Primary Structure and Coding Genes of Two Pheromones from the Antarctic Psychrophilic Ciliate, Euplotes focardii
Abstract
:1. Introduction
2. Materials and Methods
2.1. Pheromone Source
2.2. Pheromone Purification and Analysis
2.3. Assaying Pheromone Activity
2.4. Gene Amplification, Cloning and Sequencing
3. Results
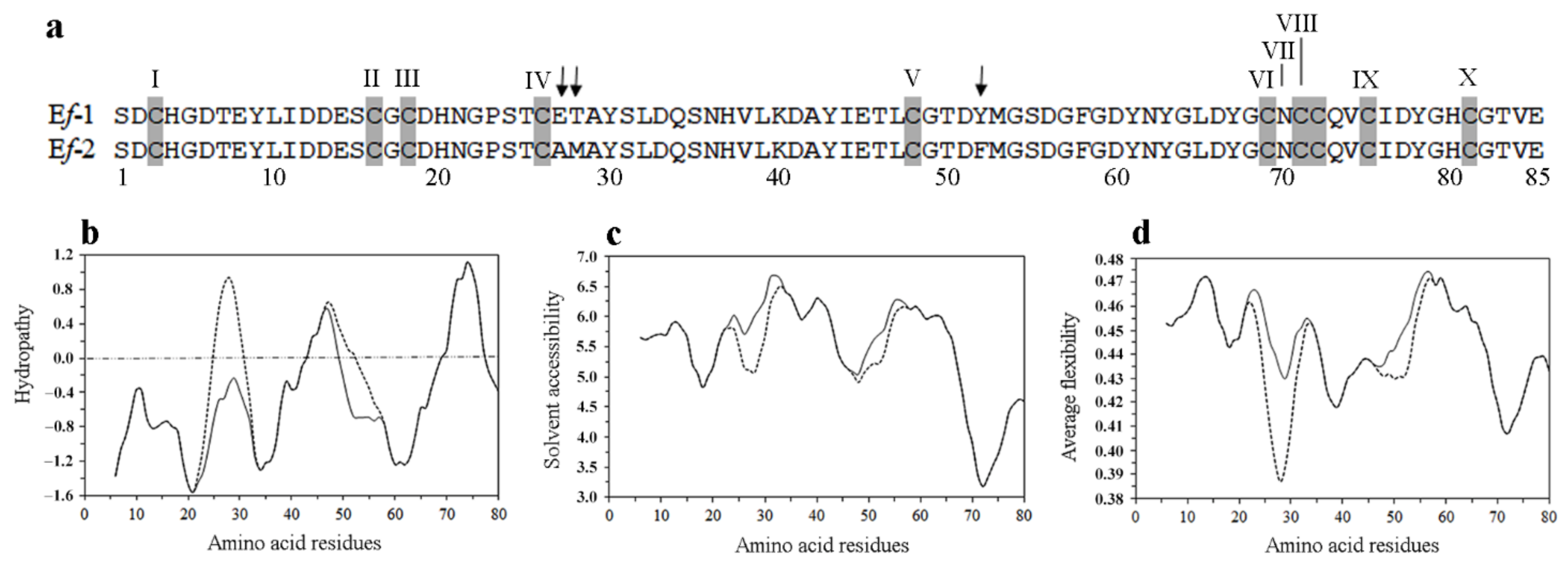
3.1. Pheromone Isolation and Structural Characterization
3.2. Pheromone Gene Structure
3.3. Pheromone Primary Structure
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stephens, K. Pheromones among the prokaryotes. Crit. Rev. Microbiol. 1986, 13, 309–334. [Google Scholar] [CrossRef] [PubMed]
- Cook, L.C.; Federle, M.J. Peptide pheromone signaling in Streptococcus and Enterococcus. FEMS Microbiol. Rev. 2014, 38, 473–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monnet, V.; Juillard, V.; Gardan, R. Peptide conversations in Gram-positive bacteria. Crit. Rev. Microbiol. 2016, 42, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Stabb, E.V. Could positive feedback enable bacterial pheromone signaling to coordinate behaviors in response to heterogeneous environmental cues? MBio 2018, 9, e00098-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyatt, T.D. Pheromones and Animal Behaviour; Cambridge University Press: Cambridge, UK, 2003; Volume 626. [Google Scholar]
- Shorey, H.H. Animal Communication by Pheromones; Academic Press: New York, NY, USA, 2013. [Google Scholar]
- Luporini, P.; Miceli, C.; Ortenzi, C.; Vallesi, A. Ciliate Pheromones. In Signaling Mechanisms in Protozoa and Invertebrates; Progress in Molecular and Subcellular Biology Series; Csaba, G., Müller, W.E.G., Eds.; Springer: Berlin/Heidelberg, Germany, 1996; Volume 17, pp. 80–104. [Google Scholar]
- Vallesi, A.; Giuli, G.; Bradshaw, R.A.; Luporini, P. Autocrine mitogenic activity of pheromones produced by the protozoan ciliate Euplotes raikovi. Nature 1995, 376, 522–524. [Google Scholar] [CrossRef]
- Nobili, R.; Luporini, P.; Dini, F. Breeding system, species relationships and evolutionary trends in some species of Euplotidae (Hypotrichida, Ciliata). In Marine Organisms; Battaglia, B., Beardmore, J., Eds.; Plenum Press: New York, NY, USA, 1978; pp. 591–616. [Google Scholar]
- Dini, F.; Nyberg, D. Sex in ciliates. In Advances in Microbial Ecology; Jones, J.G., Ed.; Plenum Press: New York, NY, USA, 1993; Volume 13, pp. 85–153. [Google Scholar]
- Miyake, A. Fertilization and sexuality in ciliates. In Ciliates, Cells and Organisms; Hausmann, K., Bradbury, P.C., Eds.; Gustav-Fisher Verlag: Stuttgard, Germany, 1996; pp. 244–290. [Google Scholar]
- Luporini, P.; Alimenti, C.; Ortenzi, C.; Vallesi, A. Ciliate mating types and their specific protein pheromones. Acta Protozool. 2005, 44, 89–101. [Google Scholar]
- Luporini, P.; Pedrini, B.; Alimenti, C.; Vallesi, A. Revisiting fifty years of research on pheromone signaling in ciliates. Eur. J. Protistol. 2016, 55, 26–38. [Google Scholar] [CrossRef]
