Application of Organic Fertilizer Changes the Rhizosphere Microbial Communities of a Gramineous Grass on Qinghai–Tibet Plateau
Abstract
:1. Introduction
2. Methods and Materials
2.1. Study Sites and Organic Fertilizer Treatment
2.2. Vegetation Measurements and Rhizosphere Soil Sampling
2.3. DNA Extraction and PCR Amplification
2.4. Processing of the Sequencing Data
2.5. Statistical Analysis
3. Results
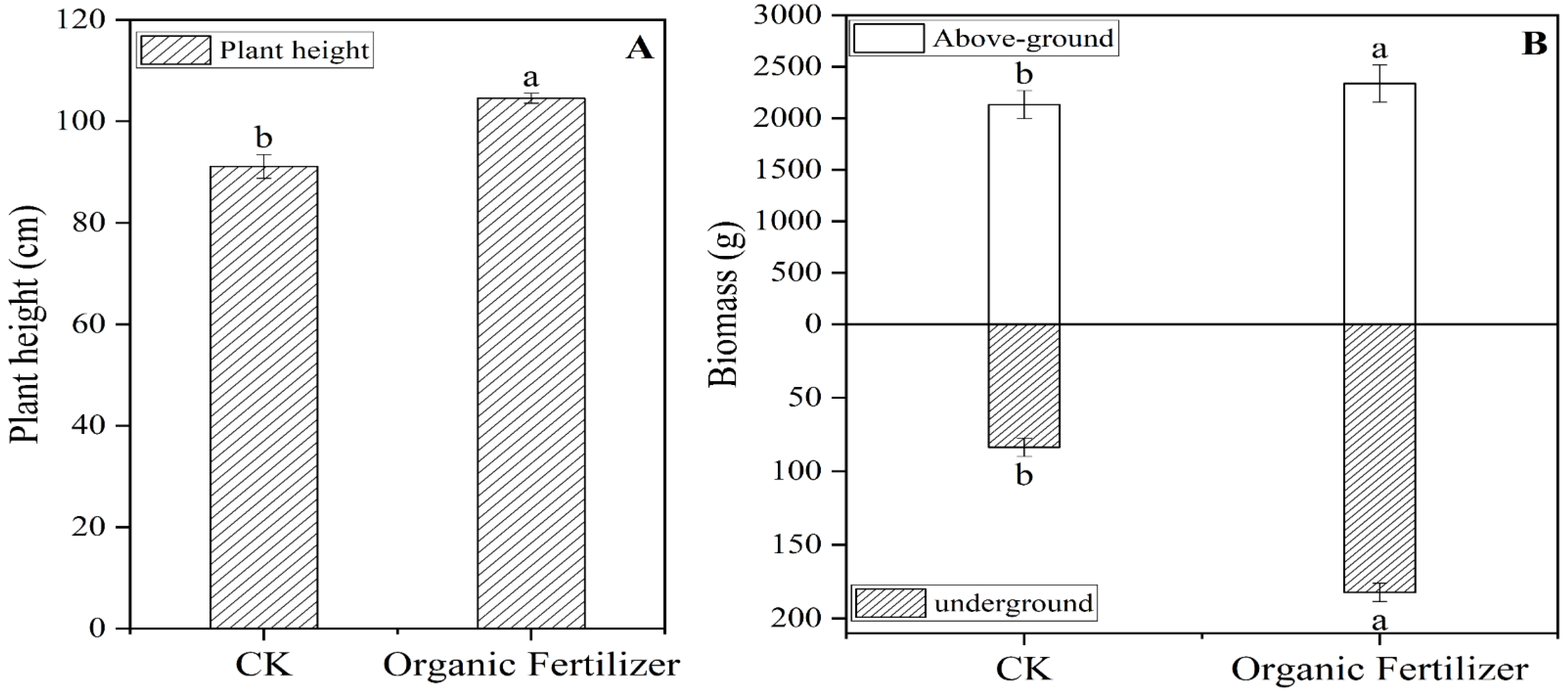
3.1. Application of Organic Fertilizer Increase the Forage Height and Biomass of Forage
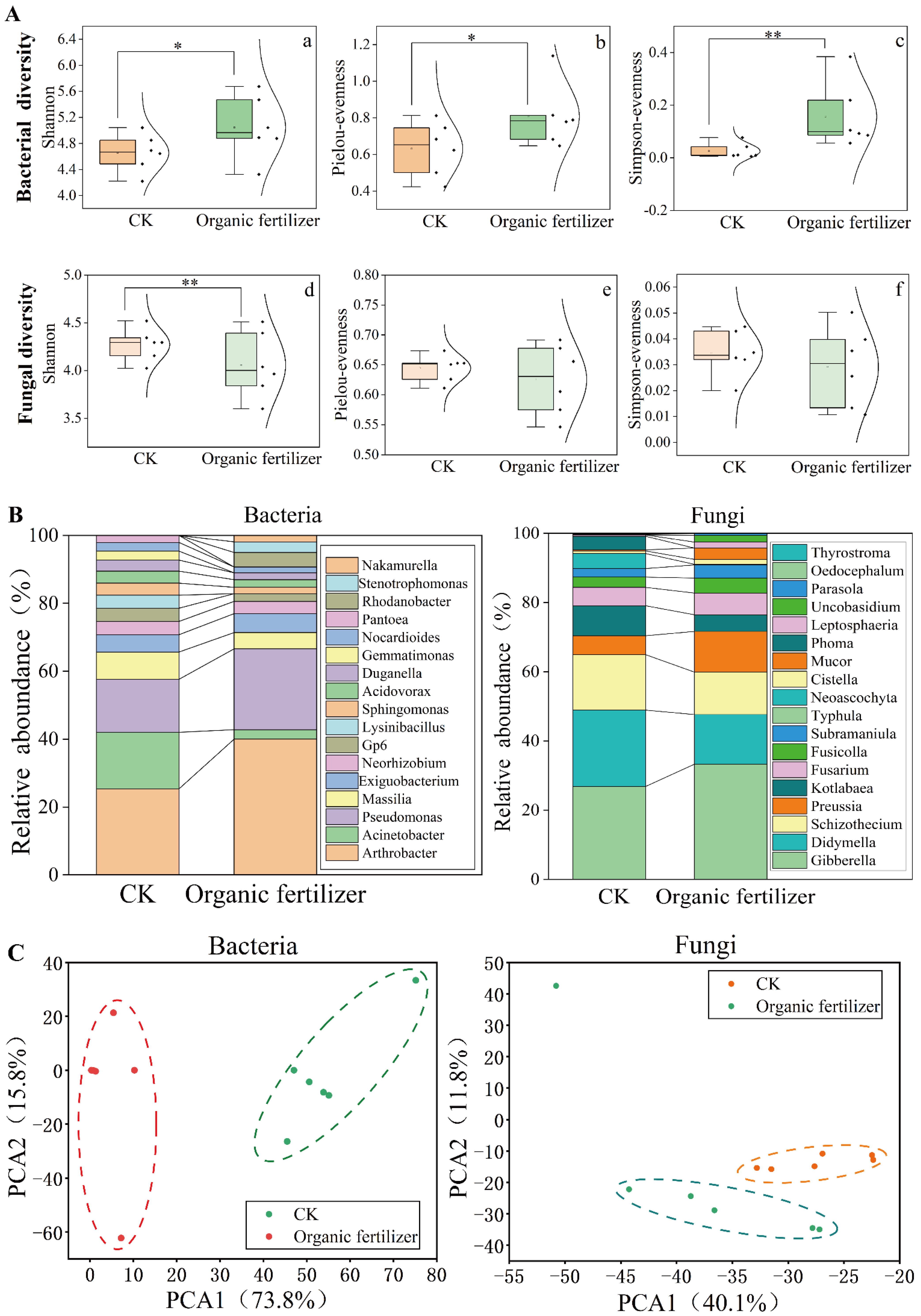
3.2. Organic Fertilizer Treatment Changed the Diversity, Composition, and Structure of Rhizosphere Soil Microbial Community
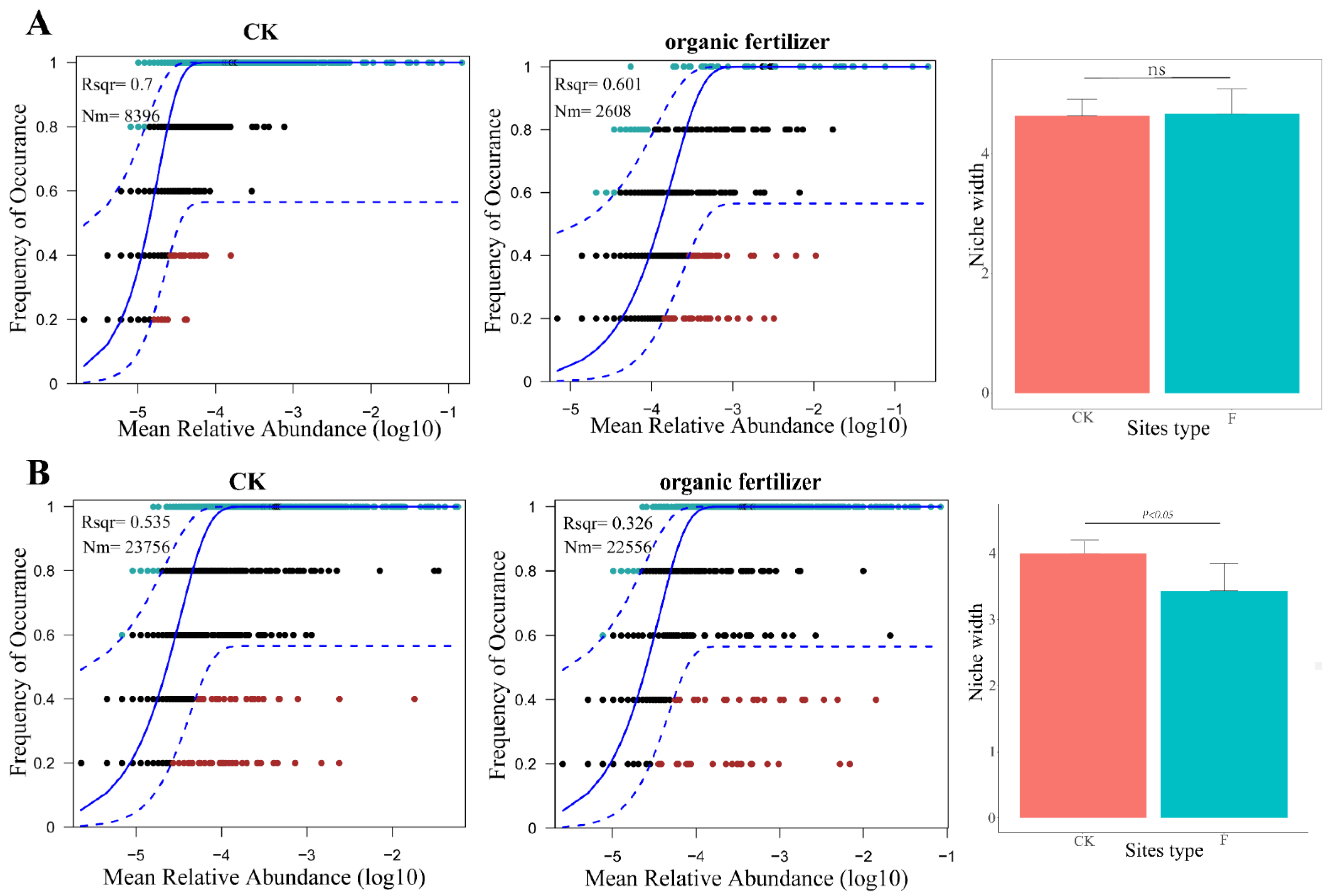
3.3. Variations of Ecological Processes in the Microbial Community Assembly under Organic Fertilizer Treatment
3.4. Effects of Fertilizer Treatments on Rhizosphere Microbial Biomarkers
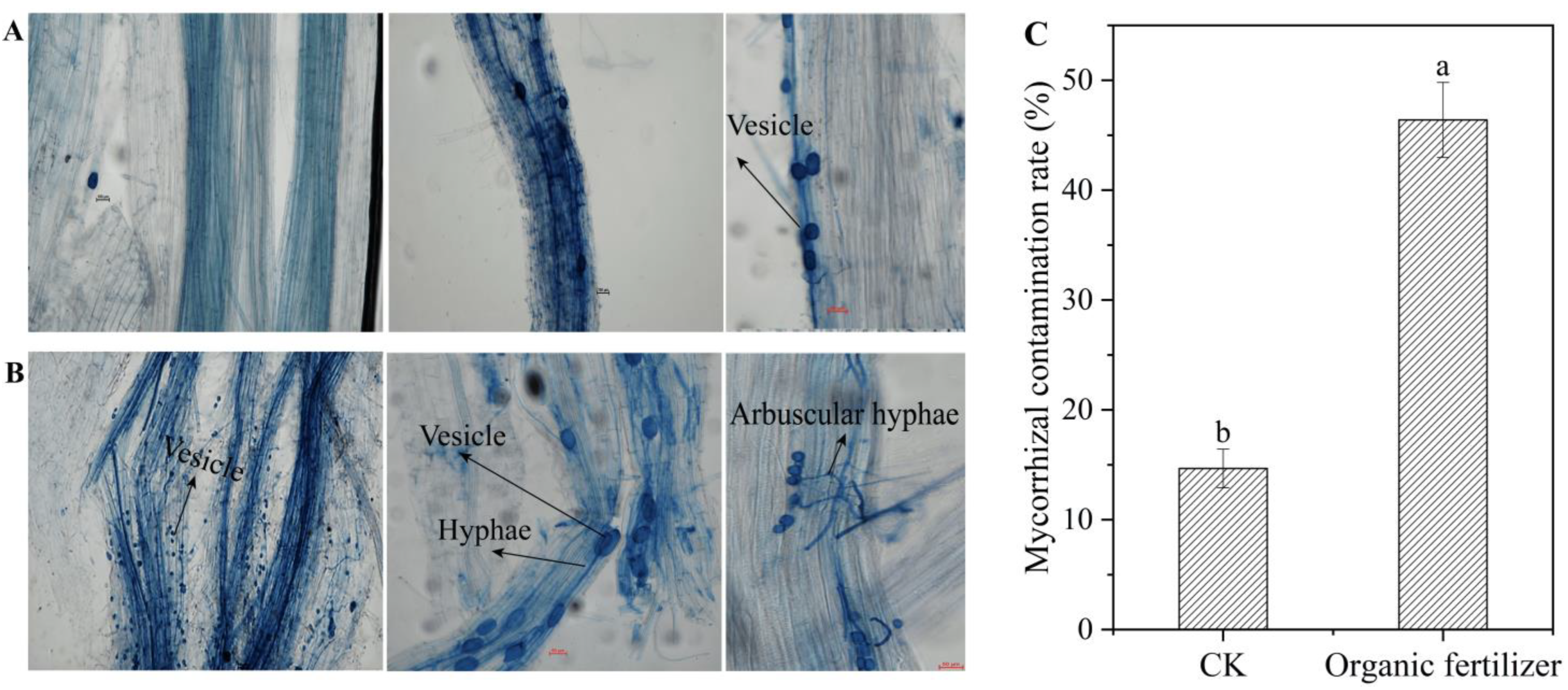
3.5. Validation of the Effect of Organic Fertilizers on the Production of Mycorrhizal Fungi Using Arbuscular Mycorrhizal Fungi (AMF)
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Jiang, Y.; Luan, L.; Hu, K.; Liu, M.; Chen, Z.; Geisen, S.; Sun, B. Trophic interactions as determinants of the arbuscular mycorrhizal fungal community with cascading plant-promoting consequences. Microbiome 2020, 8, 142. [Google Scholar] [CrossRef]
- Li, H.; Song, W. Spatiotemporal Distribution and Influencing Factors of Ecosystem Vulnerability on Qinghai-Tibet Plateau. Int. J. Environ. Res. Public Health 2021, 18, 6508. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Wang, X.; Guo, Q. Pyrosequencing investigation into the Prokaryotes communities in the Qinghai-Tibet Plateau soils associated with soil characteristic factors. Icel. Agric. Sci. 2020, 33, 57–71. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, G.; Yu, H.; Du, X.; He, Q.; Yao, S.; Zhao, L.; Huang, C.; Wen, X.; Deng, Y. Meadow degradation increases spatial turnover rates of the fungal community through both niche selection and dispersal limitation. Sci. Total Environ. 