Novel Mobile Integrons and Strain-Specific Integrase Genes within Shewanella spp. Unveil Multiple Lateral Genetic Transfer Events within The Genus
Abstract
1. Introduction
2. Materials and Methods
2.1. Shewanella Genomes Dataset and ST Assignment
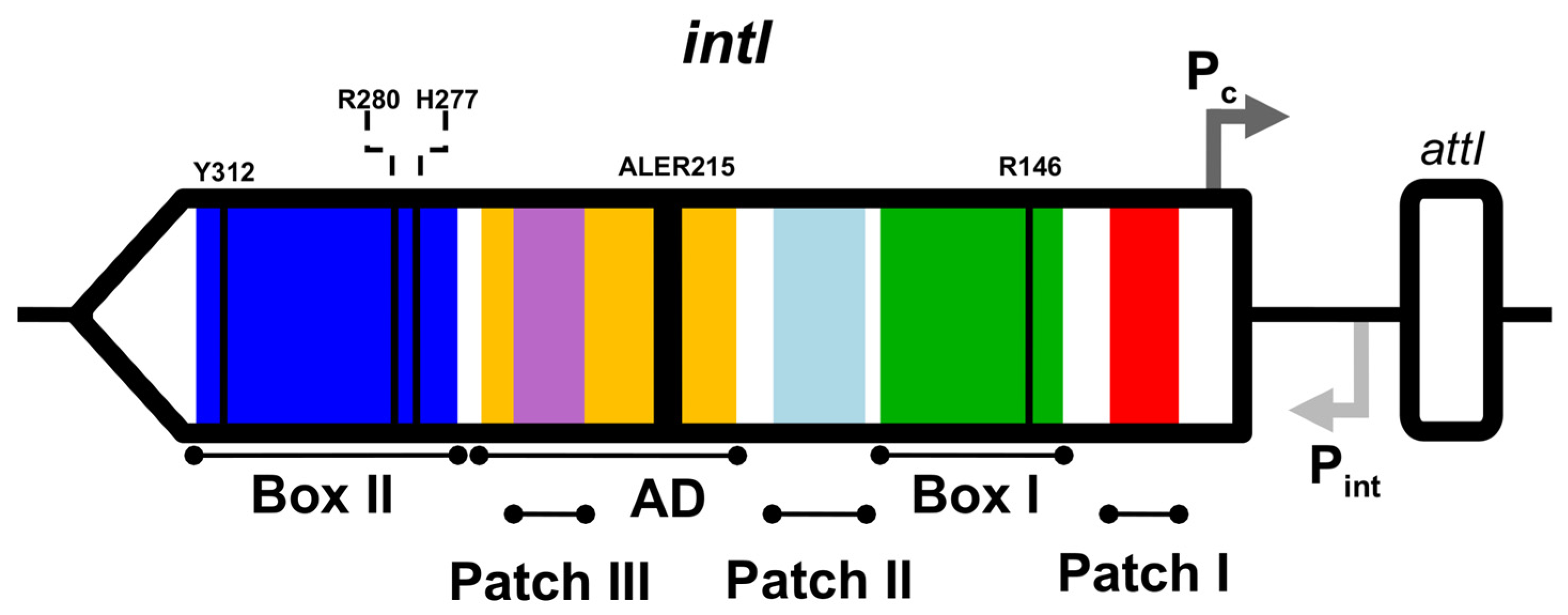
2.2. Identification of Integron Integrase Genes and attc Sites
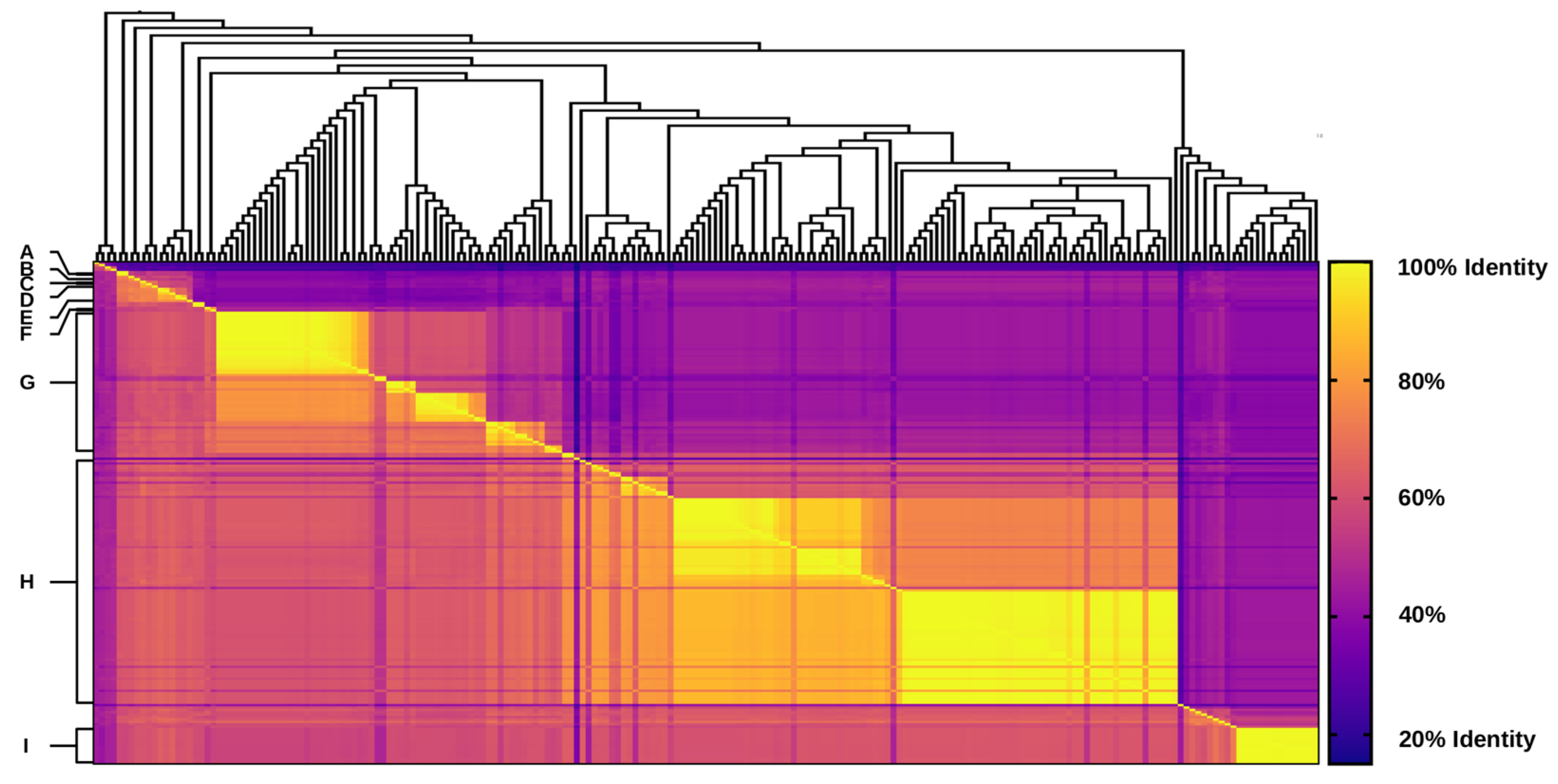
2.3. Pairwise Similarity and Identity Analyses of Integron Integrases
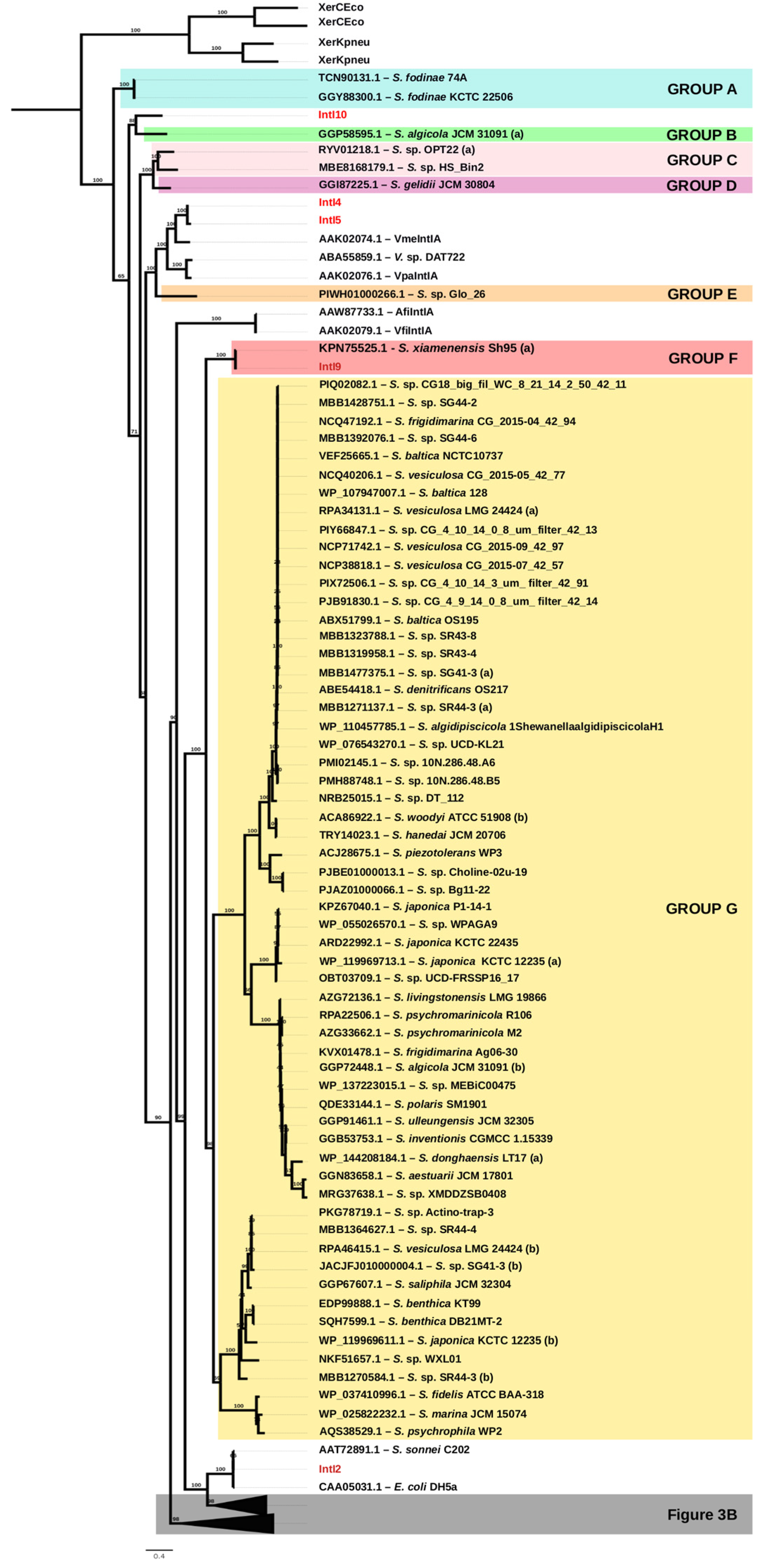
2.4. Phylogeny and Sequence Analyses
2.5. Genetic Context of Integron Analyses
3. Results
3.1. Identification of Integrase Genes Encoded in Shewanella spp.
3.2. Unique Distribution of Integrase Genes among Shewanella spp. Genomes
3.3. Integrase Genes from Shewanella spp. Are Not Niche Dependent
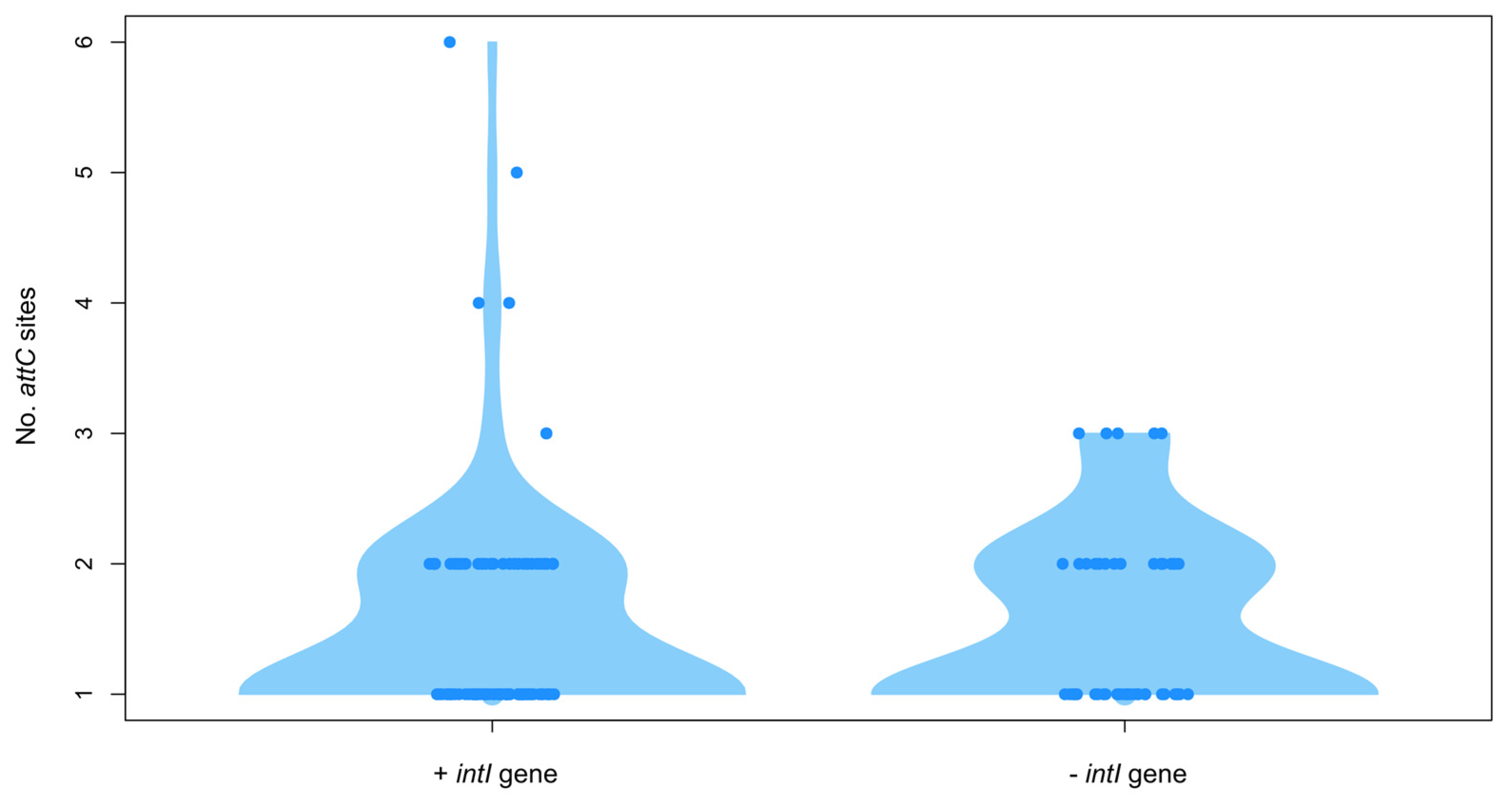
3.4. Analysis of Integron/Cassette Systems in Shewanella spp. Genomes
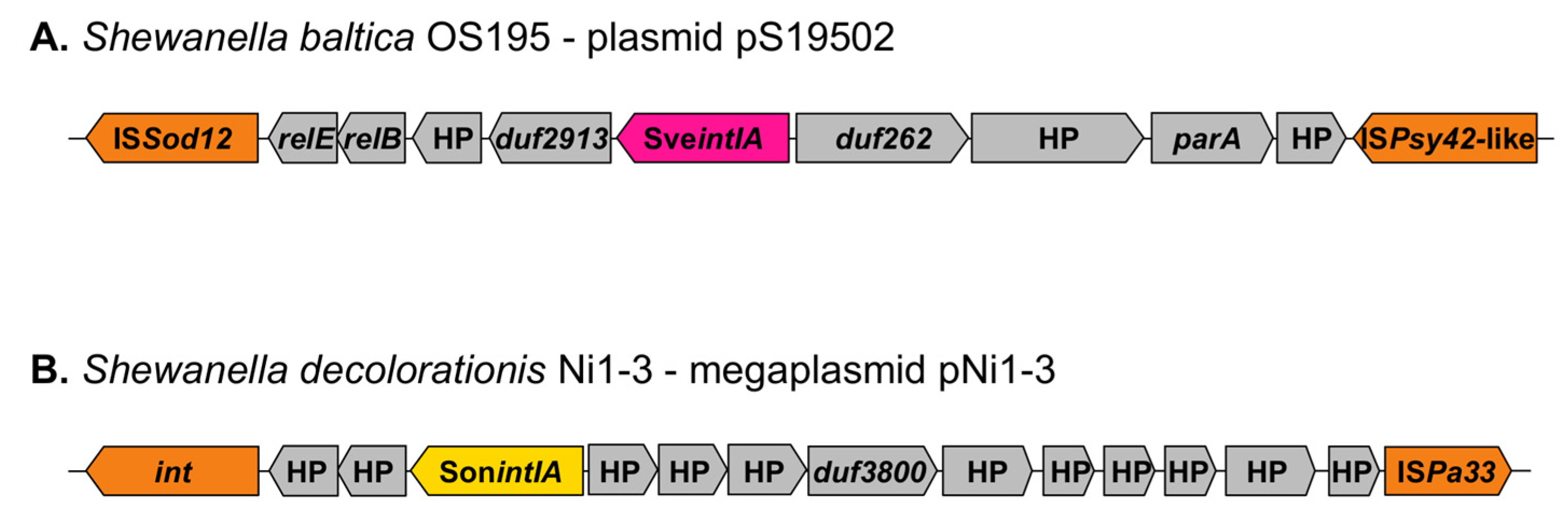
3.5. Genetic Context of Shewanella spp. Integrons
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lemaire, O.N.; Méjean, V.; Iobbi-Nivol, C. The Shewanella genus: Ubiquitous organisms sustaining and preserving aquatic ecosystems. FEMS Microbiol. Rev. 2020, 44, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Janda, J.M.; Abbott, S.L. The genus Shewanella: From the briny depths below to human pathogen. Crit. Rev. Microbiol. 2014, 40, 293–312. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, M.S.; Merkier, A.K.; Almuzara, M.; Vay, C.; Centrón, D. Reservoir of antimicrobial resistance determinants associated with horizontal gene transfer in clinical isolates of the genus Shewanella. Antimicrob. Agents Chemother. 2010, 54, 4516–4517. [Google Scholar] [CrossRef] [PubMed]
