Do Triclosan Sutures Modify the Microbial Diversity of Surgical Site Infections? A Systematic Review and Meta-Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Question Framing and Eligibility Criteria
2.2. Search Strategy
2.3. Eligibility Criteria
2.4. Study Selection, Data Extraction, and Risk of Bias Assessment
2.5. Extracted Data
- Study characteristics: Design, committee approval and informed consent, study registration, statistical methods including power calculation, screening methods, treatment allocation and blinding details, sponsor details, enrollment period and sites, inclusion sites, patient inclusion and exclusion criteria, patient demographics, clinical indication, type of surgery, suture material by suture group, SSI prevention details, and additional patient groups.
- Detailed patient disposition.
- Number of patients with a ccSSI by suture type and list of microorganisms per culture or the aggregate count of each microbial designation. When microbial percentages were reported, counts were calculated using the corresponding total number.
2.6. Microbial Data Analysis
2.7. Consistency with Clinical Outcomes
3. Results
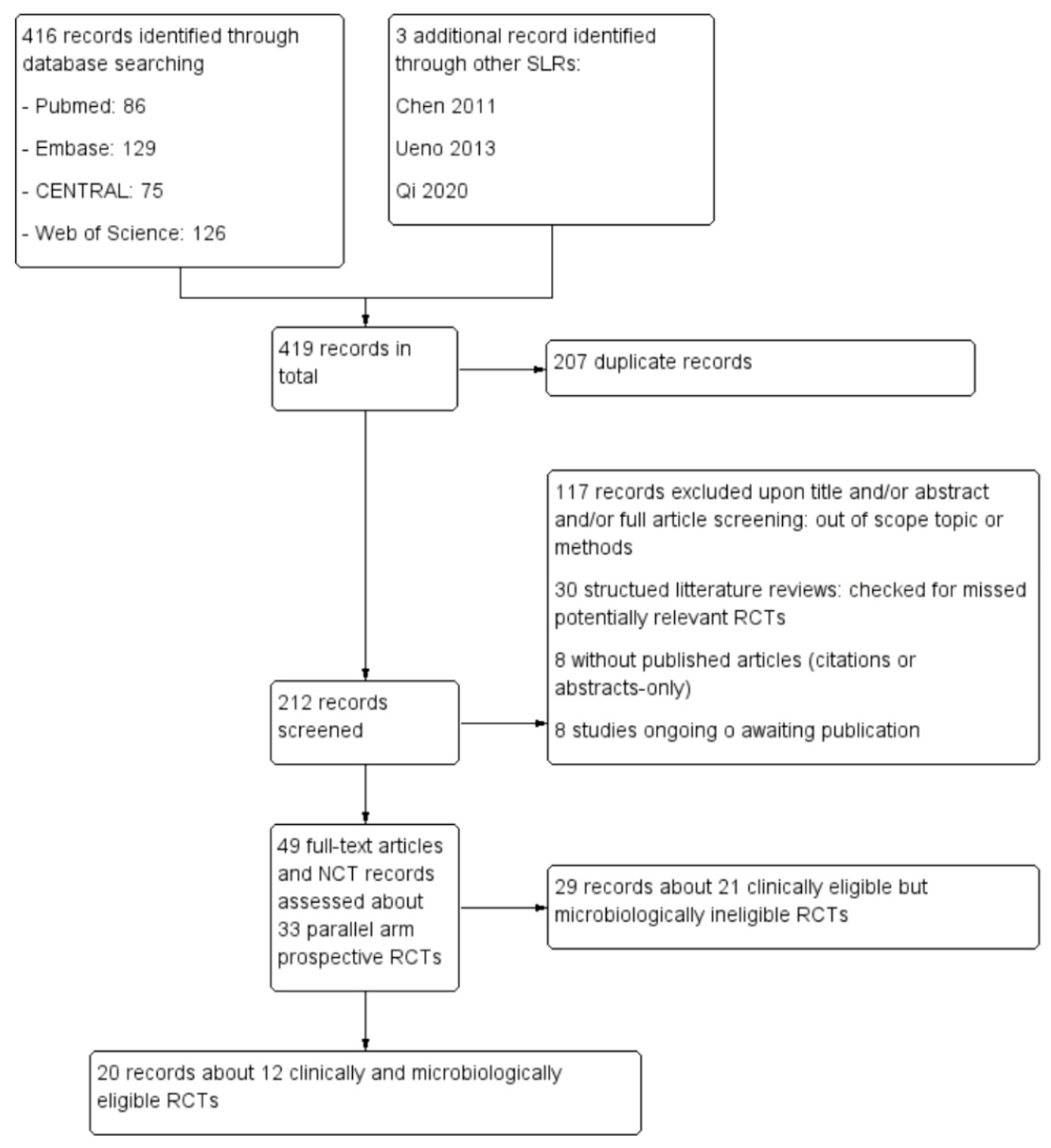
3.1. Study Identification and Selection
3.2. Characteristics of Eligible Studies and Risk of Bias
3.3. Microbial Diversity
3.4. Clinical Outcomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board
Informed Consent
Data Availability Statement
Conflicts of Interest
Appendix A. Executed Search Strategies
| Query | |
|---|---|
| (“triclosan”[MeSH Terms] OR “triclosan”[All Fields]) AND (“suturability”[All Fields] OR “suturable”[All Fields] OR “sutural”[All Fields] OR “suturation”[All Fields] OR “suture s”[All Fields] OR “sutured”[All Fields] OR “sutures”[MeSH Terms] OR “sutures”[All Fields] OR “suture”[All Fields] OR “suturing”[All Fields] OR (“suturability”[All Fields] OR “suturable”[All Fields] OR “sutural”[All Fields] OR “suturation”[All Fields] OR “suture s”[All Fields] OR “sutured”[All Fields] OR “sutures”[MeSH Terms] OR “sutures”[All Fields] OR “suture”[All Fields] OR “suturing”[All Fields]) OR (“ligate”[All Fields] OR “ligated”[All Fields] OR “ligates”[All Fields] OR “ligating”[All Fields] OR “ligation”[MeSH Terms] OR “ligation”[All Fields] OR “ligations”[All Fields]) OR (“ligate”[All Fields] OR “ligated”[All Fields] OR “ligates”[All Fields] OR “ligating”[All Fields] OR “ligation”[MeSH Terms] OR “ligation”[All Fields] OR “ligations”[All Fields])) AND (“surgery”[MeSH Subheading] OR “surgery”[All Fields] OR “surgical procedures, operative”[MeSH Terms] OR (“surgical”[All Fields] AND “procedures”[All Fields] AND “operative”[All Fields]) OR “operative surgical procedures”[All Fields] OR “general surgery”[MeSH Terms] OR (“general”[All Fields] AND “surgery”[All Fields]) OR “general surgery”[All Fields] OR “surgery s”[All Fields] OR “surgerys”[All Fields] OR “surgeries”[All Fields] OR (“surgery”[MeSH Subheading] OR “surgery”[All Fields] OR “surgical procedures, operative”[MeSH Terms] OR (“surgical”[All Fields] AND “procedures”[All Fields] AND “operative”[All Fields]) OR “operative surgical procedures”[All Fields] OR “general surgery”[MeSH Terms] OR (“general”[All Fields] AND “surgery”[All Fields]) OR “general surgery”[All Fields] OR “surgery s”[All Fields] OR “surgerys”[All Fields] OR “surgeries”[All Fields]) OR (“surgical procedures, operative”[MeSH Terms] OR (“surgical”[All Fields] AND “procedures”[All Fields] AND “operative”[All Fields]) OR “operative surgical procedures”[All Fields] OR “surgical”[All Fields] OR “surgically”[All Fields] OR “surgicals”[All Fields]) OR (“operability”[All Fields] OR “operable”[All Fields] OR “operate”[All Fields] OR “operated”[All Fields] OR “operates”[All Fields] OR “operating”[All Fields] OR “operation s”[All Fields] OR “operational”[All Fields] OR “operative”[All Fields] OR “operatively”[All Fields] OR “operatives”[All Fields] OR “operator”[All Fields] OR “operator s”[All Fields] OR “operators”[All Fields] OR “surgery”[MeSH Subheading] OR “surgery”[All Fields] OR “operations”[All Fields] OR “surgical procedures, operative”[MeSH Terms] OR (“surgical”[All Fields] AND “procedures”[All Fields] AND “operative”[All Fields]) OR “operative surgical procedures”[All Fields] OR “operation”[All Fields]) OR (“operability”[All Fields] OR “operable”[All Fields] OR “operate”[All Fields] OR “operated”[All Fields] OR “operates”[All Fields] OR “operating”[All Fields] OR “operation s”[All Fields] OR “operational”[All Fields] OR “operative”[All