Serological Hendra Virus Diagnostics Using an Indirect ELISA-Based DIVA Approach with Recombinant Hendra G and N Proteins
Abstract
1. Introduction
2. Materials and Methods
2.1. Serum Samples
2.2. Expression of Viral Proteins
2.3. Generation of a Rabbit Polyclonal Antibody Raised against HeV-N
2.4. Development of an Indirect ELISA (FLI HeV DIVA ELISA)
2.5. Evaluation of HeV-N and HeV-G Assays Using Frequentist Approach and Determination of the Provisional Cut-Off Value
2.6. Confirmation of ELISA Positive Results
2.7. Western Blot
2.8. Immunofluorescence Analysis (IF Analysis)
2.9. Virus Neutralization Test (VNT)
2.10. ACDP HeV DIVA ELISA Used as Reference Test
2.11. Determination of Analytical Sensitivity (ASe) and Specificity (ASp)
2.12. Evaluation of HeV-N and HeV-G Assays Using Bayesian Latent Class Modelling
3. Results
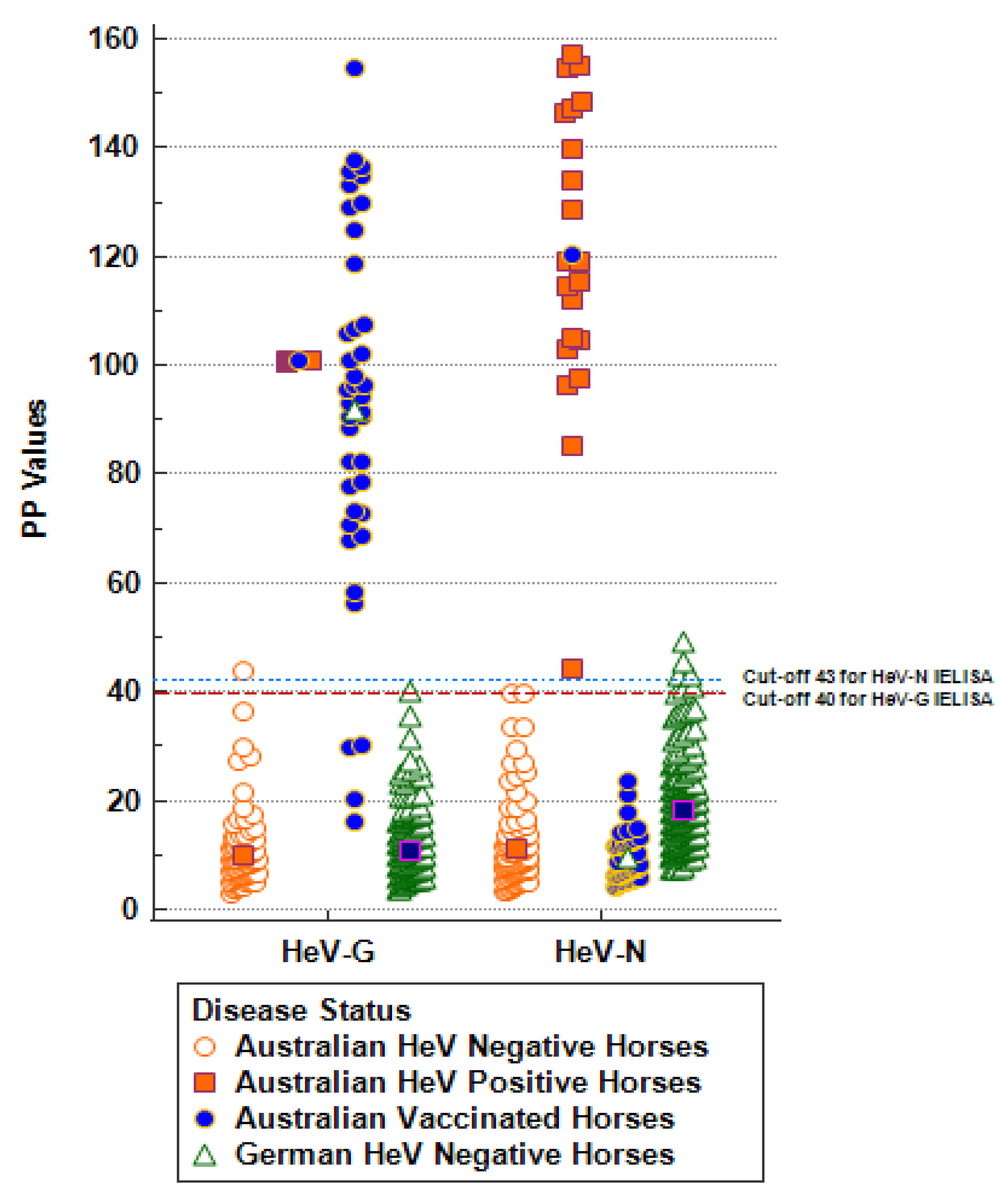
3.1. Provisional Cut-Off Value Determination for the Indirect DIVA ELISA
3.2. Cut-Off by ROC Analysis and Diagnostic Test Sensitivity (DSe) and Diagnostic Test Specificity (DSp) by Frequentist Approach
3.3. Relative Diagnostic Test Sensitivity (DSe) and Diagnostic Test Specificity (DSp) by BLCM Approach
3.4. Re-Testing of Reactive Samples by Other Serological Methods
3.5. Determination of Analytical Sensitivity (ASe) and Specificity (ASp)
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Selvey, L.A.; Wells, R.M.; McCormack, J.G.; Ansford, A.J.; Murray, K.; Rogers, R.J.; Lavercombe, P.S.; Selleck, P.; Sheridan, J.W. Infection of humans and horses by a newly described morbillivirus. Med. J. Aust. 1995, 162, 642–645. [Google Scholar] [CrossRef] [PubMed]
- Chua, K.B.; Goh, K.J.; Wong, K.T.; Kamarulzaman, A.; Tan, P.S.; Ksiazek, T.G.; Zaki, S.R.; Paul, G.; Lam, S.K.; Tan, C.T. Fatal encephalitis due to Nipah virus among pig-farmers in Malaysia. Lancet 1999, 354, 1257–1259. [Google Scholar] [CrossRef]
- Marsh, G.A.; de Jong, C.; Barr, J.A.; Tachedjian, M.; Smith, C.; Middleton, D.; Yu, M.; Todd, S.; Foord, A.J.; Haring, V.; et al. Cedar virus: A novel Henipavirus isolated from Australian bats. PLoS Pathog. 2012, 8, e1002836. [Google Scholar] [CrossRef] [PubMed]
- Drexler, J.F.; Corman, V.M.; Gloza-Rausch, F.; Seebens, A.; Annan, A.; Ipsen, A.; Kruppa, T.; Müller, M.A.; Kalko, E.K.; Adu-Sarkodie, Y.; et al. Henipavirus RNA in African bats. PLoS ONE 2009, 4, e6367. [Google Scholar] [CrossRef] [PubMed]
- Mbu’u, C.M.; Mbacham, W.F.; Gontao, P.; Sado Kamdem, S.L.; Nlôga, A.M.N.; Groschup, M.H.; Wade, A.; Fischer, K.; Balkema-Buschmann, A. Henipaviruses at the Interface Between Bats, Livestock and Human Population in Africa. Vector Borne Zoonotic Dis. 2019, 19, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Yang, L.; Yang, F.; Ren, X.; Jiang, J.; Dong, J.; Sun, L.; Zhu, Y.; Zhou, H.; Jin, Q. Novel Henipa-like virus, Mojiang Paramyxovirus, in rats, China, 2012. Emerg. Infect. Dis. 2014, 20, 1064–1066. [Google Scholar] [CrossRef]
