A.; Pfeiffer, W.; Schwartz, T. Creating the CIPRES Science Gateway for Inference of Large Phylogenetic Trees. In Proceedings of the 2010 Gateway Computing Environments Workshop (GCE 2010), New Orleans, LA, USA, 14 November 2010. [Google Scholar]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-High-Throughput Microbial Community Analysis on the Illumina HiSeq and MiSeq Platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA Hybridization Values and Their Relationship to Whole-Genome Sequence Similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Richter, M.; Rosselló-Móra, R. Shifting the Genomic Gold Standard for the Prokaryotic Species Definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Oh, H.S.; Park, S.C.; Chun, J. Towards a Taxonomic Coherence between Average Nucleotide Identity and 16S RRNA Gene Sequence Similarity for Species Demarcation of Prokaryotes. Int. J. Syst. Evol. Microbiol. 2014, 64, 346–351. [Google Scholar] [CrossRef] [PubMed]
- Bergsten, P.; Vannier, P.; Mougeolle, A.; Rigaud, L.; Marteinsson, V.T. Rhodothermus Bifroesti Sp. Nov., a Thermophilic Bacterium Isolated from the Basaltic Subsurface of the Volcanic Island Surtsey. Int. J. Syst. Evol. Microbiol. 2022, 72, 005214. [Google Scholar] [CrossRef] [PubMed]
- Oshima, T.; Imahori, K. Description of Thermus Thermophilus (Yoshida and Oshima) Comb. Nov., a Nonsporulating Thermophilic Bacterium from a Japanese Thermal Spa. Int. J. Syst. Bacteriol. 1974, 24, 102. [Google Scholar] [CrossRef]
- Nazina, T.N.; Tourova, T.P.; Poltaraus, A.B.; Novikova, E.V.; Grigoryan, A.A.; Ivanova, A.E.; Lysenko, A.M.; Petrunyaka, V.V.; Osipov, G.A.; Belyaev, S.S.; et al. Taxonomic Study of Aerobic Thermophilic Bacilli. Int. J. Syst. Evol. Microbiol. 2001, 51, 433–446. [Google Scholar] [CrossRef]
- Elumalai, P.; Parthipan, P.; Narenkumar, J.; Anandakumar, B.; Madhavan, J.; Oh, B.T.; Rajasekar, A. Role of Thermophilic Bacteria (Bacillus and Geobacillus) on Crude Oil Degradation and Biocorrosion in Oil Reservoir Environment. 3 Biotech 2019, 9, 79. [Google Scholar] [CrossRef]
- Zhang, Y.X.; Dong, C.; Biao, S. Planifilum Yunnanense Sp. Nov., a Thermophilic Thermoactinomycete Isolated from a Hot Spring. Int. J. Syst. Evol. Microbiol. 2007, 57, 1851–1854. [Google Scholar] [CrossRef]
- Zhao, W.; Zhang, C.L.; Romanek, C.S.; Wiegel, J. Description of Caldalkalibacillus Uzonensis Sp. Nov. and Emended Description of the Genus Caldalkalibacillus. Int. J. Syst. Evol. Microbiol. 2008, 58, 1106–1108. [Google Scholar] [CrossRef]
- Duckworth, A.W.; Grant, S.; Grant, W.D.; Jones, B.E.; Meijer, D. Dietzia Natronolimnaios Sp. Nov., a New Member of the Genus Dietzia Isolated from an East African Soda Lake. Extremophiles 1998, 2, 359–366. [Google Scholar] [CrossRef]