2021, 798, 149362. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Li, T.; Sivakumar, B.; Sharma, A.; Albertson, J.D.; Zhang, L.; Wang, G. Combined Effects of Warming and Grazing on Rangeland Vegetation on the Qinghai-Tibet Plateau. Front. Environ. Sci. 2021, 9, 566. [Google Scholar] [CrossRef]
- Xie, J.; Liang, C.; Li, Z.; Ji, Z.; Ou, W.; Liu, H.; Wang, H.; Kazhuocairang; Hou, Z. Characteristics of complete chloroplast genome of a high-quality forage on Qinghai-Tibet Plateau, Medicago archiducis-nicolai Sirj. (FabaceaeTrifolieae). Mitochondrial DNA B 2021, 6, 217–219. [Google Scholar] [CrossRef]
- Tian, J.; Bu, L.; Zhang, M.; Yuan, J.; Zhang, Y.; Wei, G.; Wang, H. Soil bacteria with distinct diversity and functions mediates the soil nutrients after introducing leguminous shrub in desert ecosystems. Glob. Ecol. Conserv. 2021, 31, e01841. [Google Scholar] [CrossRef]
- Zhang, M.; Zhang, X.; Zhang, L.; Zeng, L.; Liu, Y.; Wang, X.; He, P.; Li, S.; Liang, G.; Zhou, W.; et al. The stronger impact of inorganic nitrogen fertilization on soil bacterial community than organic fertilization in short-term condition. Geoderma 2021, 382, 114752. [Google Scholar] [CrossRef]
- Wołejko, E.; Jabłońska-Trypuć, A.; Wydro, U.; Butarewicz, A.; Łozowicka, B. Soil biological activity as an indicator of soil pollution with pesticides–A review. Appl. Soil Ecol. 2020, 147, 103356. [Google Scholar] [CrossRef]
- Zhang, X.; Li, Y.; Ouyang, D.; Lei, J.; Tan, Q.; Xie, L.; Li, Z.; Liu, T.; Xiao, Y.; Farooq, T.H.; et al. Systematical review of interactions between microplastics and microorganisms in the soil environment. J. Hazard. Mater. 2021, 418, 126288. [Google Scholar] [CrossRef]
