A Novel Organophosphorus Acid Anhydrolase from Deep Sea Sediment with High Degradation Efficiency for Organophosphorus Pesticides and Nerve Agent
Abstract
1. Introduction
2. Materials and Methods
2.1. Gene and Chemicals
2.2. Protein Expression and Purification
2.3. Degradation Efficiency of OPAA114644 for Different OP Pesticides and Nerve Agent
2.4. Enzyme Assay
2.5. Biochemical Characterization of OPAA114644
2.6. Metal Ions Analysis
2.7. Circular Dichroism Analysis
2.8. Decontamination of Paraoxon on Glasses, Cotton Tissues, and Apples by OPAA114644
2.8.1. Glasses
2.8.2. Cotton Tissues
2.8.3. Apples
3. Results and Discussion
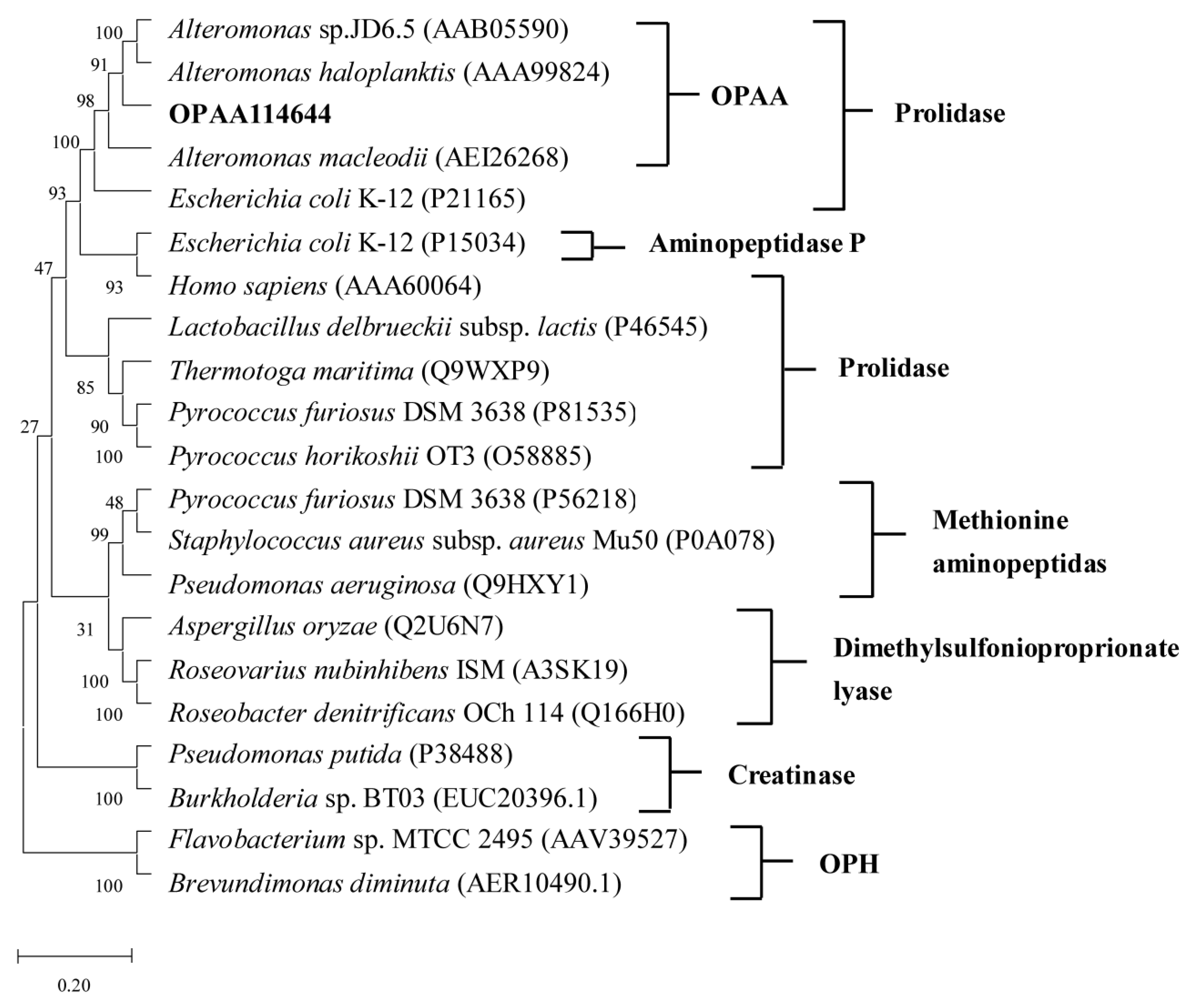
3.1. Sequence Analysis
3.2. Protein Expression and Purification
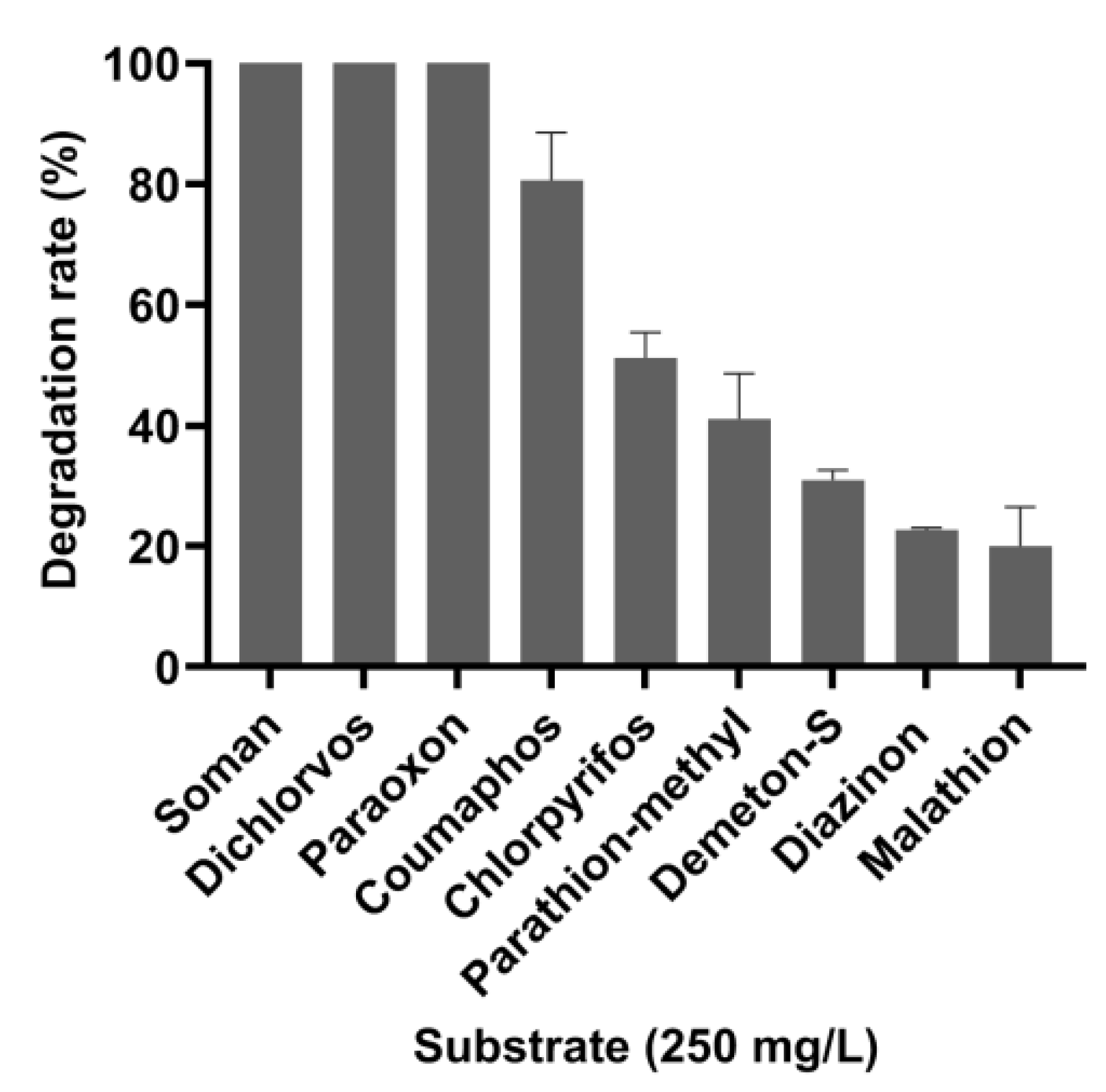
3.3. Degradation Efficiency of OPAA114644 for Different OP Pesticides and Nerve Agent
3.4. Enzyme Assay
3.5. Biochemical Characterization of OPAA114644
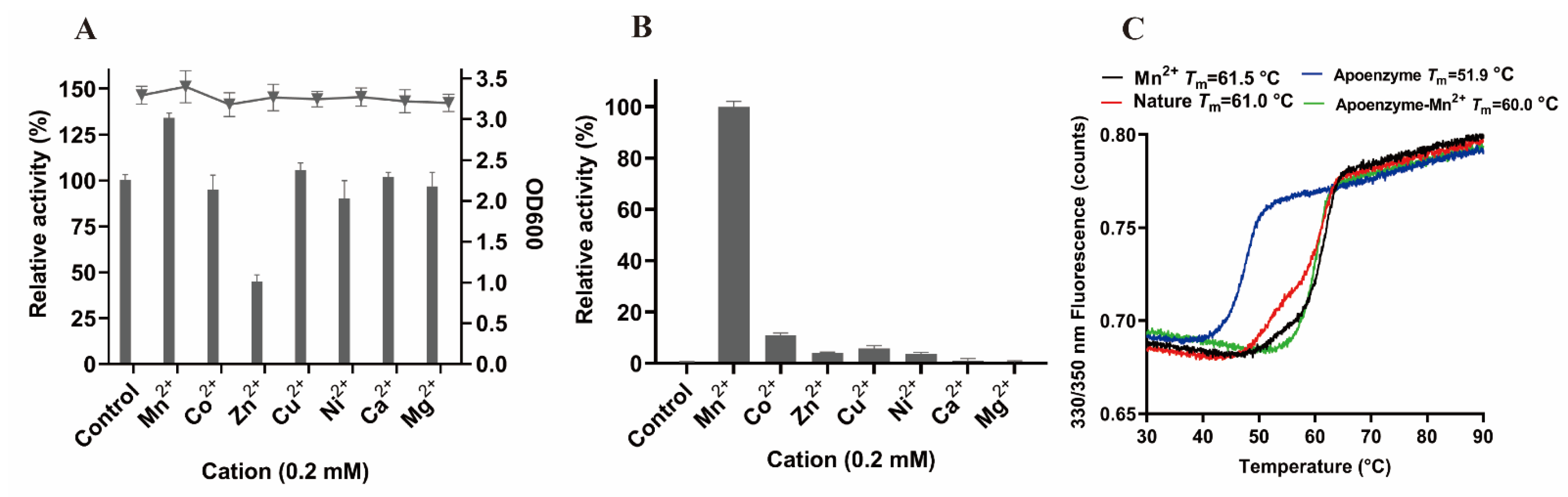
3.6. Metal Ions Analysis
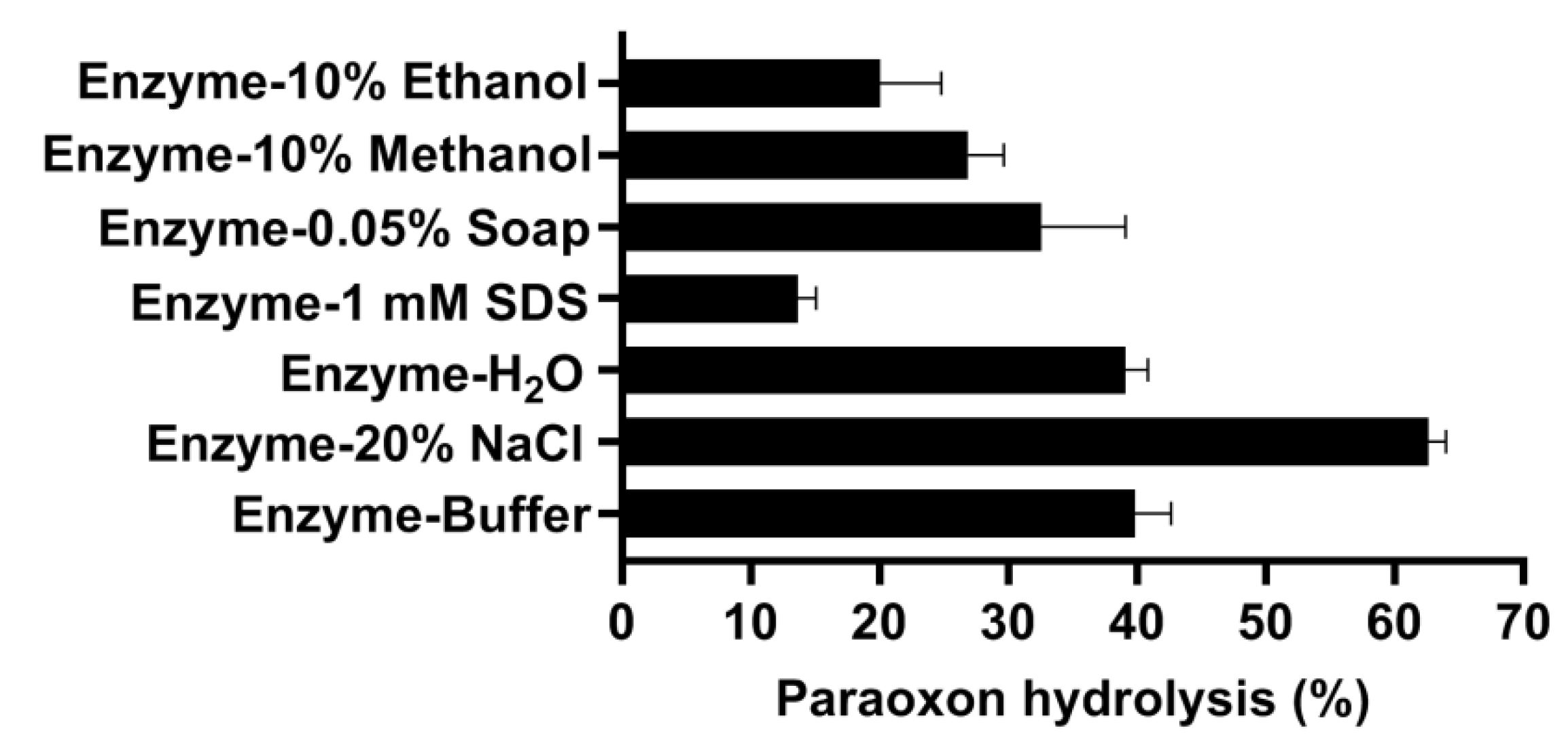
3.7. Decontamination of Paraoxon on Glasses, Cotton Tissues, and Apples by OPAA114644
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Costa, L.G. Organophosphorus Compounds at 80: Some Old and New Issues. Toxicol. Sci. 2018, 162, 24–35. [Google Scholar] [CrossRef]
- Mukherjee, S.; Gupta, R.D. Organophosphorus Nerve Agents: Types, Toxicity, and Treatments. J. Toxicol. 2020, 2020, 3007984. [Google Scholar] [CrossRef] [PubMed]