- Halpin, K.; Young, P.L.; Field, H.; Mackenzie, J.S. Newly discovered viruses of flying foxes. Vet. Microbiol. 1999, 68, 83–87. [Google Scholar] [CrossRef]
- Yob, J.M.; Field, H.; Rashdi, A.M.; Morrissy, C.; van der Heide, B.; Rota, P.; bin Adzhar, A.; White, J.; Daniels, P.; Jamaluddin, A.; et al. Nipah virus infection in bats (order Chiroptera) in peninsular Malaysia. Emerg. Infect. Dis. 2001, 7, 439–441. [Google Scholar] [CrossRef]
- Barr, J.; Smith, C.; Smith, I.; de Jong, C.; Todd, S.; Melville, D.; Broos, A.; Crameri, S.; Haining, J.; Marsh, G.; et al. Isolation of multiple novel paramyxoviruses from pteropid bat urine. J. Gen. Virol. 2015, 96 Pt 1, 24–29. [Google Scholar] [CrossRef]
- Middleton, D.J.; Weingartl, H.M. Henipaviruses in their natural animal hosts. Curr. Top Microbiol. Immunol. 2012, 359, 105–121. [Google Scholar]
- Plowright, R.K.; Field, H.E.; Smith, C.; Divljan, A.; Palmer, C.; Tabor, G.; Daszak, P.; Foley, J.E. Reproduction and nutritional stress are risk factors for Hendra virus infection in little red flying foxes (Pteropus scapulatus). Proc. Biol. Sci. 2008, 275, 861–869. [Google Scholar] [CrossRef] [PubMed]
- Plowright, R.K.; Peel, A.J.; Streicker, D.G.; Gilbert, A.T.; McCallum, H.; Wood, J.; Baker, M.L.; Restif, O. Transmission or Within-Host Dynamics Driving Pulses of Zoonotic Viruses in Reservoir-Host Populations. PLoS Negl. Trop. Dis. 2016, 10, e0004796. [Google Scholar] [CrossRef] [PubMed]
- Páez, D.J.; Giles, J.; McCallum, H.; Field, H.; Jordan, D.; Peel, A.J.; Plowright, R.K. Conditions affecting the timing and magnitude of Hendra virus shedding across pteropodid bat populations in Australia. Epidemiol. Infect. 2017, 145, 3143–3153. [Google Scholar] [CrossRef] [PubMed]
- Queensland Government. Summary of Hendra Virus Incidents in Horses. 2022. Available online: https://www.business.qld.gov.au/industries/service-industries-professionals/service-industries/veterinary-surgeons/guidelines-hendra/incident-summary (accessed on 19 April 2022).
- Middleton, D.; Pallister, J.; Klein, R.; Feng, Y.R.; Haining, J.; Arkinstall, R.; Frazer, L.; Huang, J.A.; Edwards, N.; Wareing, M.; et al. Hendra virus vaccine, a one health approach to protecting horse, human, and environmental health. Emerg. Infect. Dis. 2014, 20, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Manyweathers, J.; Field, H.; Longnecker, N.; Agho, K.; Smith, C.; Taylor, M. “Why won’t they just vaccinate?” Horse owner risk perception and uptake of the Hendra virus vaccine. BMC Vet. Res. 2017, 13, 103. [Google Scholar] [CrossRef]
- Daniels, P.; Ksiazek, T.; Eaton, B.T. Laboratory diagnosis of Nipah and Hendra virus infections. Microbes. Infect. 2001, 3, 289–295. [Google Scholar] [CrossRef]