- Cheng, Z.; Lei, S.; Li, Y. Revealing the Variation and Stability of Bacterial Communities in Tomato Rhizosphere Microbiota. Microorganisms 2020, 8, 170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chamkhi, I.; El Omari, N.; Balahbib, A.; El Menyiy, N.; Benali, T.; Ghoulam, C. Is the rhizosphere a source of applicable multi-beneficial microorganisms for plant enhancement? Saudi J. Biol. Sci. 2022, 29, 1246–1259. [Google Scholar] [CrossRef] [PubMed]
- Pande, A.; Mun, B.G.; Lee, D.S.; Khan, M.; Lee, G.M.; Hussain, A.; Yun, B.W. NO Network for Plant-Microbe Communication Underground: A Review. Front. Plant Sci. 2021, 12, 431. [Google Scholar] [CrossRef] [PubMed]
- Fei, S.; Kivlin, S.N.; Domke, G.M.; Jo, I.; LaRue, E.A.; Phillips, R.P. Coupling of plant and mycorrhizal fungal diversity: Its occurrence, relevance, and possible implications under global change. New Phytol. 2022, 234, 1960–1966. [Google Scholar] [CrossRef] [PubMed]
- Bei, S.; Zhang, Y.; Li, T.; Christie, P.; Li, X.; Zhang, J. Response of the soil microbial community to different fertilizer inputs in a wheat-maize rotation on a calcareous soil. Agric. Ecosyst. Environ. 2018, 260, 58–69. [Google Scholar] [CrossRef]
- Zhang, B.; Niu, L.; Jia, T.; Yu, X.; She, D. Spatial variability of soil organic matter and total nitrogen and the influencing factors in Huzhu County of Qinghai Province, China. Acta Agric. Scand. Sect. B Soil Plant Sci. 2022, 72, 576–588. [Google Scholar] [CrossRef]
- Ji, L.; Si, H.; He, J.; Fan, L.; Li, L. The shifts of maize soil microbial community and networks are related to soil properties under different organic fertilizers. Rhizosphere 2021, 19, 100388. [Google Scholar] [CrossRef]
- Sun, R.; Chen, Y.; Han, W.; Dong, W.; Zhang, Y.; Hu, C.; Liu, B.; Wang, F. Different contribution of species sorting and exogenous species immigration from manure to soil fungal diversity and community assemblage under long-term fertilization. Soil Biol. Biochem. 2020, 151, 108049. [Google Scholar] [CrossRef]
- Yang, Y.; Li, X.; Liu, J.; Zhou, Z.; Zhang, T.; Wang, X. Fungal community structure in relation to manure rate in red soil in southern China. Appl. Soil Ecol. 2020, 147, 103442. [Google Scholar] [CrossRef]