- Patel, S.; Gupta, R.S. A Phylogenomic and Comparative Genomic Framework for Resolving the Polyphyly of the Genus Bacillus: Proposal for Six New Genera of Bacillus Species, Peribacillus Gen. Nov., Cytobacillus Gen. Nov., Mesobacillus Gen. Nov., Neobacillus Gen. Nov., Metabacillu. Int. J. Syst. Evol. Microbiol. 2020, 70, 406–438. [Google Scholar] [CrossRef] [PubMed]
- Ettoumi, B.; Chouchane, H.; Guesmi, A.; Mahjoubi, M.; Brusetti, L.; Neifar, M.; Borin, S.; Daffonchio, D.; Cherif, A. Diversity, Ecological Distribution and Biotechnological Potential of Actinobacteria Inhabiting Seamounts and Non-Seamounts in the Tyrrhenian Sea. Microbiol. Res. 2016, 180, 366–376. [Google Scholar] [CrossRef] [PubMed]
- Hugenholtz, P.; Pitulle, C.; Hershberger, K.L.; Pace, N.R. Novel Division Level Bacterial Diversity in a Yellowstone Hot Spring. J. Bacteriol. 1998, 186–187, 71–80. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, Y.; Pan, J.; Wang, F.; Li, M. Perspectives on Cultivation Strategies of Archaea. Microb. Ecol. 2020, 73, 770–784. [Google Scholar] [CrossRef]
- Hahn, M.W.; Koll, U.; Schmidt, J. Isolation and Cultivation of Bacteria. In The Structure and Function of Aquatic Microbial Communities; Springer: Cham, Switzerland, 2019. [Google Scholar]
- Hungate, R.E. The Anaerobic Mesophilic Cellulolytic Bacteria. Bacteriol. Rev. 1950, 14, 1–49. [Google Scholar] [CrossRef] [PubMed]
- Clark, H. Culturing Anaerobes. Nat. Res. 2019, 2019. [Google Scholar]
- Cho, J.C.; Giovannoni, S.J. Cultivation and Growth Characteristics of a Diverse Group of Oligotrophic Marine Gammaproteobacteria. Appl. Environ. Microbiol. 2004, 70, 432–440. [Google Scholar] [CrossRef]
- D’Hondt, S.; Pockalny, R.; Fulfer, V.M.; Spivack, A.J. Subseafloor Life and Its Biogeochemical Impacts. Nat. Commun. 2019, 10, 1–13. [Google Scholar] [CrossRef]
- Demko, A.M.; Patin, N.V.; Jensen, P.R. Microbial Diversity in Tropical Marine Sediments Assessed Using Culture-Dependent and Culture-Independent Techniques. Environ. Microbiol. 2021, 23, 6859–6875. [Google Scholar] [CrossRef]
- Tang, L. Culturing Uncultivated Bacteria. Nat. Methods 2019, 16, 1078. [Google Scholar] [CrossRef]
- Jung, D.; Liu, L.; He, S. Application of in Situ Cultivation in Marine Microbial Resource Mining. Mar. Life Sci. Technol. 2021, 3, 148–161. [Google Scholar] [CrossRef]
- Jung, D.; Machida, K.; Nakao, Y.; Kindaichi, T.; Ohashi, A.; Aoi, Y. Triggering Growth via Growth Initiation Factors in Nature: A Putative Mechanism for in Situ Cultivation of Previously Uncultivated Microorganisms. Front. Microbiol. 2021, 12, 1107. [Google Scholar] [CrossRef] [PubMed]
- Berdy, B.; Spoering, A.L.; Ling, L.L.; Epstein, S.S. In Situ Cultivation of Previously Uncultivable Microorganisms Using the Ichip. Nat. Protoc. 2017, 12, 2232–2242. [Google Scholar] [CrossRef] [PubMed]
- Crespo, B.G.; Wallhead, P.J.; Logares, R.; Pedrós-Alió, C. Probing the Rare Biosphere of the North-West Mediterranean Sea: An Experiment with High Sequencing Effort. PLoS ONE 2016, 11, e0159195. [Google Scholar] [CrossRef] [PubMed]