- Martín-Rodríguez, A.J.; Martín-Pujol, O.; Artiles-Campelo, F.; Bolaños-Rivero, M.; Römling, U. Shewanella spp. Infections in Gran Canaria, Spain: Retrospective analysis of 31 cases and a literature review. JMM Case Rep. 2017, 4, e005131. [Google Scholar] [CrossRef]
- Ullah, S.; Mehmood, H.; Pervin, N.; Zeb, H.; Kamal, K.R.; Liaqat, S. Shewanella putrefaciens: An emerging cause of nosocomial pneumonia. J. Investig. Med. High Impact Case Rep. 2018, 6, 232470961877544. [Google Scholar] [CrossRef]
- Zong, Z. Discovery of blaOXA-199, a chromosome-based blaOXA-48-like variant, in Shewanella xiamenensis. PLoS ONE 2012, 7, e48280. [Google Scholar] [CrossRef]
- Parmeciano Di Noto, G.; Jara, E.; Iriarte, A.; Centrón, D.; Quiroga, C. Genome analysis of a clinical isolate of Shewanella sp. uncovered an active hybrid integrative and conjugative element carrying an integron platform inserted in a novel genomic locus. Microbiology 2016, 162, 1335–1345. [Google Scholar] [CrossRef]
- Araújo, S.; Azenha, S.R.; Henriques, I.; Tacão, M. QnrA Gene Diversity in Shewanella spp. Microbiology 2021, 167, 001118. [Google Scholar] [CrossRef]
- Yousfi, K.; Bekal, S.; Usongo, V.; Touati, A. Current trends of human infections and antibiotic resistance of the genus Shewanella. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 1353–1362. [Google Scholar] [CrossRef]
- Chia-Wei, L.; Cheng, J.F.; Tung, K.C.; Hong, Y.K.; Lin, J.H.; Lin, Y.H.; Tsai, C.A.; Lin, S.P.; Chen, Y.C.; Shi, Z.Y.; et al. Evolution of trimethoprim/sulfamethoxazole resistance in Shewanella algae from the perspective of comparative genomics and global phylogenic analysis. J. Microbiol. Immunol. Infect. 2021, 13, S1684-1182(21)00202-4. [Google Scholar] [CrossRef]
- Nandi, S.; Maurer, J.J.; Hofacre, C.; Summers, A.O. Gram-positive bacteria are a major reservoir of class 1 antibiotic resistance integrons in poultry litter. Proc. Natl. Acad. Sci. USA 2004, 101, 7118–7122. [Google Scholar] [CrossRef] [PubMed]