- Singh, B.K.; Walker, A. Microbial degradation of organophosphorus compounds. FEMS Microbiol. Rev. 2006, 30, 428–471. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Zhao, S.; Zhang, W.; Wu, H.; Guang, C.; Mu, W. Recent advances and future prospective of organophosphorus-degrading enzymes: Identification, modification, and application. Crit. Rev. Biotechnol. 2021, 41, 1093–1113. [Google Scholar] [CrossRef] [PubMed]
- Matula, M.; Kucera, T.; Soukup, O.; Pejchal, J. Enzymatic Degradation of Organophosphorus Pesticides and Nerve Agents by EC: 3.1.8.2. Catalysts 2020, 10, 1365. [Google Scholar] [CrossRef]
- Wang, L.; Sun, Y. Engineering organophosphate hydrolase for enhanced biocatalytic performance: A review. Biochem. Eng. J. 2021, 168, 107945. [Google Scholar] [CrossRef]
- Jeyaratnam, J. World Health Statistics Quarterly—Acute Pesticide Poisoning: A Major Global Health Problem. World Health Stat. Q. 1990, 43, 139–144. [Google Scholar]
- Jacquet, P.; Daude, D.; Bzdrenga, J.; Masson, P.; Elias, M.; Chabriere, E. Current and emerging strategies for organophosphate decontamination: Special focus on hyperstable enzymes. Environ. Sci. Pollut. Res. 2016, 23, 8200–8218. [Google Scholar] [CrossRef]
- Cheng, T.C.; Defrank, J.J. Hydrolysis of organophosphorus compounds by bacterial. In Enzymes in Action; Springer: Berlin/Heidelberg, Germany, 2000; pp. 243–261. [Google Scholar] [CrossRef]
- Vyas, N.K.; Nickitenko, A.; Rastogi, V.K.; Shah, S.S.; Quiocho, F.A. Structural insights into the dual activities of the nerve agent degrading organophosphate anhydrolase/prolidase. Biochemistry 2010, 49, 547–559. [Google Scholar] [CrossRef]
- Ghosh, M.; Grunden, A.M.; Dunn, D.M.; Weis, R.; Adams, M. Characterization of native and recombinant forms of an unusual proline dipeptidase. J. Bacteriol. 1998, 180, 4781–4789. [Google Scholar] [CrossRef]