- Pédron, J.; Guyon, L.; Lecomte, A.; Blottière, L.; Chandeysson, C.; Rochelle-Newall, E.; Raynaud, X.; Berge, O.; Barny, M.A. Comparison of Environmental and Culture-Derived Bacterial Communities through 16S Metabarcoding: A Powerful Tool to Assess Media Selectivity and Detect Rare Taxa. Microorganisms 2020, 8, 1129. [Google Scholar] [CrossRef]
- Kallmeyer, J. Contamination Control for Scientific Drilling Operations. Adv. Appl. Microbiol. 2017, 98, 61–91. [Google Scholar] [CrossRef]
- Sheik, C.S.; Reese, B.K.; Twing, K.I.; Sylvan, J.B.; Grim, S.L.; Schrenk, M.O.; Sogin, M.L.; Colwell, F.S. Identification and Removal of Contaminant Sequences from Ribosomal Gene Databases: Lessons from the Census of Deep Life. Front. Microbiol. 2018, 9, 840. [Google Scholar] [CrossRef]
- Orsi, W.D.; Edgcomb, V.P.; Christman, G.D.; Biddle, J.F. Gene Expression in the Deep Biosphere. Nature 2013, 499, 205–208. [Google Scholar] [CrossRef]
- Inagaki, F.; Hinrichs, K.U.; Kubo, Y.; Bowles, M.W.; Heuer, V.B.; Hong, W.L.; Hoshino, T.; Ijiri, A.; Imachi, H.; Ito, M.; et al. Exploring Deep Microbial Life in Coal-Bearing Sediment down to ~2.5 Km below the Ocean Floor. Science 2015, 349, 420–424. [Google Scholar] [CrossRef]
- Reese, B.K.; Zinke, L.A.; Sobol, M.S.; LaRowe, D.E.; Orcutt, B.N.; Zhang, X.; Jaekel, U.; Wang, F.; Dittmar, T.; Defforey, D.; et al. Nitrogen Cycling of Active Bacteria within Oligotrophic Sediment of the Mid-Atlantic Ridge Flank. Geomicrobiol. J. 2018, 35, 468–483. [Google Scholar] [CrossRef]
- Salter, S.J.; Cox, M.J.; Turek, E.M.; Calus, S.T.; Cookson, W.O.; Moffatt, M.F.; Turner, P.; Parkhill, J.; Loman, N.J.; Walker, A.W. Reagent and Laboratory Contamination Can Critically Impact Sequence-Based Microbiome Analyses. BMC Biol. 2014, 12, 87. [Google Scholar] [CrossRef] [PubMed]
- Degryse, E.; Glansdorff, N.; Piérard, A. A Comparative Analysis of Extreme Thermophilic Bacteria Belonging to the Genus Thermus. Arch. Microbiol. 1978, 117, 189–196. [Google Scholar] [CrossRef] [PubMed]

| Sample ID | Sample Type | Sampling Date | Collection Depth (m b.s.) | Collection Temperature (°C) |
|---|---|---|---|---|
| 16.2 | Borehole fluid | 9 June 2016 | 166 | 54 |
| 16.7 | Borehole fluid | 9 June 2016 | 160 | 60 |
| 16.8 | Borehole fluid | 9 June 2016 | mix | n.a. |
| 17.1 | Borehole fluid | 3 August 2017 | 58 | 85 |
| 17.2 | Borehole fluid | 3 August 2017 | 120 | 116 |
| 17.3 | Borehole fluid | 3 August 2017 | 150 | 76 |
| 17.4 | Borehole fluid | 3 August 2017 | 160 | 52 |
| 17.5 | Fumarole | 5 August 2017 | 0 | 64.2–82.3 |
| 17.6 | Fumarole | 4 August 2017 | 0 | 40.8 |
| 17.8, 17.9, 17.F | Fumarole | 4 August 2017 | 0 | 56.1–74.6 |
| 17.11 | Borehole fluid | 6 September 2017 | 140 | 116 |
| 17.13 | Borehole fluid | 6 September 2017 | 280 | 58 |
| 17.14 | Borehole fluid | 6 September 2017 | mix | n.a. |
| 17.15 | Borehole fluid | 6 September 2017 | 75 | 98 |