- Mungall, B.A.; Middleton, D.; Crameri, G.; Bingham, J.; Halpin, K.; Russell, G.; Green, D.; McEachern, J.; Pritchard, L.I.; Eaton, B.T.; et al. Feline model of acute nipah virus infection and protection with a soluble glycoprotein-based subunit vaccine. J. Virol. 2006, 80, 12293–12302. [Google Scholar] [CrossRef]
- Feldman, K.S.; Foord, A.; Heine, H.G.; Smith, I.L.; Boyd, V.; Marsh, G.A.; Wood, J.L.; Cunningham, A.A.; Wang, L.F. Design and evaluation of consensus PCR assays for henipaviruses. J. Virol. Methods 2009, 161, 52–57. [Google Scholar] [CrossRef]
- Chen, J.M.; Yaiw, K.C.; Yu, M.; Wang, L.F.; Wang, Q.H.; Crameri, G.; Wang, Z.L. Expression of truncated phosphoproteins of Nipah virus and Hendra virus in Escherichia coli for the differentiation of henipavirus infections. Biotechnol. Lett. 2007, 29, 871–875. [Google Scholar] [CrossRef]
- Wang, L.F.; Daniels, P. Diagnosis of henipavirus infection: Current capabilities and future directions. Curr. Top Microbiol. Immunol. 2012, 359, 179–196. [Google Scholar]
- Colling, A.; Lunt, R.; Bergfeld, J.; McNabb, L.; Halpin, K.; Juzva, S.; Newberry, K.; Morrissy, C.; Loomes, C.; Warner, S.; et al. A network approach for provisional assay recognition of a Hendra virus antibody ELISA: Test validation with low sample numbers from infected horses. J. Vet. Diagn. Invest 2018, 30, 362–369. [Google Scholar] [CrossRef] [PubMed]
- Di Rubbo, A.; McNabb, L.; Klein, R.; White, J.R.; Colling, A.; Dimitrov, D.S.; Broder, C.C.; Middleton, D.; Lunt, R.A. Optimization and diagnostic evaluation of monoclonal antibody-based blocking ELISA formats for detection of neutralizing antibodies to Hendra virus in mammalian sera. J. Virol. Methods 2019, 274, 113731. [Google Scholar] [CrossRef] [PubMed]
- McNabb, L.; Andiani, A.; Bulavaite, A.; Zvirbliene, A.; Sasnauskas, K.; Lunt, R. Development and validation of an IgM antibody capture ELISA for early detection of Hendra virus. J. Virol. Methods 2021, 298, 114296. [Google Scholar] [CrossRef] [PubMed]
- Bossart, K.N.; McEachern, J.A.; Hickey, A.C.; Choudhry, V.; Dimitrov, D.S.; Eaton, B.T.; Wang, L.F. Neutralization assays for differential henipavirus serology using Bio-Plex protein array systems. J. Virol. Methods 2007, 142, 29–40. [Google Scholar] [CrossRef]
- Hayman, D.T.; Suu-Ire, R.; Breed, A.C.; McEachern, J.A.; Wang, L.; Wood, J.L.; Cunningham, A.A. Evidence of henipavirus infection in West African fruit bats. PLoS ONE 2008, 3, e2739. [Google Scholar] [CrossRef]
- Peel, A.J.; McKinley, T.J.; Baker, K.S.; Barr, J.A.; Crameri, G.; Hayman, D.T.; Feng, Y.R.; Broder, C.C.; Wang, L.F.; Cunningham, A.A.; et al. Use of cross-reactive serological assays for detecting novel pathogens in wildlife: Assessing an appropriate cutoff for henipavirus assays in African bats. J. Virol. Methods 2013, 193, 295–303. [Google Scholar] [CrossRef]