- Cury, J.; Jové, T.; Touchon, M.; Néron, B.; Rocha, E.P. Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic Acids Res. 2016, 44, 4539–4550. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.M. Integrons and gene cassettes: Hotspots of diversity in bacterial genomes. Ann. N. Y. Acad. Sci. 2012, 1267, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Cambray, G.; Guerout, A.-M.; Mazel, D. Integrons. Annu. Rev. Genet. 2010, 44, 141–166. [Google Scholar] [CrossRef] [PubMed]
- Ghaly, T.M.; Geoghegan, J.L.; Tetu, S.G.; Gillings, M.R. The peril and promise of integrons: Beyond antibiotic resistance. Trends Microbiol. 2020, 28, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Larouche, A.; Roy, P.H. Effect of attC structure on cassette excision by integron integrases. Mob. DNA 2011, 2, 3–13. [Google Scholar] [CrossRef]
- Esposito, D.; Scocca, J.J. The integrase family of tyrosine recombinases: Evolution of a conserved active site domain. Nucleic Acids Res. 1997, 25, 3605–3614. [Google Scholar] [CrossRef]
- Nunes-Duby, S.E.; Kwon, H.J.; Tirumalai, R.S.; Ellenberger, T.; Landy, A. Similarities and differences among 105 members of the Int family of site-specific recombinases. Nucleic Acids Res. 1998, 26, 391–406. [Google Scholar] [CrossRef]
- Messier, N.; Roy, P.H. Integron integrases possess a unique additional domain necessary for activity. J. Bacteriol. 2001, 183, 6699–6706. [Google Scholar] [CrossRef]
- Mazel, D. Integrons: Agents of bacterial evolution. Nat. Rev. Microbiol. 2006, 4, 608–620. [Google Scholar] [CrossRef]