| 17.16 | Borehole fluid | 5 September 2017 | 60 | 90 |
| 17.17 | Borehole fluid | 5 September 2017 | 80 | 116 |
| 17.18 | Borehole fluid | 5 September 2017 | 90 | 122 |
| 17.19 | Borehole fluid | 5 September 2017 | 100 | 124 |
| 17.22 | Borehole fluid | 5 September 2017 | 160 | 61 |
| 17.23 | Borehole fluid | 5 September 2017 | mix | n.a. |
| 18.1 | Borehole fluid | 19 September 2018 | mix | n.a. |
| 18.2 | Borehole fluid | 19 September 2018 | mix | n.a. |
| 18.3 | Borehole fluid | 19 September 2018 | mix | n.a. |
| B3 | Drill core | 10 August 2017 | 15 | 15.3 |
| B9 | Drill core | 11 August 2017 | 32 | 30 |
| B24 | Drill core | 12 August 2017 | 70 | 109 |
| B30 | Drill core | 13 August 2017 | 87 | 121 |
| B36 | Drill core | 14 August 2017 | 105 | 123 |
| C55 | Drill core | 25 August 2017 | 156 | 64 |
| C59 | Drill core | 25 August 2017 | 167 | 55 |
| C62 | Drill core | 25 August 2017 | 176 | 44.5 |
| C65 | Drill core | 25 August 2017 | 181 | 37 |
| Phylogenetic Phylum or Class | Family or Genus | Sample Origin | Culture Conditions | Number of Strains Isolated | Borehole Fluid | Fumarole | Drill Core |
|---|---|---|---|---|---|---|---|
| Actinobacteriota | Arthrobacter | 17.9, 18.1 | 166, O2, 22 °C | 3 | 1 | 2 | 0 |
| Cryobacterium | 16.8 | M, O2, 40 °C | 1 | 1 | 0 | 0 | |
| Frigoribacterium | 16.8 | M, O2, 40 °C | 1 | 1 | 0 | 0 | |
| Microbacterium | B3, C59 | 166, MB, SO, O2, 22 °C | 4 | 0 | 0 | 4 | |
| Dietzia | B9 | MB, O2, 22 °C | 1 | 0 | 0 | 1 | |
| Georgenia | B24 | MB, O2, 22 °C | 1 | 0 | 0 | 1 | |
| Glutamicibacter | 17.16 | 166 and MB, O2, 22 °C | 4 | 4 | 0 | 0 | |
| Janibacter | 17.3, 16.7 | 166, O2, 22 °C | 4 | 4 | 0 | 0 | |
| Leifsonia | 16.8 | M, O2, 40 °C | 1 | 1 | 0 | 0 | |
| Intrasporangiaceae | 17.4 | 166, O2, 22 °C | 2 | 2 | 0 | 0 | |
| Kocuria | 17.15, 17.16 | 166, O2, 22 °C | 2 | 2 | 0 | 0 | |
| Micrococcus | 16.8, B3 | M and SO, O2, 22 and 40 °C | 2 | 1 | 0 | 1 | |
| Paeniglutamicibacter | 17.17, 17.19, 18.2, 18.3 | 166 and MB, O2, 22 °C | 7 | 7 | 0 | 0 | |
| Rhodococcus group 1 | 17.17 | MB, O2, 22 °C | 1 | 1 | 0 | 0 | |
| Rhodococcus group 2 | 17.2, 17.17, 17.22, 18.1 | 166 and MB, O2, 22 °C | 5 | 5 | 0 | 0 | |
| Rubrobacter | 17.15, 17.2, 17.22, B24 | 166, O2, 60 °C | 4 | 3 | 0 | 1 | |
| 43 | 33 | 2 | 8 | ||||
| Bacteroidota | Rhodothermus | 17.15, 17.2, 17.22, B24 | 166, O2, 60 °C | 8 | 6 | 0 | 2 |
| 8 | 6 | 0 | 2 | ||||
| Deinococcota | Thermus | 17.5, 17.8, 17.9 | 166, O2, 80 °C | 4 | 0 | 4 | 0 |
| 4 | 0 | 4 | 0 | ||||
| Firmicutes | Bacillus group 1 | 17.11, 17.14, 17.15, B9 | MB, O2, 22 °C | 5 | 5 | 0 | 0 |
| Bacillus group 2 | 17.1 | 166, O2, 22 °C | 1 | 0 | 0 | 1 | |
| Bacillus (para)licheniformis | 17.2, 17.5, 17.8, 17.9, 17.F | 166, with and without O2, 22, 50 and 60 °C | 15 | 2 | 13 | 0 | |
| Bacillus cereus group | 17.8, 17.15, 17.F, C55, C65 | 166, with and without O2, 22 and 37 °C | 5 | 1 | 2 | 2 | |