- Gao, Y.; Pallister, J.; Lapierre, F.; Crameri, G.; Wang, L.F.; Zhu, Y. A rapid assay for Hendra virus IgG antibody detection and its titre estimation using magnetic nanoparticles and phycoerythrin. J. Virol. Methods 2015, 222, 170–177. [Google Scholar] [CrossRef]
- McNabb, L.; Barr, J.; Crameri, G.; Juzva, S.; Riddell, S.; Colling, A.; Boyd, V.; Broder, C.; Wang, L.F.; Lunt, R. Henipavirus microsphere immuno-assays for detection of antibodies against Hendra virus. J. Virol. Methods 2014, 200, 22–28. [Google Scholar] [CrossRef]
- Fischer, K.; dos Reis, V.P.; Finke, S.; Sauerhering, L.; Stroh, E.; Karger, A.; Maisner, A.; Groschup, M.H.; Diederich, S.; Balkema-Buschmann, A. Expression, characterisation and antigenicity of a truncated Hendra virus attachment protein expressed in the protozoan host Leishmania tarentolae. J. Virol. Methods 2016, 228, 48–54. [Google Scholar] [CrossRef][Green Version]
- Jacobson, R.H. Validation of serological assays for diagnosis of infectious diseases. Rev. Sci. Tech. 1998, 17, 469–526. [Google Scholar] [CrossRef]
- Fischer, K.; Diederich, S.; Smith, G.; Reiche, S.; Pinho Dos Reis, V.; Stroh, E.; Groschup, M.H.; Weingartl, H.M.; Balkema-Buschmann, A. Indirect ELISA based on Hendra and Nipah virus proteins for the detection of henipavirus specific antibodies in pigs. PLoS ONE 2018, 13, e0194385. [Google Scholar] [CrossRef]
- Bossart, K.N.; Crameri, G.; Dimitrov, A.S.; Mungall, B.A.; Feng, Y.R.; Patch, J.R.; Choudhary, A.; Wang, L.F.; Eaton, B.T.; Broder, C.C. Receptor binding, fusion inhibition, and induction of cross-reactive neutralizing antibodies by a soluble G glycoprotein of Hendra virus. J. Virol. 2005, 79, 6690–6702. [Google Scholar] [CrossRef]
- Stevenson, M.; Nunes, T.; Sanchez, J.; Thornton, R.; Reiczigel, J.; Robison-Cox, J.; Sebastiani, P. EpiR: An R Package for the Analysis of Epidemiological Data; R Package Version 0.9-43; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Cheung, A.; Dufour, S.; Jones, G.; Kostoulas, P.; Stevenson, M.A.; Singanallur, N.B.; Firestone, S.M. Bayesian latent class analysis when the reference test is imperfect. Rev. Sci. Tech. 2021, 40, 271–286. [Google Scholar] [CrossRef] [PubMed]
- Branscum, A.; Gardner, I.; Johnson, W. Estimation of diagnostic-test sensitivity and specificity through Bayesian modeling. Prev. Vet. Med. 2005, 68, 145–163. [Google Scholar] [CrossRef] [PubMed]
- Christensen, R.; Branscum, A.; Hanson, T.E.; Johnson, W. Bayesian Ideas and Data Analysis: An Introduction for Scientists and Statisticians/by Ronald Christensen, Wesley Johnson, Adam Branscum and Timothy E Hanson, 1st ed.; CRC Press, An Imprint of Taylor and Francis: Boca Raton, FL, USA, 2010. [Google Scholar]