Fields] OR “operatively”[All Fields] OR “operatives”[All Fields] OR “operator”[All Fields] OR “operator s”[All Fields] OR “operators”[All Fields] OR “surgery”[MeSH Subheading] OR “surgery”[All Fields] OR “operations”[All Fields] OR “surgical procedures, operative”[MeSH Terms] OR (“surgical”[All Fields] AND “procedures”[All Fields] AND “operative”[All Fields]) OR “operative surgical procedures”[All Fields] OR “operation”[All Fields])) AND (((“classification”[MeSH Terms] OR “classification”[All Fields] OR “systematic”[All Fields] OR “classification”[MeSH Subheading] OR “systematics”[All Fields] OR “systematical”[All Fields] OR “systematically”[All Fields] OR “systematisation”[All Fields] OR “systematise”[All Fields] OR “systematised”[All Fields] OR “systematization”[All Fields] OR “systematizations”[All Fields] OR “systematize”[All Fields] OR “systematized”[All Fields] OR “systematizes”[All Fields] OR “systematizing”[All Fields]) AND (“review”[Publication Type] OR “review literature as topic”[MeSH Terms] OR “review”[All Fields])) OR “random*”[All Fields] OR “RCT”[All Fields] OR “guide*”[All Fields] OR “recom*”[All Fields] OR “meta analy*”[All Fields] OR “metaanaly*”[All Fields]) Translations triclosan: “triclosan”[MeSH Terms] OR “triclosan”[All Fields] suture: “suturability”[All Fields] OR “suturable”[All Fields] OR “sutural”[All Fields] OR “suturation”[All Fields] OR “suture’s”[All Fields] OR “sutured”[All Fields] OR “sutures”[MeSH Terms] OR “sutures”[All Fields] OR “suture”[All Fields] OR “suturing”[All Fields] sutures: “suturability”[All Fields] OR “suturable”[All Fields] OR “sutural”[All Fields] OR “suturation”[All Fields] OR “suture’s”[All Fields] OR “sutured”[All Fields] OR “sutures”[MeSH Terms] OR “sutures”[All Fields] OR “suture”[All Fields] OR “suturing”[All Fields] ligation: “ligate”[All Fields] OR “ligated”[All Fields] OR “ligates”[All Fields] OR “ligating”[All Fields] OR “ligation”[MeSH Terms] OR “ligation”[All Fields] OR “ligations”[All Fields] ligations: “ligate”[All Fields] OR “ligated”[All Fields] OR “ligates”[All Fields] OR “ligating”[All Fields] OR “ligation”[MeSH Terms] OR “ligation”[All Fields] OR “ligations”[All Fields] surgery: “surgery”[Subheading] OR “surgery”[All Fields] OR “surgical procedures, operative”[MeSH Terms] OR (“surgical”[All Fields] AND “procedures”[All Fields] AND “operative”[All Fields]) OR “operative surgical procedures”[All Fields] OR “general surgery”[MeSH Terms] OR (“general”[All Fields] AND “surgery”[All Fields]) OR “general surgery”[All Fields] OR “surgery’s”[All Fields] OR “surgerys”[All Fields] OR “surgeries”[All Fields] surgeries: “surgery”[Subheading] OR “surgery”[All Fields] OR “surgical procedures, operative”[MeSH Terms] OR (“surgical”[All Fields] AND “procedures”[All Fields] AND “operative”[All Fields]) OR “operative surgical procedures”[All Fields] OR “general surgery”[MeSH Terms] OR (“general”[All Fields] AND “surgery”[All Fields]) OR “general surgery”[All Fields] OR “surgery’s”[All Fields] OR “surgerys”[All Fields] OR “surgeries”[All Fields] surgical: “surgical procedures, operative”[MeSH Terms] OR (“surgical”[All Fields] AND “procedures”[All Fields] AND “operative”[All Fields]) OR “operative surgical procedures”[All Fields] OR “surgical”[All Fields] OR “surgically”[All Fields] OR “surgicals”[All Fields] operation: “operability”[All Fields] OR “operable”[All Fields] OR “operate”[All Fields] OR “operated”[All Fields] OR “operates”[All Fields] OR “operating”[All Fields] OR “operation’s”[All Fields] OR “operational”[All Fields] OR “operative”[All Fields] OR “operatively”[All Fields] OR “operatives”[All Fields] OR “operator”[All Fields] OR “operator’s”[All Fields] OR “operators”[All Fields] OR “surgery”[Subheading] OR “surgery”[All Fields] OR “operations”[All Fields] OR “surgical procedures, operative”[MeSH Terms] OR (“surgical”[All Fields] AND “procedures”[All Fields] AND “operative”[All Fields]) OR “operative surgical procedures”[All Fields] OR “operation”[All Fields] operations: “operability”[All Fields] OR “operable”[All Fields] OR “operate”[All Fields] OR “operated”[All Fields] OR “operates”[All Fields] OR “operating”[All Fields] OR “operation’s”[All Fields] OR “operational”[All Fields] OR “operative”[All Fields] OR “operatively”[All Fields] OR “operatives”[All Fields] OR “operator”[All Fields] OR “operator’s”[All Fields] OR “operators”[All Fields] OR “surgery”[Subheading] OR “surgery”[All Fields] OR “operations”[All Fields] OR “surgical procedures, operative”[MeSH Terms] OR (“surgical”[All Fields] AND “procedures”[All Fields] AND “operative”[All Fields]) OR “operative surgical procedures”[All Fields] OR “operation”[All Fields] systematic: “classification”[MeSH Terms] OR “classification”[All Fields] OR “systematic”[All Fields] OR “classification”[Subheading] OR “systematics”[All Fields] OR “systematical”[All Fields] OR “systematically”[All Fields] OR “systematisation”[All Fields] OR “systematise”[All Fields] OR “systematised”[All Fields] OR “systematization”[All Fields] OR “systematizations”[All Fields] OR “systematize”[All Fields] OR “systematized”[All Fields] OR “systematizes”[All Fields] OR “systematizing”[All Fields] review: “review”[Publication Type]. or. “review literature as topic”[MeSH Terms]. or. “review”[All Fields] | |
| Query | |
|---|---|
| (‘triclosan’/exp OR triclosan) AND (‘suture’/exp OR suture OR ‘sutures’/exp OR sutures OR ‘ligation’/exp OR ligation OR ligations) AND (‘surgery’/exp OR surgery OR surgeries OR surgical OR ‘operation’/exp OR operation OR operations) AND (systematic AND (‘review’/exp OR review) OR random* OR rct OR guide* OR recom* OR ‘meta analy*’ OR metaanaly*) | |
| Query | |
|---|---|
| triclosan AND (suture OR sutures OR ligation OR ligations) AND (surgery OR surgeries OR surgical OR operation OR operations) AND ((systematic AND review) OR random* OR RCT OR guide* OR recom* OR meta-analy* OR metaanaly*) (All Fields) | |
| Query | |
|---|---|
| triclosan AND (suture OR sutures OR ligation OR ligations) AND (surgery OR surgeries OR surgical OR operation OR operations) AND ((systematic AND review) OR random* OR RCT OR guide* OR recom* OR meta-analy* OR metaanaly*) in Title Abstract Keyword | |
Appendix B. Risk of Bias (RoB) of Included Studies
| Bias | Author’s Judgement | Support for Judgement |
|---|---|---|
| Random sequence generation (selection bias) | Unclear risk | Not reported |
| Allocation concealment (selection bias) | Unclear risk | Not reported |
| Blinding of participants and personnel (performance bias) | High risk | No |
| Blinding of outcome assessment (detection bias) | High risk | No |
| Incomplete outcome data (attrition bias) | High risk | Patient disposition: no patient lost to follow-up reported. Excluded patients after randomisation and use of allocated sutures due to postoperative administration of antibiotics or use of drains caused a risk of bias. |
| Selective reporting (reporting bias) | Low risk | Not with respect to ccSSIs |
| Other bias | Unclear risk | Calculated sample size was not justified with respect to the primary endpoint. |
| Bias | Author’s Judgement | Support for Judgement |
|---|---|---|
| Random sequence generation (selection bias) | Low risk | Permuted block (size 2) randomisation, although generation process was not described |
| Allocation concealment (selection bias) | Low risk | Envelope with randomisation code delivered the allocated sutures to the operating room |
| Blinding of participants and personnel (performance bias) | Low risk | Yes |
| Blinding of outcome assessment (detection bias) | Low risk | Yes |
| Incomplete outcome data (attrition bias) | Low risk | Patient disposition: no, as described in details of patient flow |
| Selective reporting (reporting bias) | High risk | Cultures collected in 22/35 and 9/30 SSIs |
| Other bias | Low risk | No |
| Bias | Author’s Judgement | Support for Judgement |
|---|---|---|
| Random sequence generation (selection bias) | Unclear risk | Not reported |
| Allocation concealment (selection bias) | Unclear risk | Not reported |
| Blinding of participants and personnel (performance bias) | Unclear risk | Reported as double blind, but proedures not described |
| Blinding of outcome assessment (detection bias) | Unclear risk | Reported as double blind, but proedures not described |
| Incomplete outcome data (attrition bias) | Unclear risk | Patient disposition: Insufficient details |
| Selective reporting (reporting bias) | High risk | Fewer data reported about cSSIs than about diagnosed SSIs: sternal TS = 4/170, NTS = 12/328 N.S. (bacteria reported in 4/4 and 8/12); leg TS = 5/142, NTS = 10/160 N.S. (bacteria reported in 2/5 and 2/10) |
| Other bias | Low risk | No |
| Bias | Author’s Judgement | Support for Judgement |
|---|---|---|
| Random sequence generation (selection bias) | Low risk | Random block sizes of 50 to 100, although the generation process was not described |
| Allocation concealment (selection bias) | Unclear risk | Reported, but without description |