- Ning, D.; Deng, Y.; Tiedje, J.M.; Zhou, J. A general framework for quantitatively assessing ecological stochasticity. Proc. Natl. Acad. Sci. USA 2019, 116, 16892–16898. [Google Scholar] [CrossRef] [Green Version]
- Ning, D.; Yuan, M.; Wu, L.; Zhang, Y.; Guo, X.; Zhou, X.; Zhou, J. A quantitative framework reveals ecological drivers of grassland microbial community assembly in response to warming. Nat. Commun. 2020, 11, 4717. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.L.; Hu, H.W.; Yan, Z.Z.; Li, C.Y.; He, J.Z. Deterministic Selection Dominates Microbial Community Assembly in Termite Mounds Across a Large Spatial Area. Soil Biol. Biochem. 2021, 152. [Google Scholar] [CrossRef]
- Gad, M.; Hou, L.; Li, J.; Wu, Y.; Rashid, A.; Chen, N.; Hu, A. Distinct mechanisms underlying the assembly of microeukaryotic generalists and specialists in an anthropogenically impacted river. Sci. Total Environ. 2020, 748, 141434. [Google Scholar] [CrossRef] [PubMed]
- Qin, Z.; Zhang, H.; Feng, G.; Christie, P.; Zhang, J.; Li, X.; Gai, J. Soil phosphorus availability modifies the relationship between AM fungal diversity and mycorrhizal benefits to maize in an agricultural soil. Soil Biol. Biochem. 2020, 144, 107790. [Google Scholar] [CrossRef]
- MTahat, M.; MAlananbeh, K.; AOthman, Y.; ILeskovar, D. Soil Health and Sustainable Agriculture. Sustainability 2020, 12, 4859. [Google Scholar] [CrossRef]
- Cruz-Paredes, C.; Diera, T.; Davey, M.; Rieckmann, M.M.; Christensen, P.; Dela Cruz, M.; Jakobsen, I. Disentangling the abiotic and biotic components of AMF suppressive soils. Soil Biol. Biochem. 2021, 159, 108305. [Google Scholar] [CrossRef]
- El Omari, B.; El Ghachtouli, N. Arbuscular mycorrhizal fungi-weeds interaction in cropping and unmanaged ecosystems: A review. Symbiosis 2021, 83, 279–292. [Google Scholar] [CrossRef]
- Lambers, H.; Mougel, C.; Jaillard, B.; Hinsinger, P. Plant-microbe-soil interactions in the rhizosphere: An evolutionary perspective. Plant Soil 2009, 321, 83–115. [Google Scholar] [CrossRef]