- Dendukuri, N.; Joseph, L. Bayesian approaches to modeling the conditional dependence between multiple diagnostic tests. Biometrics 2001, 57, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Enøe, C.; Georgiadis, M.P.; Johnson, W.O. Estimation of sensitivity and specificity of diagnostic tests and disease prevalence when the true disease state is unknown. Prev. Vet. Med. 2000, 45, 61–81. [Google Scholar] [CrossRef]
- RC Team. R: A Language and Environment for Statistical Computing. 2017. Available online: https://www.R-project.org/ (accessed on 19 April 2022).
- Georgiadis, M.P.; Johnson, W.O.; Gardner, I.A.; Singh, R. Correlation-Adjusted Estimation of Sensitivity and Specificity of Two Diagnostic Tests. J. R. Stat. Society. Ser. C (Appl. Stat.) 2003, 52, 63–76. [Google Scholar] [CrossRef]
- Lunn, D.; Spiegelhalter, D.; Thomas, A.; Best, N. The BUGS project: Evolution, critique and future directions. Stat. Med. 2009, 28, 3049–3067. [Google Scholar] [CrossRef]
- Saatkamp, H.W.; Berentsen, P.B.; Horst, H.S. Economic aspects of the control of classical swine fever outbreaks in the European Union. Vet. Microbiol. 2000, 73, 221–237. [Google Scholar] [CrossRef]
- Galvin, P.; Gildea, S.; Arkins, S.; Walsh, C.; Cullinane, A. The evaluation of a nucleoprotein ELISA for the detection of equine influenza antibodies and the differentiation of infected from vaccinated horses (DIVA). Influenza Other Respir Viruses 2013, 7 (Suppl. S4), 73–80. [Google Scholar] [CrossRef]
- Avellaneda, G.; Mundt, E.; Lee, C.W.; Jadhao, S.; Suarez, D.L. Differentiation of infected and vaccinated animals (DIVA) using the NS1 protein of avian influenza virus. Avian Dis. 2010, 54 (Suppl. S1), 278–286. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Feng, J.; Quan, K.; Sun, Z.; Yin, Y.; Yin, Y.; Chen, S.; Qin, T.; Peng, D.; Liu, X. Generation of an avian influenza DIVA vaccine with a H3-peptide replacement located at HA2 against both highly and low pathogenic H7N9 virus. Virulence 2022, 13, 530–541. [Google Scholar] [CrossRef] [PubMed]
- Uddowla, S.; Hollister, J.; Pacheco, J.M.; Rodriguez, L.L.; Rieder, E. A Safe Foot-and-Mouth Disease Vaccine Platform with Two Negative Markers for Differentiating Infected from Vaccinated Animals. J. Virol. 2012, 86, 11675–11685. [Google Scholar] [CrossRef] [PubMed]
- Silva-Junior, L.C.; Fontes, K.; Nascimento, S.A.; Rodriguez, M.C.; Camargos, M.F.; Freitas, A.C.; Castro, R.S.; Jesus, A.L.S. Development of a DIVA ELISA for diagnosis of Aujeszky’s disease using recombinant gE fused to thioredoxin as antigen. Vet. J. 2020, 257, 105448. [Google Scholar] [CrossRef]
- Li, M.; Embury-Hyatt, C.; Weingartl, H.M. Experimental inoculation study indicates swine as a potential host for Hendra virus. Vet. Res. 2010, 41, 33. [Google Scholar] [CrossRef]