| Brevibacillus | 17.8 | 166, O2, 60 °C | 2 | 0 | 2 | 0 | |
| Brevibacillus thermoruber | 17.1, 17.5, 17.9 | 166, O2, 60 °C | 4 | 1 | 3 | 0 | |
| Caldalkalibacillus | 17.1, 17.4 | 166, O2, 60 °C | 6 | 6 | 0 | 0 | |
| Geobacillus | 17.16 | 166, O2, 22 and 60 °C | 4 | 4 | 0 | 0 | |
| Geobacillus thermoleovorans group | 17.5, 17.8, 17.F | 166, O2, 60 and 65 °C | 20 | 2 | 18 | 0 | |
| Paenibacillus | B3, C55, C65 | 166, O2, 22 °C | 8 | 0 | 0 | 8 | |
| Planifilum | 18.3 | 166, O2, 60 °C | 1 | 1 | 0 | 0 | |
| Planococcaceae | 18.3 | MB, O2, 22 °C | 1 | 1 | 0 | 0 | |
| Planomicrobium | 18.3 | 166, O2, 22 °C | 2 | 2 | 0 | 0 | |
| Ureibacillus | 17.4, 17.6 | 166, O2, 60 °C | 4 | 1 | 3 | 0 | |
| 78 | 26 | 41 | 11 | ||||
| Alpha-proteobacteria | Brevundimonas | 17.16 | 166, O2, 22 °C | 1 | 1 | 0 | 0 |
| Paracoccus | C65 | MB, O2, 22 °C | 1 | 0 | 0 | 1 | |
| Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium | B9 | MB, O2, 22 °C | 2 | 0 | 0 | 2 | |
| Sphingomonadaceae | B3 | YPS, O2, 22 °C | 1 | 0 | 0 | 1 | |
| Rhodobacteraceae | B3 | SO, O2, 22 °C | 1 | 0 | 0 | 1 | |
| 6 | 1 | 0 | 5 | ||||
| Gamma-proteobacteria | Acinetobacter | 16.8 | M, O2, 40 °C | 1 | 1 | 0 | 0 |
| Halomonas | 17.15, 17.23, 18.2, 18.3 | 166 and MB, O2, 22 °C | 11 | 11 | 0 | 0 | |
| Marinomonas | 17.13, 17.15 | 166 and MB, O2, 22 °C | 2 | 2 | 0 | 0 | |
| Enterobacter | 16.2, 16.7, 17.5, B3, C55, C65 | 166, M, I and SO, without O2, 22 °C | 18 | 5 | 1 | 12 | |
| Enterobacteriaceae | 17.5 | 166, O2, 22 °C | 1 | 0 | 1 | 0 | |
| Pseudoalteromonas | 17.23 | 166 and MB, O2, 22 °C | 7 | 7 | 0 | 0 | |
| Pseudomonas group 1 | B30, B36, B9, C62 | 166 and MB, O2, 22 °C | 8 | 0 | 0 | 8 | |
| Pseudomonas group 2 | 18.2, 18.3 | 166, O2, 22 °C | 3 | 3 | 0 | 0 | |
| Pseudomonas group 3 | 17.8 | 166, O2, 22 °C | 1 | 0 | 1 | 0 | |
| Serratia | 17.5 | 166, O2, 22 °C | 1 | 0 | 1 | 0 | |
| Shewanella | B9 | MB, O2, 22 °C | 2 | 0 | 0 | 2 | |
| Polaromonas | 16.8 | M, O2, 40 °C | 1 | 1 | 0 | 0 | |
| 56 | 30 | 4 | 22 | ||||
| Total number of isolated strains | 195 | 96 | 51 | 48 |
| Phylogenetic Class | Genus | F | BF | DC | Control | Closest Sequence Similarity Percentage (Megablast) | Isolated From |
|---|---|---|---|---|---|---|---|
| Actinobacteriota | Arthrobacter | 0 | 1 | 0 | 0 | 97.794 | BF and F |
| Dietzia | 0 | 2 | 2 | 0 | 99.265–100 | DC | |
| Georgenia | 0 | 0 | 0 | 0 | DC | ||
| Glutamicibacter | 0 | 0 | 0 | 1 | 100 | BF | |
| Intrasporangiaceae | 3 | 7 | 6 | 1 | 100 | BF | |
| Kocuria | 0 | 1 | 1 | 0 | 99.259 | BF | |
| Leifsonia | 0 | 2 | 2 | 0 | 98.684 | BF | |
| Microbacterium lacus | 0 | 0 | 2 | 1 | 99.029 | DC | |
| Micrococcus | 1 | 2 | 4 | 2 | 99.457–100 | BF and DC | |
| Paeniglutamicibacter | 1 | 1 | 2 | 0 | 100 | BF | |
| Rhodococcus group 1 | 1 | 1 | 2 | 1 | 100 | BF | |
| Rhodococcus group 2 | 1 | 2 | 2 | 0 | 100 | BF | |
| Rubrobacter | 2 | 0 | 1 | 0 | 93.605 | BF and DC | |
| Bacteroidota | Rhodothermus | 0 | 2 | 0 | 0 | 95.588 | BF and DC |