- Schenk, G.; Mateen, I.; Ng, T.-K.; Pedroso, M.M.; Mitić, N.; Jafelicci, M.; Marques, R.F.C.; Gahan, L.R.; Ollis, D.L. Organophosphate-degrading metallohydrolases: Structure and function of potent catalysts for applications in bioremediation. Coord. Chem. Rev. 2016, 317, 122–131. [Google Scholar] [CrossRef]
- Bazan, J.F.; Weaver, L.H.; Roderick, S.L.; Huber, R.; Matthews, B.W. Sequence and structure comparison suggest that methionine aminopeptidase, prolidase, aminopeptidase P, and creatinase share a common fold. Proc. Natl. Acad. Sci. USA 1994, 91, 2473–2477. [Google Scholar] [CrossRef]
- Theriot, C.M.; Tove, S.R.; Grunden, A.M. Biotechnological Applications of Recombinant Microbial Prolidases. Adv. Appl. Microbiol. 2009, 68, 99–132. [Google Scholar] [CrossRef]
- Xiao, Y.; Yang, J.; Tian, X.; Wang, X.; Li, J.; Zhang, S.; Long, L. Biochemical basis for hydrolysis of organophosphorus by a marine bacterial prolidase. Process Biochem. 2017, 52, 141–148. [Google Scholar] [CrossRef]
- Daczkowski, C.M.; Pegan, S.D.; Harvey, S.P. Engineering the organophosphorus acid anhydrolase enzyme for increased catalytic efficiency and broadened stereospecificity on Russian VX. Biochemistry 2015, 54, 6423–6433. [Google Scholar] [CrossRef]
- Bae, S.Y.; Myslinski, J.M.; McMahon, L.R.; Height, J.J.; Bigley, A.N.; Raushel, F.M.; Harvey, S.P. An OPAA enzyme mutant with increased catalytic efficiency on the nerve agents sarin, soman, and GP. Enzyme Microb. Technol. 2018, 112, 65–71. [Google Scholar] [CrossRef]