- Boucher, Y.; Labbate, M.; Koenig, J.E.; Stokes, H.W. Integrons: Mobilizable platforms that promote genetic diversity in bacteria. Trends Microbiol. 2007, 15, 301–309. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.M.; Holmes, A.J.; Roy, P.H.; Stokes, H.W. What are superintegrons? Nat. Rev. Microbiol. 2007, 5, 1–2. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Partridge, S.R.; Tsafnat, G.; Coiera, E.; Iredell, J.R. Gene cassettes and cassette arrays in mobile resistance integrons. FEMS Microbiol. Rev. 2009, 33, 757–784. [Google Scholar] [CrossRef] [PubMed]
- Loot, C.; Nivina, A.; Cury, J.; Escudero, J.A.; Ducos-Galand, M.; Bikard, D.; Rocha, E.P.C.; Mazel, D. Differences in integron cassette excision dynamics shape a trade-off between evolvability and genetic capacitance. mBio 2017, 8, e02296-16. [Google Scholar] [CrossRef] [PubMed]
- Roy, P.H.; Partridge, S.R.; Hall, R.M. Comment on “Conserved phylogenetic distribution and limited antibiotic resistance of class 1 integrons revealed by assessing the bacterial genome and plasmid collection” by A.N. Zhang et al. Microbiome 2021, 9, 3. [Google Scholar] [CrossRef] [PubMed]
- Ghaly, T.M.; Gillings, M.R.; Penesyan, A.; Qi, Q.; Rajabal, V.; Tetu, S.G. The natural history of integrons. Microorganisms 2021, 9, 2212. [Google Scholar] [CrossRef]
- Poirel, L.; Carrër, A.; Pitout, J.D.; Nordmann, P. Integron mobilization unit as a source of mobility of antibiotic resistance genes. Antimicrob. Agents Chemother. 2009, 53, 2492–2498. [Google Scholar] [CrossRef]
- Arduino, S.M.; Roy, P.H.; Jacoby, G.A.; Orman, B.E.; Pineiro, S.A.; Centron, D. blaCTX-M-2 is located in an unusual class 1 integron (In35) which includes orf513. Antimicrob. Agents Chemother. 2002, 46, 2303–2306. [Google Scholar] [CrossRef]
- Leverstein-van Hall, M.A.; Blok, H.E.M.; Donders, A.R.T.; Paauw, A.; Fluit, A.C.; Verhoef, J. Multidrug resistance among enterobacteriaceae is strongly associated with the presence of integrons and is independent of species or isolate origin. J. Infect. Dis. 2003, 187, 251–259. [Google Scholar] [CrossRef]