| Deinococcota | Thermus | 0 | 4 | 5 | 1 | 98.693–100 | F |
| Firmicutes | Bacillus (para)licheniformis | 0 | 1 | 3 | 0 | 99.495–100 | BF and F |
| Bacillus cereus | 2 | 2 | 2 | 1 | 100 | BF, F and DC | |
| Bacillus group 1 | 0 | 0 | 1 | 0 | 97.024 | BF and DC | |
| Bacillus group 2 | 0 | 0 | 0 | 0 | / | BF | |
| Brevibacillus | 0 | 1 | 8 | 0 | 99.052–100 | BF and F | |
| Caldalkalibacillus | 0 | 2 | 0 | 1 | 98.529 | BF | |
| Geobacillus | 0 | 2 | 2 | 0 | 99.074–99.537 | BF | |
| Geobacillus thermoleovorans group | 0 | 2 | 5 | 0 | 99.487–100 | F | |
| Paenibacillus | 0 | 0 | 2 | 0 | 100 | DC | |
| Planifilum | 0 | 0 | 0 | 0 | / | BF | |
| Planococcaceae | 0 | 2 | 2 | 0 | 97.674 | BF | |
| Planomicrobium | 0 | 1 | 0 | 0 | 98.897 | BF | |
| Ureibacillus | 0 | 0 | 2 | 0 | 100 | F and BF | |
| Alphaproteobacteria | Allorhizobium-Neorhizobium-Pararhizobium-Rhizobium | 0 | 1 | 1 | 0 | 99.034 | DC |
| Brevundimonas | 1 | 1 | 5 | 1 | 99.457–100 | BF | |
| Paracoccus | 0 | 0 | 1 | 0 | 100 | DC | |
| Rhodobacteraceae | 0 | 0 | 3 | 0 | 100 | DC | |
| Sphingobium | 2 | 1 | 1 | 0 | 99.425–100 | DC | |
| Gammaproteobacteria | Acinetobacter | 4 | 4 | 5 | 2 | 99.254–100 | BF |
| Enterobacteriaceae | 2 | 3 | 4 | 2 | 99.533–100 | F, BF and DC | |
| Halomonas | 0 | 4 | 0 | 0 | 99.265–100 | BF | |
| Marinomonas | 0 | 1 | 3 | 0 | 99.052–100 | BF | |
| Polaromonas | 0 | 2 | 3 | 1 | 98.276 | BF | |
| Pseudoalteromonas | 2 | 4 | 4 | 1 | 100 | BF | |
| Pseudomonas group 1 | 2 | 7 | 3 | 0 | 99.306–100 | DC | |
| Pseudomonas group 2 | 0 | 2 | 2 | 2 | 98.529–98.897 | F | |
| Pseudomonas group 3 | 3 | 4 | 7 | 0 | 98.907–100 | BF | |
| Serratia | 0 | 0 | 0 | 1 | 97.619 | F | |
| Shewanella | 0 | 0 | 1 | 0 | 96.691 | DC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bergsten, P.; Vannier, P.; Frion, J.; Mougeolle, A.; Marteinsson, V.Þ. Culturable Bacterial Diversity from the Basaltic Subsurface of the Young Volcanic Island of Surtsey, Iceland. Microorganisms 2022, 10, 1177. https://doi.org/10.3390/microorganisms10061177
Bergsten P, Vannier P, Frion J, Mougeolle A, Marteinsson VÞ. Culturable Bacterial Diversity from the Basaltic Subsurface of the Young Volcanic Island of Surtsey, Iceland. Microorganisms. 2022; 10(6):1177. https://doi.org/10.3390/microorganisms10061177
Chicago/Turabian StyleBergsten, Pauline, Pauline Vannier, Julie Frion, Alan Mougeolle, and Viggó Þór Marteinsson. 2022. "Culturable Bacterial Diversity from the Basaltic Subsurface of the Young Volcanic Island of Surtsey, Iceland" Microorganisms 10, no. 6: 1177. https://doi.org/10.3390/microorganisms10061177
APA StyleBergsten, P., Vannier, P., Frion, J., Mougeolle, A., & Marteinsson, V. Þ. (2022). Culturable Bacterial Diversity from the Basaltic Subsurface of the Young Volcanic Island of Surtsey, Iceland. Microorganisms, 10(6), 1177. https://doi.org/10.3390/microorganisms10061177