- Goud, K.Y.; Teymourian, H.; Sandhu, S.S.; Tostado, N.; Mishra, R.K.; Moore, L.C.; Harvey, S.P.; Wang, J. OPAA/fluoride biosensor chip towards field detection of G-type nerve agents. Sens. Actuators B Chem. 2020, 320, 128344. [Google Scholar] [CrossRef]
- Jain, M.; Yadav, P.; Joshi, B.; Joshi, A.; Kodgire, P. A novel biosensor for the detection of organophosphorus (OP)-based pesticides using organophosphorus acid anhydrolase (OPAA)-FL variant. Appl. Microbiol. Biotechnol. 2021, 105, 389–400. [Google Scholar] [CrossRef]
- Song, T.; Wang, F.; Xiong, S.; Jiang, H. Surface display of organophosphorus-degrading enzymes on the recombinant spore of Bacillus subtilis. Biochem. Biophys. Res. Commun. 2019, 510, 13–19. [Google Scholar] [CrossRef]
- Dick, G.J. The microbiomes of deep-sea hydrothermal vents: Distributed globally, shaped locally. Nat. Rev. Microbiol. 2019, 17, 271–283. [Google Scholar] [CrossRef]
- Chen, P.; Zhang, L.; Guo, X.; Dai, X.; Liu, L.; Xi, L.; Wang, J.; Song, L.; Wang, Y.; Zhu, Y.; et al. Diversity, biogeography, and biodegradation potential of Actinobacteria in the deep-sea sediments along the Southwest Indian Ridge. Front. Microbiol. 2016, 7, 1340. [Google Scholar] [CrossRef] [PubMed]
- Theriot, C.M.; Grunden, A.M. Hydrolysis of organophosphorus compounds by microbial enzymes. Appl. Microbiol. Biotechnol. 2011, 89, 35–43. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Xu, W.; Zhang, W.; Wu, H.; Guang, C.; Mu, W. In-depth biochemical identification of a novel methyl parathion hydrolase from Azohydromonas australica and its high effectiveness in the degradation of various organophosphorus pesticides. Bioresour. Technol. 2021, 323, 124641. [Google Scholar] [CrossRef] [PubMed]