- Liang, J.; Xue, Z.; Yang, Z.; Chai, Z.; Niu, J.; Shi, Z. Effects of microbial organic fertilizers on Astragalus membranaceus growth and rhizosphere microbial community. Ann. Microbiol. 2021, 71, 11. [Google Scholar] [CrossRef]
- Little, N.G.; DiTommaso, A.; Westbrook, A.S.; Ketterings, Q.M.; Mohler, C.L. Effects of fertility amendments on weed growth and weed–crop competition: A review. Weed Sci. 2021, 69, 132–146. [Google Scholar] [CrossRef]
- Tao, R.; Hu, B.; Chu, G. Impacts of organic fertilization with a drip irrigation system on Prokaryotes and fungal communities in cotton field. Agric. Syst. 2020, 182, 102820. [Google Scholar] [CrossRef]
- Kowal, J.; Arrigoni, E.; Serra, J.; Bidartondo, M. Prevalence and phenology of fine root endophyte colonization across populations of Lycopodiella inundata. Mycorrhiza 2020, 30, 577–587. [Google Scholar] [CrossRef]
- Blažková, A.; Jansa, J.; Püschel, D.; Vosátka, M.; Janoušková, M. Is mycorrhiza functioning influenced by the quantitative composition of the mycorrhizal fungal community? Soil Biol. Biochem. 2021, 157, 108249. [Google Scholar] [CrossRef]
- Li, S.; Deng, Y.; Wang, Z.; Zhang, Z.; Kong, X.; Zhou, W.; Yi, Y.; Qu, Y. Exploring the accuracy of amplicon-based internal transcribed spacer (ITS) markers for fungal community. Mol. Ecol. Resour. 2020, 20, 170–184. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Zhang, Z.; Cai, W.; Liu, W.; Xu, M.; Yin, H.; Wang, A.; He, Z.; Deng, Y. Biodiversity and species competition regulate the resilience of microbial biofilm community. Mol. Ecol. 2017, 26, 6170–6182. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Peng, X.; Zhang, Z.; Gu, S.; He, Q.; Shen, W.; Wang, Z.; Wang, D.; Hu, Q.; Li, Y.; et al. iNAP: An integrated network analysis pipeline for microbiome studies. iMeta 2022, 2, e13. [Google Scholar] [CrossRef]