| Blinding of participants and personnel (performance bias) | Low risk | Yes |
| Blinding of outcome assessment (detection bias) | Low risk | Yes |
| Incomplete outcome data (attrition bias) | High risk | Patient disposition: number of patients excluded after randomisation was much larger than the number of SSIs (111 > 73), especially in the TS group, which had twice as many excluded than the NTS group |
| Selective reporting (reporting bias) | Unclear risk | The number of patients with culture results and isolated microorganisms compared to the number of SSIs was unclear |
| Other bias | Unclear risk | Identified bacteria reported as percentages that, when multiplied by the number of SSIs, resulted in numbers with a decimal instead of being integers |
| Bias | Author’s Judgement | Support for Judgement |
|---|---|---|
| Random sequence generation (selection bias) | Unclear risk | Suggested, but mechanisms were not reported |
| Allocation concealment (selection bias) | Low risk | Sealed envelopes |
| Blinding of participants and personnel (performance bias) | Low risk | Yes |
| Blinding of outcome assessment (detection bias) | Low risk | Yes |
| Incomplete outcome data (attrition bias) | Low risk | Patient disposition: all randomised patients completed study in their group and were included in the analysis |
| Selective reporting (reporting bias) | Low risk | Not with respect to ccSSIs |
| Other bias | Unclear risk | Calculated sample size was not justified with respect to the primary endpoint |
| Bias | Author’s Judgement | Support for Judgement |
|---|---|---|
| Random sequence generation (selection bias) | Low risk | Computer-generated list |
| Allocation concealment (selection bias) | Low risk | Seaed envelopes |
| Blinding of participants and personnel (performance bias) | High risk | Operators not blinded, although nonoperating staff and patients were blinded |
| Blinding of outcome assessment (detection bias) | Low risk | Assessor-blinded |
| Incomplete outcome data (attrition bias) | Low risk | Patient disposition: detailed. Discontinuations explained and not related to SSIs. |
| Selective reporting (reporting bias) | High risk | Number of cultures less than the number of diagnosed SSIs |
| Other bias | Unclear risk | Recruited sample size could not be checked against the calculated sample size |
| Bias | Author’s Judgement | Support for Judgement |
|---|---|---|
| Random sequence generation (selection bias) | Low risk | Random number tables |
| Allocation concealment (selection bias) | Unclear risk | Insufficiently described |
| Blinding of participants and personnel (performance bias) | Unclear risk | Insufficiently described |
| Blinding of outcome assessment (detection bias) | Unclear risk | Insufficiently described |
| Incomplete outcome data (attrition bias) | Unclear risk | Patient disposition: inconsistencies in flowchart |
| Selective reporting (reporting bias) | High risk | Inconsistencies in flowchart and ccSSI reporting |
| Other bias | High risk | Discontinuation after 7.4% of calculated sample size |
| Bias | Author’s Judgement | Support for Judgement |
|---|---|---|
| Random sequence generation (selection bias) | Unclear risk | Not reported |
| Allocation concealment (selection bias) | Unclear risk | Envelope method without further detail |
| Blinding of participants and personnel (performance bias) | Low risk | No |
| Blinding of outcome assessment (detection bias) | Low risk | Yes |
| Incomplete outcome data (attrition bias) | Low risk | Patient disposition: detailed. No losses to follow-up or dropouts |
| Selective reporting (reporting bias) | Low risk | Not with respect to ccSSIs |
| Other bias | High risk | Insufficient sample size to reach target power |
| Bias | Author’s Judgement | Support for Judgement |
|---|---|---|
| Random sequence generation (selection bias) | Low risk | Described |
| Allocation concealment (selection bias) | Low risk | Described |
| Blinding of participants and personnel (performance bias) | Low risk | Described |
| Blinding of outcome assessment (detection bias) | Low risk | Described |
| Incomplete outcome data (attrition bias) | Unclear risk | Patient disposition: no flowchart, but no loss to follow-up reported |
| Selective reporting (reporting bias) | Low risk | Not with respect to ccSSIs |
| Other bias | Unclear risk | No sample-size calculation. 37.7% of patients (23/61) were included twice; i.e., 27.4% (23/84) of procedures. The distribution of those 23 dual-inclusions between the two suture groups was not accurately reported, and two observations in the same patient were not statistically independent. |