- Merone, L.; Mandrich, L.; Rossi, M.; Manco, G. A thermostable phosphotriesterase from the archaeon Sulfolobus solfataricus: Cloning, overexpression and properties. Extremophiles 2005, 9, 297–305. [Google Scholar] [CrossRef]
- Omburo, G.A.; Kuo, J.M.; Mullins, L.S.; Raushel, F.M. Characterization of the zinc binding site of bacterial phosphotriesterase. J. Biol. Chem. 1992, 267, 13278–13283. [Google Scholar] [CrossRef]
- Del Giudice, I.; Coppolecchia, R.; Merone, L.; Porzio, E.; Carusone, T.M.; Mandrich, L.; Worek, F.; Manco, G. An efficient thermostable organophosphate hydrolase and its application in pesticide decontamination. Biotechnol. Bioeng. 2016, 113, 724–734. [Google Scholar] [CrossRef]
- Robert, X.; Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucleic. Acids. Res. 2014, 42, W320–W324. [Google Scholar] [CrossRef]
- Grunden, A.M.; Ghosh, M.; Adams, M. Proline dipeptidase from Pyrococcus furiosus. Methods Enzymol. 2001, 330, 433–445. [Google Scholar] [CrossRef]
- Theriot, C.M.; Tove, S.R.; Grunden, A.M. Characterization of two proline dipeptidases (prolidases) from the hyperthermophilic archaeon Pyrococcus horikoshii. Appl. Microbiol. Biotechnol. 2010, 86, 177–188. [Google Scholar] [CrossRef]
- Du, X.; Tove, S.; Kast-Hutcheson, K.; Grunden, A.M. Characterization of the dinuclear metal center of Pyrococcus furiosus prolidase by analysis of targeted mutants. FEBS Lett. 2005, 579, 6140–6146. [Google Scholar] [CrossRef]