- Magoc, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Kong, Y. Btrim: A fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 2011, 98, 152–153. [Google Scholar] [CrossRef] [Green Version]
- Sabir, M.S.; Shahzadi, F.; Ali, F.; Shakeela, Q.; Niaz, Z.; Ahmed, S. Comparative Effect of Fertilization Practices on Soil Microbial Diversity and Activity: An Overview. Curr. Microbiol. 2021, 78, 3644–3655. [Google Scholar] [CrossRef]
- Wan, L.-J.; Tian, Y.; He, M.; Zheng, Y.-Q.; Lyu, Q.; Xie, R.-J.; Ma, Y.-Y.; Deng, L.; Yi, S.-L. Effects of Chemical Fertilizer Combined with Organic Fertilizer Application on Soil Properties, Citrus Growth Physiology, and Yield. Agriculture 2021, 11, 1207. [Google Scholar] [CrossRef]
- Ruibo Sun, X.; Zhang, X.; Guo, C. Prokaryotes diversity in soils subjected to long-term chemical fertilization can be more stably maintained with the addition of livestock manure than wheat straw. Soil Biol. Biochem. 2015, 88, 9–18. [Google Scholar]
- Meng, Q.J.; Yan-Li, X.U.; Chun-Jie, L.I.; Han, X.Z.; Pei, X.C. Effects of Different Fertilization and Land Use History on the Bacterial Diversity in Black Soils. Soybean Sci. 2008, 27, 480–486. [Google Scholar]
- Kamaa, M.; Mburu, H.; Blanchart, E.; Chibole, L.; Chotte, J.L.; Kibunja, C.N.; Lesueur, D. Effects of organic and inorganic fertilization on soil Prokaryotes and fungal microbial diversity in the Kabete long-term trial, Kenya. Can. J. Public Health Revue Canadienne Santé Publique 2011, 97, 69–71. [Google Scholar]
- Wu, X.; Wang, R.; Hu, H.; Xiu, W.M.; Wang, X.Y. Response of Bacterial and Fungal Communities to Chemical Fertilizer Reduction Combined with Organic Fertilizer and Straw in Fluvo-aquic Soil. Huan Jing Xue = Huanjing Kexue 2020, 41, 4669–4681. [Google Scholar]
- Falagas, M.E.; Bliziotis, I.A.; Siempos, I.I. Attributable mortality of Acinetobacter baumannii infections in critically ill patients: A systematic review of matched cohort and case-control studies. Crit. Care 2006, 10, 1–8. [Google Scholar]