| Bias | Author’s Judgement | Support for Judgement |
|---|---|---|
| Random sequence generation (selection bias) | Unclear risk | Not reported |
| Allocation concealment (selection bias) | Low risk | Sequentially numbered container method |
| Blinding of participants and personnel (performance bias) | High risk | Randomisation performed by the surgeon without blinding |
| Blinding of outcome assessment (detection bias) | Low risk | Nurse in charge of diagnosing SSIs was blinded |
| Incomplete outcome data (attrition bias) | Low risk | Patient disposition flowchart available showed no attrition. Exclusions from SSI incidence comparison were deaths before SSIs. |
| Selective reporting (reporting bias) | Low risk | Not with respect to ccSSIs |
| Other bias | Unclear risk | Insufficient information |
| Bias | Author’s Judgement | Support for Judgement |
|---|---|---|
| Random sequence generation (selection bias) | Unclear risk | Not reported |
| Allocation concealment (selection bias) | Unclear risk | Not reported |
| Blinding of participants and personnel (performance bias) | Unclear risk | Operator blinded until the last minute. Operator should have been blinded to the presence of triclosan until the operation was completed. |
| Blinding of outcome assessment (detection bias) | Low risk | Nurse in charge of SSI diagnosis was blinded as well |
| Incomplete outcome data (attrition bias) | Low risk | Patient disposition CONSORT flowchart available. No patients lost to follow-up or dropout. Patients excluded due to reoperation or mortality within 30 days were counted. Their exclusions were explainable given the change in risk, and an analysis on an intention-to-treat basis was not performed |
| Selective reporting (reporting bias) | Low risk | Not with respect to incisional ccSSIs reported, for both incisional and organ/space |
| Other bias | Unclear risk | Uncertain whether deep and incisional SSIs were in the same patients or different patients. No culture report for deep SSIs. |
| Bias | Author’s Judgement | Support for Judgement |
|---|---|---|
| Random sequence generation (selection bias) | Unclear risk | Not reported, although some details were provided |
| Allocation concealment (selection bias) | Low risk | Yes |
| Blinding of participants and personnel (performance bias) | Low risk | Yes |
| Blinding of outcome assessment (detection bias) | Low risk | Yes |
| Incomplete outcome data (attrition bias) | Low risk | Patient disposition detailed flowchart showed a small number of patients lost to follow-up or unreachable minor compared to the number of SSIs |
| Selective reporting (reporting bias) | Low risk | Results reported for all outcome variables described in the methods |
| Other bias | Low risk | Assuming a one-sided test was planned |
References
- European Centre for Disease Prevention and Control. Surveillance of Surgical Site Infections in European Hospitals–HAISSI protocol. Version 1.02. Stockholm: ECDC. 2012. Available online: http://ecdc.europa.eu/en/publications/Publications/120215_TED_SSI_protocol.pdf (accessed on 17 December 2019).
- WHO. Protocol for Surgical Site Infection Surveillance with a Focus on Settings with Limited Resources; Licence: CC BY-NC-SA 3.0 IGO; World Health Organization: Geneva, Switzerland, 2018; Available online: https://www.who.int/infection-prevention/tools/surgical/SSI-surveillance-protocol.pdf (accessed on 17 December 2019).
- European Centre for Disease Prevention and Control. Healthcare-Associated Infections: Surgical Site Infections; Annual Epidemiological Report for 2017; ECDC: Stockholm, Sweden, 2019; Available online: https://www.ecdc.europa.eu/sites/default/files/documents/AER_for_2017-SSI.pdf (accessed on 17 December 2019).
- Mangram, A.J.; Horan, T.C.; Pearson, M.L.; Silver, L.C.; Jarvis, W.R. The Hospital Infection Control Practices Advisory Committee Guideline for Prevention of Surgical Site Infection, 1999. Infect. Control Hosp. Epidemiol. 1999, 20, 250–278. [Google Scholar] [CrossRef] [PubMed]
- Centers for Disease Control and Prevention (CDC). Principles of Epidemiology in Public Health Practice. In An Introduction to Applied Epidemiology and Biostatistic, 3rd ed.; Uptated May 2020; Centers for Disease Control and Prevention: Atlanta, GA, USA. Available online: https://www.cdc.gov/csels/dsepd/ss1978/index.html (accessed on 20 March 2021).