- Roderick, S.L.; Matthews, B.W. Structure of the cobalt-dependent methionine aminopeptidase from Escherichia coli a new type of proteolytic enzyme. Biochemistry 1993, 32, 3907–3912. [Google Scholar] [CrossRef]
- Cheng, T.C.; Rastogi, V.K.; Defrank, J.J.; Sawiris, G.P. G-type nerve agent decontamination by Alteromonas prolidase. Ann. N. Y. Acad. Sci. 2010, 864, 253–258. [Google Scholar] [CrossRef]
- Cheng, T.C.; Liu, L.; Wang, B.; Wu, J.; Defrank, J.J.; Anderson, D.M.; Rastogi, V.K.; Hamilton, A.B. Nucleotide sequence of a gene encoding an organophosphorus nerve agent degrading enzyme from Alteromonas haloplanktis. J. Ind. Microbiol. Biot. 1997, 18, 49–55. [Google Scholar] [CrossRef]
- Defrank, J.J.; Cheng, T.C. Purification and properties of an organophosphorus acid anhydrase from a halophilic bacterial isolate. J. Bacteriol. 1991, 173, 1938–1943. [Google Scholar] [CrossRef][Green Version]
- Manavathi, B.; Pakala, S.B.; Gorla, P.; Merrick, M.; Siddavattam, D. Influence of zinc and cobalt on expression and activity of parathion hydrolase from Flavobacterium sp. ATCC27551. Pestic. Biochem. Phys. 2005, 83, 37–45. [Google Scholar] [CrossRef]
- Johnson, C.M. Differential scanning calorimetry as a tool for protein folding and stability. Arch. Biochem. Biophys. 2013, 531, 100–109. [Google Scholar] [CrossRef]
- Lupi, A.; Della Torre, S.; Campari, E.; Tenni, R.; Cetta, G.; Rossi, A.; Forlino, A. Human recombinant prolidase from eukaryotic and prokaryotic sources. Expression, purification, characterization and long-term stability studies. FEBS J. 2006, 273, 5466–5478. [Google Scholar] [CrossRef]