- Rowe-Magnus, D.A.; Guerout, A.-M.; Biskri, L.; Bouige, P.; Mazel, D. Comparative analysis of superintegrons: Engineering extensive genetic diversity in the Vibrionaceae. Genome Res. 2003, 13, 428–442. [Google Scholar] [CrossRef]
- Hochhut, B.; Lotfi, Y.; Mazel, D.; Faruque, S.M.; Woodgate, R.; Waldor, M.K. Molecular analysis of antibiotic resistance gene clusters in Vibrio cholerae O139 and O1 SXT Constins. Antimicrob. Agents Chemother. 2001, 45, 2991–3000. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.-W.; Doak, T.G.; Ye, Y. The gain and loss of chromosomal integron systems in the Treponema species. BMC Evol. Biol. 2013, 13, 16. [Google Scholar] [CrossRef] [PubMed]
- Escudero, J.A.; Loot, C.; Nivina, A.; Mazel, D. The integron: Adaptation on demand. Microbiol. Spectr. 2015, 3, 139–161. [Google Scholar] [CrossRef] [PubMed]
- Drouin, F.; Mélançon, J.; Roy, P.H. The IntI-like tyrosine recombinase of Shewanella oneidensis is active as an integron integrase. J. Bacteriol. 2002, 184, 1811–1815. [Google Scholar] [CrossRef] [PubMed]
- Léon, G.; Roy, P.H. Excision and integration of cassettes by an integron integrase of Nitrosomonas europaea. J. Bacteriol. 2003, 185, 2036–2041. [Google Scholar] [CrossRef]
- Nemergut, D.R.; Martin, A.P.; Schmidt, S.K. Integron diversity in heavy-metal-contaminated mine tailings and inferences about integron evolution. Appl. Environ. Microbiol. 2004, 70, 1160–1168. [Google Scholar] [CrossRef]
- Gillings, M.R.; Holley, M.P.; Stokes, H.W.; Holmes, A.J. Integrons in Xanthomonas: A source of species genome diversity. Proc. Natl. Acad. Sci. USA 2005, 102, 4419–4424. [Google Scholar] [CrossRef]