- Kerry, O.; Nirenberg, H.I.; Takayuki, A.; Cigelnik, E. A multigene phylogeny of theGibberella fujikuroi species complex: Detection of additional phylogenetically distinct species. Mycoscience 2000, 41, 61–78. [Google Scholar]
- Arenal, F.; Platas, G.; Peláez, F. Two new Preussia species defined based on morphological and molecular evidence. Fungal Divers. 2004, 20, 1–15. [Google Scholar]
- Li, Y.; Gao, Y.; Zhang, W.; Wang, C.; Wang, P.; Niu, L.; Wu, H. Homogeneous selection dominates the microbial community assembly in the sediment of the Three Gorges Reservoir. Sci. Total Environ. 2019, 690, 50–60. [Google Scholar] [CrossRef]
- Cao, M.; Cui, L.; Sun, H.; Zhang, X.; Jiang, J. Effects of Spartina alterniflora Invasion on Soil Microbial Community Structure and Ecological Functions. Microorganisms 2021, 9, 138. [Google Scholar] [CrossRef]
- Lidbury, I.; Scanlan, D.; Murphy, A.; Bo Tt Rill, A.; Wellington, E. Novel mechanisms for phosphate acquisition in abundant rhizosphere-dwelling Bacteroidetes 2. bioRxiv 2019. [Google Scholar] [CrossRef]
- Kim, S.J.; Kim, J.H.; Kim, J.K.; Chang, K.J.; Choi, J.N. Comparative Study of Infection Effects with AMF (Arbuscular-Mycorrhizal Fungi) Isolated from Upland Plants. Korean J. Plant Resour. 2014, 27, 194–201. [Google Scholar] [CrossRef] [Green Version]
- Cruz, A.F.; Martins, M.A. Effects of arbuscular mycorrhizal fungi (AMF) and N fertilization on plants under intercrop system conditions. Ceres 1998, 45, 41–54. [Google Scholar]
- Jing, L.I.; Chen, R.; Lan, Z.; Lin, D. Effect of Organic Fertilizer Substituting Part of Inorganic Fertilizer on Physical and Chemical Properties, Enzyme Activity and Fruit Nutrient of Garden Soil. Chin. J. Trop. Crop. 2018, 39, 656–660. [Google Scholar]
- Cheng, X.; Wu, H.N.; Guo, Q.; Hou, B.N.; Xia, L.Y.; Wang, H.J.; Al, E. Paleomagnetic results of Late Paleozoic rocks from northern Qiangtang Block in Qinghai-Tibet Plateau, China. Sci. China Earth Sci. 2012, 15, 67–75. [Google Scholar] [CrossRef]