- Luporini, P.; Alimenti, C.; Pedrini, B.; Vallesi, A. Ciliate communication via water-borne pheromones. In Biocommunication of Ciliates; Witzany, G., Nowacki, M., Eds.; Springer: Berlin/Heidelberg, Germany, 2016; pp. 159–174. [Google Scholar]
- Grell, K.G. Die Konjugation von Ephelota gemmipara R. Hertwig. Arch Protistenk 1953, 98, 287–326. [Google Scholar]
- Sonneborn, T.M. Genetics of cell–cell interaction in ciliates. In The Molecular Basis of Cell–Cell Interaction; Lerner, R.A., Bergsma, D., Eds.; Alan R. Liss: New York, NY, USA, 1978; pp. 417–427. [Google Scholar]
- Kubota, T.; Tokoroyama, T.; Tsukuda, Y.; Koyama, H.; Miyake, A. Isolation and structure determination of blepharismin, a conjugation initiating gamone in the ciliate Blepharisma. Science 1973, 179, 400–402. [Google Scholar] [CrossRef]
- Sugiura, M.; Harumoto, T. Identification, characterization, and complete amino acid sequence of the conjugation-inducing glycoprotein (blepharmone) in the ciliate Blepharisma japonicum. Proc. Natl. Acad. Sci. USA 2001, 98, 14446–14451. [Google Scholar] [CrossRef] [Green Version]
- Pedrini, B.; Suter-Stahel, T.; Vallesi, A.; Alimenti, C.; Luporini, P. Molecular structures and coding genes of the water-borne protein pheromones of Euplotes petzi, an early diverging polar species of Euplotes. J. Eukaryot. Microbiol. 2017, 64, 164–172. [Google Scholar] [CrossRef] [Green Version]
- Dieckhoff, H.S.; Freiburg, M.; Heckmann, K. The isolation of gamones 3 and 4 of Euplotes octocarinatus. Eur. J. Biochem. 1987, 168, 89–94. [Google Scholar] [CrossRef]
- Brünen-Nieveler, C.; Weiligmann, J.C.; Hansen, B.; Kuhlmann, H.W.; Möllenbeck, M.; Heckmann, K. The pheromones and pheromone genes of new stocks of Euplotes octocarinatus species complex. Eur. J. Protistol. 1998, 34, 124–132. [Google Scholar] [CrossRef]
- Alimenti, C.; Vallesi, A.; Federici, S.; Di Giuseppe, G.; Dini, F.; Carratore, V.; Luporini, P. Isolation and structural characterization of two water-borne pheromones from Euplotes crassus, a ciliate commonly known to carry membrane-bound pheromones. J. Eukaryot. Microbiol. 2011, 58, 234–241. [Google Scholar] [CrossRef]
- Vallesi, A.; Alimenti, C.; Federici, S.; Di Giuseppe, G.; Dini, F.; Guella, G.; Luporini, P. Evidence for gene duplication and allelic codominance (not hierarchical dominance) at the mating-type locus of the ciliate, Euplotes crassus. J. Eukaryot. Microbiol. 2014, 61, 620–629. [Google Scholar] [CrossRef]
- Mronga, S.; Luginbühl, P.; Brown, L.R.; Ortenzi, C.; Luporini, P.; Bradshaw, R.A.; Wüthrich, K. The NMR solution structure of the pheromone Er-1 from the ciliated protozoan Euplotes raikovi. Protein Sci. 1994, 3, 1527–1536. [Google Scholar] [CrossRef]
- Luginbühl, P.; Ottiger, M.; Mronga, S.; Wüthrich, K. Structure comparison of the pheromones Er-1, Er-10, and Er-2 from Euplotes raikovi. Protein Sci. 1994, 3, 1537–1546. [Google Scholar] [CrossRef]