- Larouche, A.; Roy, P.H. Analysis by mutagenesis of a chromosomal integron integrase from Shewanella amazonensis SB2BT. J. Bacteriol. 2009, 191, 1933–1940. [Google Scholar] [CrossRef][Green Version]
- UNAFold. Mfold Web Server for Nucleic Acid Folding and Hybridization Prediction. Nucleic. Acids. Res. 2003, 31, 3406–3415. [Google Scholar] [CrossRef]
- Campanella, J.J.; Bitincka, L.; Smalley, J. MatGAT: An application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinform. 2003, 4, 29. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Elsaied, H.; Stokes, H.W.; Kitamura, K.; Kurusu, Y.; Kamagata, Y.; Maruyama, A. Marine integrons containing novel integrase genes, attachment sites, attI, and associated gene cassettes in polluted sediments from Suez and Tokyo Bays. ISME J. 2011, 5, 1162–1177. [Google Scholar] [CrossRef]
- Pong, C.H. Evolution of IS26-bounded pseudo-compound transposons carrying the Tet(C) tetracycline resistance determinant. Plasmid 2020, 112, 102541. [Google Scholar] [CrossRef]
- Gillings, M.R. Integrons: Past, present, and future. Microbiol. Mol. Biol. Rev. 2014, 78, 257–277. [Google Scholar] [CrossRef]
- Centrón, D.; Roy, P.H. Presence of a group II intron in a multiresistant Serratia marcescens strain that harbors three integrons and a novel gene fusion. Antimicrob. Agents Chemother. 2002, 46, 1402–1409. [Google Scholar] [CrossRef]
- Quiroga, C.; Roy, P.H.; Centrón, D. The S.ma.I2 class C group II intron inserts at integron attC sites. Microbiology 2008, 154, 1341–1353. [Google Scholar] [CrossRef]
- Post, V.; Hall, R.M. Insertion sequences in the IS1111 family that target the attC recombination sites of integron-associated gene cassettes. FEMS Microbiol. Lett. 2008, 290, 182–187. [Google Scholar] [CrossRef]