- Halpin, K.; Graham, K.; Durr, P.A. Sero-Monitoring of Horses Demonstrates the Equivac® HeV Hendra Virus Vaccine to Be Highly Effective in Inducing Neutralising Antibody Titres. Vaccines 2021, 9, 731. [Google Scholar] [CrossRef]
- Kirkland, P.D.; Gabor, M.; Poe, I.; Neale, K.; Chaffey, K.; Finlaison, D.S.; Gu, X.; Hick, P.M.; Read, A.J.; Wright, T.; et al. Hendra Virus Infection in Dog, Australia, 2013. Emerg. Infect. Dis. 2015, 21, 2182–2185. [Google Scholar] [CrossRef]
- Chowdhury, S.; Khan, S.U.; Crameri, G.; Epstein, J.H.; Broder, C.C.; Islam, A.; Peel, A.J.; Barr, J.; Daszak, P.; Wang, L.F.; et al. Serological evidence of henipavirus exposure in cattle, goats and pigs in Bangladesh. PLoS Negl. Trop. Dis. 2014, 8, e3302. [Google Scholar] [CrossRef]
- Hayman, D.T.; Wang, L.F.; Barr, J.; Baker, K.S.; Suu-Ire, R.; Broder, C.C.; Cunningham, A.A.; Wood, J.L. Antibodies to henipavirus or henipa-like viruses in domestic pigs in Ghana, West Africa. PLoS ONE 2011, 6, e25256. [Google Scholar] [CrossRef]
- Pernet, O.; Schneider, B.S.; Beaty, S.M.; LeBreton, M.; Yun, T.E.; Park, A.; Zachariah, T.T.; Bowden, T.A.; Hitchens, P.; Ramirez, C.M.; et al. Evidence for henipavirus spillover into human populations in Africa. Nat. Commun. 2014, 5, 5342. [Google Scholar] [CrossRef]


| Panel | Purpose, Determination of | Source of Serum | Assays Performed |
|---|---|---|---|
| 1 | Cut-off DSp | German negative horses (n = 288); These samples had been submitted from different clinics to the National Reference Laboratory for West Nile Virus (WNV) between 2009 and 2012 for WNV screening. None of the horses had a history of travelling to Australia or being vaccinated against HeV, and these samples were therefore considered HeV-negative | Preliminary Cut-off determination ROC curve analysis FLI HeV DIVA ELISA, ACDP HeV DIVA ELISA, HeV VNT |
| 2 | Cut-off DSp | Australian negative horses (n = 105) | Cut-off determination ROC curve analysis BLCM FLI HeV DIVA ELISA, ACDP HeV DIVA ELISA, HeV VNT |
| 3 | Cut-off DSe, DSp | Australian vaccinated horses (n = 40); diagnostic field samples | FLI HeV DIVA ELISA, ACDP HeV DIVA ELISA BLCM |
| 4 | Cut-off DSe ASe | Australian HeV-positive samples (n = 21) from outbreak episodes (QLD) and follow-up testing | Cut-off determination and ROC curve analysis BLCM FLI HeV DIVA ELISA, ACDP HeV DIVA ELISA, HeV VNT |
| 5 | ASp | Serum samples originating from different species (n = 17) including horses, guinea pigs, pigs, rabbits, and goats, containing antibodies against different paramyxoviruses (peste des petits ruminants virus, rinderpest virus, canine distemper virus, Newcastle disease virus, parainfluenzavirus type 1–4, mumps virus, Nariva virus, Tioman virus, Menangle virus, blue eye rubulavirus, Mossman virus) | FLI HeV DIVA ELISA |
| Population | HeV-G Assay | HeV-N Assay | ||||