- Zahn, R.; Damberger, F.; Ortenzi, C.; Luporini, P.; Wüthrich, K. NMR structure of the Euplotes raikovi pheromone Er-23 and identification of its five disulfide bonds. J. Mol. Biol. 2001, 313, 923–931. [Google Scholar] [CrossRef] [Green Version]
- Placzek, W.J.; Etezady-Esfarjani, T.; Herrmann, T.; Pedrini, B.; Peti, W.; Alimenti, C.; Luporini, P.; Wüthrich, K. Cold-adapted signal proteins: NMR structures of pheromones from the Antarctic ciliate Euplotes nobilii. IUBMB Life 2007, 59, 578–585. [Google Scholar] [CrossRef]
- Vallesi, A.; Alimenti, C.; Pedrini, B.; Di Giuseppe, G.; Dini, F.; Wüthrich, K.; Luporini, P. Coding genes and molecular structures of the diffusible signaling proteins (pheromones) of the polar ciliate, Euplotes nobilii. Mar. Genom. 2012, 8, 9–13. [Google Scholar] [CrossRef]
- Weiss, M.S.; Anderson, D.H.; Raffioni, S.; Bradshaw, R.A.; Ortenzi, C.; Luporini, P.; Eisenberg, D. A cooperative model for ligand recognition and cell adhesion: Evidence from the molecular packing in the 1.6 Å crystal structure of the pheromone Er-1 from the ciliate protozoan Euplotes raikovi. Proc. Natl. Acad. Sci. USA 1995, 92, 10172–10176. [Google Scholar] [CrossRef] [Green Version]
- Pedrini, B.; Finke, A.D.; Marsh, M.; Luporini, P.; Vallesi, A.; Alimenti, C. Crystal structure of the pheromone Er-13 from the ciliate Euplotes raikovi, with implications for a protein–protein association model in pheromone/receptor interactions. J. Struct. Biol. 2002, 214, 107812. [Google Scholar] [CrossRef]
- Ortenzi, C.; Luporini, P. Competition among homologous polypeptide pheromones of the ciliate Euplotes raikovi for binding to each other’s receptors. J. Eukaryot. Microbiol. 1995, 42, 242–248. [Google Scholar] [CrossRef]
- Kimball, R.F. The nature and inheritance of mating types in Euplotes patella. Genetics 1942, 27, 269–285. [Google Scholar] [CrossRef]
- Heckmann, K. Paarungssystem und genabhängige paarungstypdifferenzierung bei dem hypotrichen ciliaten Euplotes vannus O. F. Müller. Arch. Protistenk. 1963, 106, 393–421. [Google Scholar]
- Dini, F.; Luporini, P. Mating-type polymorphic variation in Euplotes minuta (Ciliophora: Hypotrichida). J. Protozool. 1985, 32, 111–118. [Google Scholar] [CrossRef]
- Hoffman, D.C.; Anderson, R.C.; DuBois, M.L.; Prescott, D.M. Macronuclear gene-sized molecules of hypotrichs. Nucleic Acids Res. 1995, 23, 1279–1283. [Google Scholar] [CrossRef] [Green Version]
- Jahn, C.L.; Klobutcher, L.A. Genome remodeling in ciliated protozoa. Annu. Rev. Microbiol. 2002, 56, 489–520. [Google Scholar] [CrossRef]
- Valbonesi, A.; Luporini, P. A new marine species of Euplotes (Ciliophora, Hypotrichhida) from Antarctica. Bull. Br. Mus. Nat. Hist. Zool. 1990, 56, 57–61. [Google Scholar]