- Nield, B.S.; Holmes, A.J.; Gillings, M.R.; Recchia, G.D.; Mabbutt, B.C.; Nevalainen, K.M.H.; Stokes, H.W. Recovery of new integron classes from environmental DNA. FEMS Microbiol. Lett. 2001, 195, 59–65. [Google Scholar] [CrossRef]
- Elsaied, H.; Stokes, H.W.; Nakamura, T.; Kitamura, K.; Fuse, H.; Maruyama, A. Novel and diverse integron integrase genes and integron-like gene cassettes are prevalent in deep-sea hydrothermal vents. Environ. Microbiol. 2007, 9, 2298–2312. [Google Scholar] [CrossRef] [PubMed]
- Abella, J.; Fahy, A.; Duran, R.; Cagnon, C. Integron diversity in bacterial communities of freshwater sediments at different contamination levels. FEMS Microbiol. Ecol. 2015, 91, fiv140. [Google Scholar] [CrossRef] [PubMed]
- Buongermino Pereira, M.; Österlund, T.; Eriksson, K.M.; Backhaus, T.; Axelson-Fisk, M.; Kristiansson, E. A comprehensive survey of integron-associated genes present in metagenomes. BMC Genom. 2020, 21, 495. [Google Scholar] [CrossRef] [PubMed]
- Antelo, V.; Giménez, M.; Azziz, G.; Valdespino-Castillo, P.; Falcón, L.I.; Ruberto, L.A.M.; Mac Cormack, W.P.; Mazel, D.; Batista, S. Metagenomic strategies identify diverse integron-integrase and antibiotic resistance genes in the Antarctic environment. MicrobiologyOpen 2021, 10, e1219. [Google Scholar] [CrossRef] [PubMed]
- Rapa, R.A.; Labbate, M. The function of integron-associated gene cassettes in Vibrio species: The tip of the iceberg. Front. Microbiol. 2013, 4, 385. [Google Scholar] [CrossRef] [PubMed]
- Rowe-Magnus, D.A.; Guerout, A.-M.; Mazel, D. Bacterial resistance evolution by recruitment of super-integron gene cassettes. Mol. Microbiol. 2002, 43, 1657–1669. [Google Scholar] [CrossRef] [PubMed]
- Yousfi, K.; Touati, A.; Lefebvre, B.; Fournier, É.; Côté, J.-C.; Soualhine, H.; Walker, M.; Bougdour, D.; Tremblay, C.; Bekal, S. A novel plasmid, pSx1, harboring a new Tn1696 derivative from extensively drug-resistant Shewanella xiamenensis Encoding OXA-416. Microb. Drug Resist. 2017, 23, 429–436. [Google Scholar] [CrossRef]