- Leitão, J.H. Microbial Virulence Factors. Int. J. Mol. Sci. 2020, 21, 5320. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.K.; Dhasmana, N.; Dubey, N.; Kumar, N.; Gangwal, A.; Gupta, M.; Singh, Y. Bacterial Virulence Factors: Secreted for Survival. Indian J. Microbiol. 2016, 57, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, S.R.; Elkinton, J.S. Pathogenicity and virulence. J. Invertebr. Pathol. 2004, 85, 146–151. [Google Scholar] [CrossRef]
- Young, P.Y.; Khadaroo, R.G. Surgical Site Infections. Surg. Clin. N. Am. 2014, 94, 1245–1264. [Google Scholar] [CrossRef]
- Krizek, T.J.; Robson, M.C. Evolution of quantitative bacteriology in wound management. Am. J. Surg. 1975, 130, 579–584. [Google Scholar] [CrossRef]
- Elek, S.D.; Conen, P.E. The virulence of Staphylococcus pyogenes for man; a study of the problems of wound infection. Br. J. Exp. Pathol. 1957, 38, 573–586. Available online: https://www.ncbi.nlm.nih.gov/pubmed/13499821 (accessed on 20 March 2021).
- James, R.C.; Macleod, C.J. Induction of staphylococcal infections in mice with small inocula introduced on sutures. Br. J. Exp. Pathol. 1961, 42, 266–277. Available online: https://www.ncbi.nlm.nih.gov/pubmed/13789320 (accessed on 20 March 2021).
- Noble, W.C. The production of subcutaneous staphylococcal skin lesions in mice. Br. J. Exp. Pathol. 1965, 46, 254–262. Available online: https://www.ncbi.nlm.nih.gov/pubmed/5829388 (accessed on 20 March 2021).
- WHO. World Health Organization Global Guidelines for the Prevention of Surgical Site Infection, 2nd ed.; Licence: CC BY-NC-SA 3.0 IGO. 2018; World Health Organization: Geneva, Switzerland, 2018; Available online: https://apps.who.int›handle›9789241550475-eng.pdf (accessed on 20 March 2021).
- Wu, X.; Kubilay, N.Z.; Ren, J.; Allegranzi, B.; Bischoff, P.; Zayed, B.; Pittet, D.; Li, J. Antimicrobial-coated sutures to decrease surgical site infections: A systematic review and meta-analysis. Eur. J. Clin. Microbiol. Infect. Dis. 2017, 36, 19–32. [Google Scholar] [CrossRef]
- Ming, X.; Nichols, M.; Rothenburger, S. In Vivo Antibacterial Efficacy of MONOCRYL Plus Antibacterial Suture (Poliglecaprone 25 with Triclosan). Surg. Infect. 2007, 8, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Ming, X.; Rothenburger, S.; Yang, D. In Vitro Antibacterial Efficacy of MONOCRYL Plus Antibacterial Suture (Poliglecaprone 25 with Triclosan). Surg. Infect. 2007, 8, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Rothenburger, S.; Spangler, D.; Bhende, S.; Burkley, D. In Vitro Antimicrobial Evaluation of Coated VICRYL* Plus Antibacterial Suture (Coated Polyglactin 910 with Triclosan) using Zone of Inhibition Assays. Surg. Infect. 2002, 3 (Suppl. S1), s79–s87. [Google Scholar] [CrossRef] [PubMed]
- Matl, F.D.; Obermeier, A.; Repmann, S.; Friess, W.; Stemberger, A.; Kuehn, K.-D. New Anti-Infective Coatings of Medical Implants. Antimicrob. Agents Chemother. 2008, 52, 1957–1963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ming, X.; Rothenburger, S.; Nichols, M.M. In Vivo and In Vitro Antibacterial Efficacy of PDS Plus (Polidioxanone with Triclosan) Suture. Surg. Infect. 2008, 9, 451–457. [Google Scholar] [CrossRef] [PubMed]
- McCagherty, J.; Yool, D.A.; Paterson, G.K.; Mitchell, S.R.; Woods, S.; Marques, A.I.; Hall, J.L.; Mosley, J.R.; Nuttall, T.J. Investigation of the in vitro antimicrobial activity of triclosan-coated suture material on bacteria commonly isolated from wounds in dogs. Am. J. Veter-Res. 2020, 81, 84–90. [Google Scholar] [CrossRef] [PubMed]
- Daoud, F.C.; Goncalves, R.; Moore, N. How Long Do Implanted Triclosan Sutures Inhibit Staphylococcus aureus in Surgical Conditions? A Pharmacological Model. Pharmaceutics 2022, 14, 539. [Google Scholar] [CrossRef]
- VICRYL Plus-389595.R04; Ethicon Inc.: Raritan, NJ, USA, 2002.
- MONOCRYL Plus-389680.R02; Ethicon Inc.: Raritan, NJ, USA, 2005.
- PDS Plus-389688.R02; Ethicon Inc.: Raritan, NJ, USA, 2006.