|---|---|---|---|---|---|---|
| Cross-Classified Counts * | Diagnostic Sensitivity (DSe) | Diagnostic Specificity (DSp) | Cross-Classified Counts * | Diagnostic Sensitivity (DSe) | Diagnostic Specificity (DSp) | |
| German negative horse sera | n.a. | n.a. | 99.04% | n.a. | n.a. | 99.30% |
| (284/288) | (286/288) | |||||
| Australian negative horse sera | 0, 1, 0, 104 | n.a. | 99.04% | 0, 0, 0, 105 | n.a. | 100% |
| (104/105) | (105/105) | |||||
| Australian vaccinated horse sera | 36, 0, 4, 0 | 90.00% | n.a. | 0, 0, 0, 40 | n.a. | 100% |
| (36/40) | (40/40) | |||||
| Australian HeV-infected horse sera | 21, 0, 0, 0 | 100% | n.a. | 21, 0, 0, 0 | 100% | n.a. |
| (21/21) | (21/21) | |||||
| Sample ID | Sample Information | PP | Result | Interpretation |
|---|---|---|---|---|
| neg control | 8.55 | neg | ||
| pos control | 100.00 | pos | ||
| RTS 1 | infected horse number 1; 1/100 | 47.72 | pos | correct |
| RTS 2 | vaccinated horse | 76.57 | pos | correct |
| RTS 3 | infected horse number 1; 1/150 | 35.28 | neg | questionable |
| RTS 4 | infected horse number 2; 1/60 | 43.59 | pos | correct |
| RTS 5 | negative horse | 8.21 | neg | correct |
| RTS 6 | infected horse number 2; 1/40 | 42.36 | pos | correct |
| RTS 7 | infected horse number 2; 1/60 | 31.43 | neg | questionable |
| RTS 8 | infected horse number 2; 1/40 | 59.40 | pos | correct |
| RTS 9 | negative horse | 10.25 | pos | correct |
| Estimates | HeV-G Assay | HeV-N Assay | DIVA Assay | |||
|---|---|---|---|---|---|---|
| PM | 95% PI | PM | 95% PI | PM | 95% PI | |
| DSeACDP | 0.919 | 0.872, 0.953 | 0.908 | 0.852, 0.950 | 0.910 | 0.854, 0.948 |
| DSpACDP | 0.942 | 0.903, 0.968 | 0.954 | 0.923, 0.976 | 0.952 | 0.915, 0.975 |
| DSeFLI | 0.985 | 0.932, 1.000 | 0.953 | 0.826, 0.998 | 0.941 | 0.810, 0.997 |
| DSpFLI | 0.994 | 0.967, 1.000 | 0.975 | 0.938, 0.994 | 0.993 | 0.965, 1.000 |
| PrAustralian Infected horses | 0.953 | 0.908, 0.981 | 0.953 | 0.907, 0.981 | 0.954 | 0.912, 0.980 |
| PrAustralian Negative horses | 0.002 | 0, 0.009 | 0.002 | 0, 0.009 | 0.030 | 0.013, 0.060 |
| PrAustralian Vaccinated horses | 0.939 | 0.877, 0.977 | 0.014 | 0.002, 0.046 | 0.057 | 0.022, 0.113 |
| Correlation coefficient negative (rhoDc) | 0.185 | −0.016, 0.678 | 0.325 | −0.027, 0.872 | 0.370 | −0.027, 0.865 |
| Correlation coefficient positive (rhoD) | 0.124 | −0.012, 0.475 | 0.094 | −0.030, 0.442 | 0.142 | 0, 0.554 |
| Sera Used for Analytical Specificity (ASp) | Anti-Horse-HRP | Protein A/G-HRP | ||
|---|---|---|---|---|
| HeV G PP Value | HeV N PP Value | HeV G PP Value | HeV N PP Value | |
| No serum control (NSC) | 1.81 | 40.44 | 1.50 | 7.80 |
| Negative horse serum | 1.05 | 34.85 | 5.50 | 6.50 |
| HeV-infected horse | 100.00 | 100.00 | 100.00 | 100.00 |
| Caprine anti-PPR 1.4.78 | n.a. | n.a. | 2.60 | 7.90 |