- Bai, Y.P.; Luo, X.J.; Zhao, Y.L.; Li, C.X.; Xu, D.S.; Xu, J.H. Efficient degradation of malathion in the presence of detergents using an engineered organophosphorus hydrolase highly expressed by Pichia pastoris without methanol induction. J. Agric. Food Chem. 2017, 65, 9094–9100. [Google Scholar] [CrossRef]




| Substrate | Km (mM) | Vmax (µmol/min/mg) | kcat (s−1) | kcat/Km (mM−1 s−1) |
|---|---|---|---|---|
| Met–Pro | 2.72 ± 0.66 | 11,295 ± 1102 | 5445 | 2002 |
| Paraoxon | 2.06 ± 0.19 | 2.22 ± 0.07 | 0.75 | 0.36 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, X.; Wang, L.; Qi, L.; Dong, Z. A Novel Organophosphorus Acid Anhydrolase from Deep Sea Sediment with High Degradation Efficiency for Organophosphorus Pesticides and Nerve Agent. Microorganisms 2022, 10, 1112. https://doi.org/10.3390/microorganisms10061112
Zheng X, Wang L, Qi L, Dong Z. A Novel Organophosphorus Acid Anhydrolase from Deep Sea Sediment with High Degradation Efficiency for Organophosphorus Pesticides and Nerve Agent. Microorganisms. 2022; 10(6):1112. https://doi.org/10.3390/microorganisms10061112
Chicago/Turabian StyleZheng, Xiaofang, Li Wang, Lihong Qi, and Zhiyang Dong. 2022. "A Novel Organophosphorus Acid Anhydrolase from Deep Sea Sediment with High Degradation Efficiency for Organophosphorus Pesticides and Nerve Agent" Microorganisms 10, no. 6: 1112. https://doi.org/10.3390/microorganisms10061112
APA StyleZheng, X., Wang, L., Qi, L., & Dong, Z. (2022). A Novel Organophosphorus Acid Anhydrolase from Deep Sea Sediment with High Degradation Efficiency for Organophosphorus Pesticides and Nerve Agent. Microorganisms, 10(6), 1112. https://doi.org/10.3390/microorganisms10061112