- Da Re, S.; Ploy, M.-C. Antibiotiques et réponse SOS bactérienne: Une voie efficace d’acquisition des résistances aux antibiotiques. Med. Sci. 2012, 28, 179–184. [Google Scholar] [CrossRef][Green Version]
- Souque, C.; Escudero, J.A.; MacLean, R.C. Integron activity accelerates the evolution of antibiotic resistance. eLife 2021, 10, e62474. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ayala Nuñez, T.; Cerbino, G.N.; Rapisardi, M.F.; Quiroga, C.; Centrón, D. Novel Mobile Integrons and Strain-Specific Integrase Genes within Shewanella spp. Unveil Multiple Lateral Genetic Transfer Events within The Genus. Microorganisms 2022, 10, 1102. https://doi.org/10.3390/microorganisms10061102
Ayala Nuñez T, Cerbino GN, Rapisardi MF, Quiroga C, Centrón D. Novel Mobile Integrons and Strain-Specific Integrase Genes within Shewanella spp. Unveil Multiple Lateral Genetic Transfer Events within The Genus. Microorganisms. 2022; 10(6):1102. https://doi.org/10.3390/microorganisms10061102
Chicago/Turabian StyleAyala Nuñez, Teolincacihuatl, Gabriela N. Cerbino, María Florencia Rapisardi, Cecilia Quiroga, and Daniela Centrón. 2022. "Novel Mobile Integrons and Strain-Specific Integrase Genes within Shewanella spp. Unveil Multiple Lateral Genetic Transfer Events within The Genus" Microorganisms 10, no. 6: 1102. https://doi.org/10.3390/microorganisms10061102
APA StyleAyala Nuñez, T., Cerbino, G. N., Rapisardi, M. F., Quiroga, C., & Centrón, D. (2022). Novel Mobile Integrons and Strain-Specific Integrase Genes within Shewanella spp. Unveil Multiple Lateral Genetic Transfer Events within The Genus. Microorganisms, 10(6), 1102. https://doi.org/10.3390/microorganisms10061102