| Rabbit anti-rinderpest 5.6.78 | n.a. | n.a. | 20.60 | 10.80 |
| Horse anti-canine-distemper-virus 8807-25-0205 | 3.23 | 21.98 | 16.40 | 11.60 |
| Rabbit anti-NDV V4 8604-28-4425 | n.a. | n.a. | 8.50 | 12.90 |
| Horse anti-parainfluenza type 1 (Sendai) 4.17.68 | 2.67 | 25.36 | 3.50 | 11.90 |
| Horse anti-parainfluenza type 2 (SV-5) 11.14.66 | 1.99 | 20.94 | 20.10 | 10.90 |
| Horse anti-parainfluenza type 3 (C-243) 1.5.64 | 3.29 | 53.97 | 1.50 | 6.70 |
| Guinea pig anti-parainfluenza type 1 (C-39) Nov 1964 | n.a. | n.a. | 2.70 | 7.80 |
| Guinea pig anti-parainfluenza type 4A (M-25) | n.a. | n.a. | 1.60 | 6.40 |
| Guinea pig anti-parainfluenza type 4B (19503) | n.a. | n.a. | 1.60 | 6.10 |
| Horse anti-mumps Enders strain 8.9.66 | 6.53 | 43.95 | 1.50 | 6.90 |
| Rabbit anti-Nariva number 1 22/11/2001 | n.a. | n.a. | 1.60 | 6.10 |
| Pig anti-Tioman-virus P298 26/09/05 | n.a. | n.a. | 1.80 | 51.80 |
| Rabbit anti-Menangle-virus number 4 | n.a. | n.a. | 3.50 | 6.40 |
| Pig anti-Menangle-virus number 1 Sept 1999 | n.a. | n.a. | 3.90 | 6.70 |
| Pig anti-blue-eye-rubulavirus 9/9/80 | n.a. | n.a. | 7.90 | 6.70 |
| Rabbit anti-Mossman-virus 27/07/2000 | n.a. | n.a. | 1.90 | 57.1 |
| cut-off | 40 | 43 | 26.1 | 55.4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Balkema-Buschmann, A.; Fischer, K.; McNabb, L.; Diederich, S.; Singanallur, N.B.; Ziegler, U.; Keil, G.M.; Kirkland, P.D.; Penning, M.; Sadeghi, B.; et al. Serological Hendra Virus Diagnostics Using an Indirect ELISA-Based DIVA Approach with Recombinant Hendra G and N Proteins. Microorganisms 2022, 10, 1095. https://doi.org/10.3390/microorganisms10061095
Balkema-Buschmann A, Fischer K, McNabb L, Diederich S, Singanallur NB, Ziegler U, Keil GM, Kirkland PD, Penning M, Sadeghi B, et al. Serological Hendra Virus Diagnostics Using an Indirect ELISA-Based DIVA Approach with Recombinant Hendra G and N Proteins. Microorganisms. 2022; 10(6):1095. https://doi.org/10.3390/microorganisms10061095
Chicago/Turabian StyleBalkema-Buschmann, Anne, Kerstin Fischer, Leanne McNabb, Sandra Diederich, Nagendrakumar Balasubramanian Singanallur, Ute Ziegler, Günther M. Keil, Peter D. Kirkland, Maren Penning, Balal Sadeghi, and et al. 2022. "Serological Hendra Virus Diagnostics Using an Indirect ELISA-Based DIVA Approach with Recombinant Hendra G and N Proteins" Microorganisms 10, no. 6: 1095. https://doi.org/10.3390/microorganisms10061095
APA StyleBalkema-Buschmann, A., Fischer, K., McNabb, L., Diederich, S., Singanallur, N. B., Ziegler, U., Keil, G. M., Kirkland, P. D., Penning, M., Sadeghi, B., Marsh, G., Barr, J., & Colling, A. (2022). Serological Hendra Virus Diagnostics Using an Indirect ELISA-Based DIVA Approach with Recombinant Hendra G and N Proteins. Microorganisms, 10(6), 1095. https://doi.org/10.3390/microorganisms10061095