- Zhao, Y.; Yi, Z.; Warren, A.; Song, W.B. Species delimitation for the molecular taxonomy and ecology of the widely distributed microbial eukaryote genus Euplotes (Alveolata, Ciliophora). Proc. R. Soc. B 2018, 285, 20172159. [Google Scholar] [CrossRef] [Green Version]
- Valbonesi, A.; Di Giuseppe, G.; Vallesi, A.; Luporini, P. Two new species of Euplotes with cirrotype-9, Euplotes foissneri sp. nov. and Euplotes warreni sp. nov. (Ciliophora, Spirotrichea, Euplotida), from the coasts of Patagonia: Implications from their distant, early and late branching in the Euplotes phylogenetic tree. Int. J. Syst. Evol. Microbiol. 2021, 71, 004568. [Google Scholar]
- Raffioni, S.; Miceli, C.; Vallesi, A.; Chowdhury, S.K.; Chait, B.T.; Luporini, P.; Bradshaw, R.A. Primary structure of Euplotes raikovi pheromones: Comparison of five sequences of pheromones from cells with variable mating interactions. Proc. Natl. Acad. Sci. USA 1992, 89, 2071–2075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valbonesi, A.; Luporini, P. Biology of Euplotes focardii, an Antarctic ciliate. Polar Biol. 1993, 13, 489–493. [Google Scholar] [CrossRef]
- Vallesi, A.; Di Pretoro, B.; Ballarini, P.; Apone, F.; Luporini, P. A novel protein kinase from the ciliate Euplotes raikovi with close structural identity to the mammalian intestinal and male-germ cell kinases: Characterization and functional implications in the autocrine pheromone signaling Loop. Protist 2010, 161, 250–263. [Google Scholar] [CrossRef]
- Mozzicafreddo, M.; Pucciarelli, S.; Swart, E.C.; Piersanti, A.; Emmerich, C.; Migliorelli, G.; Ballarini, P.; Miceli, C. The macronuclear genome of the Antarctic psychrophilic marine ciliate Euplotes focardii reveals new insights on molecular cold adaptation. Sci. Rep. 2021, 11, 18782. [Google Scholar] [CrossRef]
- Ricci, F.; Candelori, A.; Brandi, A.; Alimenti, C.; Luporini, P.; Vallesi, A. The sub-chromosomic macronuclear pheromone genes of the ciliate Euplotes raikovi: Comparative structural analysis and insights into the mechanism of expression. J. Eukaryot. Microbiol. 2019, 66, 376–384. [Google Scholar] [CrossRef]
- Pace, C.N.; Scholtz, J.M.; Grimsley, G.R. Forces stabilizing proteins. FEBS Lett. 2014, 588, 2177–2184. [Google Scholar] [CrossRef] [Green Version]
- Alimenti, C.; Vallesi, A.; Pedrini, B.; Wüthrich, K.; Luporini, P. Molecular cold-adaptation: Comparative analysis of two homologous families of psychrophilic and mesophilic signal proteins of the protozoan ciliate, Euplotes. IUBMB Life 2009, 61, 838–845. [Google Scholar] [CrossRef]
- Metpally, R.P.R.; Reddy, B.V.B. Comparative proteome analysis of psychrophilic versus mesophilic bacterial species: Insights into the molecular basis of cold adaptation of proteins. BMC Genomics 2009, 10, 11. [Google Scholar] [CrossRef] [Green Version]