- Trisilawati, O.; Hartoyo, B.; Bermawie, N.; Pribadi, E.R. Application of AMF (Arbuscular Mycorrhizal Fungi) and Organic Fertilizer to Increase the Growth, Biomass and Bioactive Content of Centella. IOP Conf. Ser. Earth Environ. Sci. 2019, 292, 012067. [Google Scholar] [CrossRef]


| Dissimilarity Test | Method | Bacteria | Fungi |
|---|---|---|---|
| Bray–Curtis | Bray–Curtis | ||
| CK vs. organic fertilizer | MRPP | 0.6157 ** | 0.4222 * |
| ANOSIM | 0.2962 ** | 0.3333 * |
| Bacteria | Fungi | |||
|---|---|---|---|---|
| CK | Organic Fertilizer | CK | Organic Fertilizer | |
| Observed dissimilarity of community | 0.666 ± 0.119 | 0.526 ± 0.166 | 0.508 ± 0.094 | 0.472 ± 0.063 |
| Normalized stochasticity ratio (NST) | 67% | 55% | 52% | 48% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ma, K.; Wang, Y.; Jin, X.; Zhao, Y.; Yan, H.; Zhang, H.; Zhou, X.; Lu, G.; Deng, Y. Application of Organic Fertilizer Changes the Rhizosphere Microbial Communities of a Gramineous Grass on Qinghai–Tibet Plateau. Microorganisms 2022, 10, 1148. https://doi.org/10.3390/microorganisms10061148
Ma K, Wang Y, Jin X, Zhao Y, Yan H, Zhang H, Zhou X, Lu G, Deng Y. Application of Organic Fertilizer Changes the Rhizosphere Microbial Communities of a Gramineous Grass on Qinghai–Tibet Plateau. Microorganisms. 2022; 10(6):1148. https://doi.org/10.3390/microorganisms10061148
Chicago/Turabian StyleMa, Kun, Yingcheng Wang, Xin Jin, Yangan Zhao, Huilin Yan, Haijuan Zhang, Xueli Zhou, Guangxin Lu, and Ye Deng. 2022. "Application of Organic Fertilizer Changes the Rhizosphere Microbial Communities of a Gramineous Grass on Qinghai–Tibet Plateau" Microorganisms 10, no. 6: 1148. https://doi.org/10.3390/microorganisms10061148
APA StyleMa, K., Wang, Y., Jin, X., Zhao, Y., Yan, H., Zhang, H., Zhou, X., Lu, G., & Deng, Y. (2022). Application of Organic Fertilizer Changes the Rhizosphere Microbial Communities of a Gramineous Grass on Qinghai–Tibet Plateau. Microorganisms, 10(6), 1148. https://doi.org/10.3390/microorganisms10061148