- Ford, H.R.; Jones, P.; Gaines, B.; Reblock, K.; Simpkins, D.L. Intraoperative Handling and Wound Healing: Controlled Clinical Trial Comparing Coated VICRYL® Plus Antibacterial Suture (Coated Polyglactin 910 Suture with Triclosan) with Coated VICRYL® Suture (Coated Polyglactin 910 Suture). Surg. Infect. 2005, 6, 313–321. [Google Scholar] [CrossRef] [Green Version]
- Chang, W.K.; Srinivasa, S.; Morton, R.; Hill, A.G. Triclosan-Impregnated Sutures to Decrease Surgical Site Infections. Ann. Surg. 2012, 255, 854–859. [Google Scholar] [CrossRef]
- Wang, Z.; Jiang, C.P.; Cao, Y.; Ding, Y.T. Systematic review and meta-analysis of triclosan-coated sutures for the prevention of surgical-site infection. Br. J. Surg. 2013, 100, 465–473. [Google Scholar] [CrossRef]
- Edmiston, C.E.; Daoud, F.C.; Leaper, D. Is there an evidence-based argument for embracing an antimicrobial (triclosan)-coated suture technology to reduce the risk for surgical-site infections?: A meta-analysis. Surgery 2013, 154, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Daoud, F.C.; Edmiston, C.E.; Leaper, D. Meta-Analysis of Prevention of Surgical Site Infections following Incision Closure with Triclosan-Coated Sutures: Robustness to New Evidence. Surg. Infect. 2014, 15, 165–181. [Google Scholar] [CrossRef] [PubMed]
- de Jonge, S.; Atema, J.J.; Solomkin, J.; Boermeester, M.A. Meta-analysis and trial sequential analysis of triclosan-coated sutures for the prevention of surgical-site infection. Br. J. Surg. 2017, 104, e118–e133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahmed, I.; Boulton, A.J.; Rizvi, S.; Carlos, W.; Dickenson, E.; A Smith, N.; Reed, M. The use of triclosan-coated sutures to prevent surgical site infections: A systematic review and meta-analysis of the literature. BMJ Open 2019, 9, e029727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horan, T.C.; Gaynes, R.P.; Martone, W.J.; Jarvis, W.R.; Emori, T.G. CDC definitions of nosocomial surgical site infections, 1992: A modification of CDC definitions of surgical wound infections. Am. J. Infect. Control 1992, 20, 271–274. [Google Scholar] [CrossRef]
- Cameron, A.; Barbieri, R.; Read, R.; Church, D.; Adator, E.H.; Zaheer, R.; McAllister, T.A. Functional screening for triclosan resistance in a wastewater metagenome and isolates of Escherichia coli and Enterococcus spp. from a large Canadian healthcare region. PLoS ONE 2019, 14, e0211144. [Google Scholar] [CrossRef] [Green Version]
- Australian Government, Department of Health and Ageing (NICNAS). Priority Existing Chemical Assessment Report No. 30-Triclosan. 2009. Available online: https://catalogue.nla.gov.au/Record/4610392/Details (accessed on 20 March 2021).
- Escalada, M.G.; Harwood, J.L.; Maillard, J.-Y.; Ochs, D. Triclosan inhibition of fatty acid synthesis and its effect on growth of Escherichia coli and Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2005, 55, 879–882. [Google Scholar] [CrossRef] [Green Version]
- Fan, F.; Yan, K.; Wallis, N.G.; Reed, S.; Moore, T.D.; Rittenhouse, S.F.; DeWolf, W.E.; Huang, J.; McDevitt, D.; Miller, W.H.; et al. Defining and Combating the Mechanisms of Triclosan Resistance in Clinical Isolates of Staphylococcus aureus. Antimicrob. Agents Chemother. 2002, 46, 3343–3347. [Google Scholar] [CrossRef] [Green Version]
- Öztürk, A.; Kalkanci, A. Investigation of Antifungal Activities of Some Disinfectants on Candida albicans. Mikrobiyoloji Bul. 2018, 52, 376–389. [Google Scholar] [CrossRef]
- Scientific Committee on Consumer Safety (SCCS); European Commission-Directorate General for Health and Consumers. Opinion on Triclosan 2010. Available online: https://op.europa.eu/en/publication-detail/-/publication/3b684b59-27f7-4d0e-853c-4a9efe0fb4cb/language-en (accessed on 3 December 2021).