- Lewis, P.N.; Go, N.; Go, M.; Kotelchuck, D.; Scheraga, H.A. Helix probability profiles of denatured proteins and their correlation with native structures. Proc. Natl. Acad. Sci. USA 1970, 65, 810–815. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.Y.; Kim, K.A.; Yu, Y.G.; Kim, K.S. Substitution of aspartic acid with glutamic acid increases the unfolding transition temperature of a protein. Biochem. Biophys. Res. Commun. 2004, 320, 900–906. [Google Scholar] [CrossRef]
- Kyte, J.; Doolittle, R.F. Hydropathicity. J. Mol. Biol. 1982, 157, 105–132. [Google Scholar] [CrossRef] [Green Version]
- Janin, J. Free energy of transfer from inside to outside of a globular protein. Nature 1979, 277, 491–492. [Google Scholar] [CrossRef]
- Bhaskaran, R.; Ponnuswanny, P.K. Average flexibility index. Int. J. Pept. Protein Res. 1988, 32, 242–255. [Google Scholar]
- Meyer, F.; Schmidt, H.J.; Heckmann, K. Pheromone 4 gene of Euplotes octocarinatus. Dev. Genet. 1992, 13, 16–25. [Google Scholar] [CrossRef]
- Pedrini, B.; Plackzek, W.J.; Koculi, E.; Alimenti, C.; La Terza, A.; Luporini, P.; Wüthrich, K. Cold-adaptation in seawater-borne signal proteins: Sequence and NMR structure of the pheromone En-6 from the Antarctic ciliate Euplotes nobilii. J. Mol. Biol. 2007, 372, 277–286. [Google Scholar] [CrossRef]
- Geralt, M.; Alimenti, C.; Vallesi, A.; Luporini, P.; Wüthrich, K. Thermodynamic stability of psychrophilic and mesophilic pheromones of the protozoan ciliate Euplotes. Biology 2013, 2, 142–150. [Google Scholar] [CrossRef] [Green Version]



| Name | Nucleotide Sequence (5′–3′) |
|---|---|
| dFW1 a | GATTGCCAYGGWGATACNGA |
| dFW2 a | GATACTGAATAYTTMATYGAYGART |
| TEL | CCCCAAAACCCCAAAACCCC |
| RV1 | TCAATACAAACTTGACAGCAGTTACA |
| RV2 | TAATCACCAAAACCGTCAGATCCCAT |
| 5′-FW1 | TGGCTAAAGCATAGAATTCTAACAT |
| 5′-FW2 | TGTTTATAAATGAGGAAGTGCTTAAG |
| 3′-RV | ACACTATTGAACCAGAATATTCCTCT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alimenti, C.; Candelori, A.; Jiang, Y.; Luporini, P.; Vallesi, A. Primary Structure and Coding Genes of Two Pheromones from the Antarctic Psychrophilic Ciliate, Euplotes focardii. Microorganisms 2022, 10, 1089. https://doi.org/10.3390/microorganisms10061089
Alimenti C, Candelori A, Jiang Y, Luporini P, Vallesi A. Primary Structure and Coding Genes of Two Pheromones from the Antarctic Psychrophilic Ciliate, Euplotes focardii. Microorganisms. 2022; 10(6):1089. https://doi.org/10.3390/microorganisms10061089
Chicago/Turabian StyleAlimenti, Claudio, Annalisa Candelori, Yaohan Jiang, Pierangelo Luporini, and Adriana Vallesi. 2022. "Primary Structure and Coding Genes of Two Pheromones from the Antarctic Psychrophilic Ciliate, Euplotes focardii" Microorganisms 10, no. 6: 1089. https://doi.org/10.3390/microorganisms10061089
APA StyleAlimenti, C., Candelori, A., Jiang, Y., Luporini, P., & Vallesi, A. (2022). Primary Structure and Coding Genes of Two Pheromones from the Antarctic Psychrophilic Ciliate, Euplotes focardii. Microorganisms, 10(6), 1089. https://doi.org/10.3390/microorganisms10061089