- Curiao, T.; Marchi, E.; Viti, C.; Oggioni, M.R.; Baquero, F.; Martinez, J.L.; Coque, T.M. Polymorphic Variation in Susceptibility and Metabolism of Triclosan-Resistant Mutants of Escherichia coli and Klebsiella pneumoniae Clinical Strains Obtained after Exposure to Biocides and Antibiotics. Antimicrob. Agents Chemother. 2015, 59, 3413–3423. [Google Scholar] [CrossRef] [Green Version]
- Aiello, A.E.; Marshall, B.; Levy, S.B.; Della-Latta, P.; Larson, E. Relationship between Triclosan and Susceptibilities of Bacteria Isolated from Hands in the Community. Antimicrob. Agents Chemother. 2004, 48, 2973–2979. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lear, J.C.; Maillard, J.-Y.; Dettmar, P.W.; A Goddard, P.; Russell, A.D. Chloroxylenol- and triclosan-tolerant bacteria from industrial sources. J. Ind. Microbiol. Biotechnol. 2002, 29, 238–242. [Google Scholar] [CrossRef] [PubMed]
- Cole, E.; Addison, R.; Rubino, J.; Leese, K.; Dulaney, P.; Newell, M.; Wilkins, J.; Gaber, D.; Wineinger, T.; Criger, D. Investigation of antibiotic and antibacterial agent cross-resistance in target bacteria from homes of antibacterial product users and nonusers. J. Appl. Microbiol. 2003, 95, 664–676. [Google Scholar] [CrossRef] [PubMed]
- Liberati, A.; Altman, D.G.; Tetzlaff, J.; Mulrow, C.; Gøtzsche, P.C.; Ioannidis, J.P.; Clarke, M.; Devereaux, P.J.; Kleijnen, J.; Moher, D. The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: Explanation and elaboration. J. Clin. Epidemiol. 2009, 62, e1–e34. [Google Scholar] [CrossRef] [Green Version]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; the PRISMA Group. Preferred reporting items for systematic reviews and meta-analyses: The PRISMA statement. BMJ 2009, 339, b2535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haynes, R.B.; Sackett, D.L.; Guyatt, G.H.; Tugwell, P. (Eds.) Clinical Epidemiology: How to do Clinical Practice Research, 3rd ed.; Lippincott Williams Wilkins: Philadelphia, PA, USA, 2006. [Google Scholar]
- Sterne, J.A.C.; Savović, J.; Page, M.J.; Elbers, R.G.; Blencowe, N.S.; Boutron, I.; Cates, C.J.; Cheng, H.Y.; Corbett, M.S.; Eldridge, S.M.; et al. RoB 2: A revised tool for assessing risk of bias in randomised trials. BMJ 2019, 366, l4898. [Google Scholar] [CrossRef] [Green Version]
- Cramér, H. Mathematical Methods of Statistic; Princeton University Press: Princeton, NJ, USA, 1946. [Google Scholar]
- Harbord, R.M.; Egger, M.; Sterne, J. A modified test for small-study effects in meta-analyses of controlled trials with binary endpoints. Stat. Med. 2006, 25, 3443–3457. [Google Scholar] [CrossRef]
- Sterne, J.A.C.; Sutton, A.J.; Ioannidis, J.P.A.; Terrin, N.; Jones, D.R.; Lau, J.; Carpenter, J.; Rücker, G.; Harbord, R.M.; Schmid, C.H.; et al. Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ 2011, 343, d4002. [Google Scholar] [CrossRef] [Green Version]
- DerSimonian, R.; Laird, N. Meta-analysis in clinical trials. Control Clin. Trials 1986, 7, 177–188. [Google Scholar] [CrossRef]
- DerSimonian, R.; Laird, N. Meta-analysis in clinical trials revisited. Contemp. Clin. Trials 2015, 45, 139–145. [Google Scholar] [CrossRef] [Green Version]
- Mantel, N.; Haenszel, W. Statistical aspects of the analysis of data from retrospective studies of disease. JNCI J. Natl. Cancer Inst. 1959, 22, 719–748. Available online: https://www.ncbi.nlm.nih.gov/pubmed/13655060 (accessed on 20 March 2021). [PubMed]
- Gavaghan, D.J.; Moore, A.R.; McQuay, H.J. An evaluation of homogeneity tests in meta-analyses in pain using simulations of individual patient data. Pain 2000, 85, 415–424. [Google Scholar] [CrossRef]
- Higgins, J.P.T.; Thompson, S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002, 21, 1539–1558. [Google Scholar] [CrossRef] [PubMed]
- Higgins, J.P.T.; Thompson, S.G.; Deeks, J.J.; Altman, D.G. Measuring inconsistency in meta-analyses. BMJ 2003, 327, 557–560. [Google Scholar] [CrossRef] [Green Version]
- Guyatt, G.H.; Oxman, A.D.; Vist, G.E.; Kunz, R.; Falck-Ytter, Y.; Alonso-Coello, P.; Schünemann, H.J. GRADE: An emerging consensus on rating quality of evidence and strength of recommendations. BMJ 2008, 336, 924–926. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Tovar, J.; Llavero, C.; Jimenez-Fuertes, M.; Duran, M.; Perez-Lopez, M.; Garcia-Marin, A. Incisional Surgical Site Infection after Abdominal Fascial Closure with Triclosan-Coated Barbed Suture vs Triclosan-Coated Polydioxanone Loop Suture vs Polydioxanone Loop Suture in Emergent Abdominal Surgery: A Randomized Clinical Trial. J. Am. Coll. Surg. 2020, 230, 766–774. [Google Scholar] [CrossRef]
- Arslan, N.C.; Atasoy, G.; Altintas, T.; Terzi, C. Effect of triclosan-coated sutures on surgical site infections in pilonidal disease: Prospective randomized study. Int. J. Color. Dis. 2018, 33, 1445–1452. [Google Scholar] [CrossRef]
- Ichida, K.; Noda, H.; Kikugawa, R.; Hasegawa, F.; Obitsu, T.; Ishioka, D.; Fukuda, R.; Yoshizawa, A.; Tsujinaka, S.; Rikiyama, T. Effect of triclosan-coated sutures on the incidence of surgical site infection after abdominal wall closure in gastroenterological surgery: A double-blind, randomized controlled trial in a single center. Surgery 2018, 164, 91–95. [Google Scholar] [CrossRef]
- Lin, S.-J.; Chang, F.-C.; Huang, T.-W.; Peng, K.-T.; Shih, H.N.; Lee, M.S. Temporal Change of Interleukin-6, C-Reactive Protein, and Skin Temperature after Total Knee Arthroplasty Using Triclosan-Coated Sutures. BioMed Res. Int. 2018, 2018, 9136208. [Google Scholar] [CrossRef]
- Mattavelli, I.; Rebora, P.; Doglietto, G.; Dionigi, P.; Dominioni, L.; Luperto, M.; La Porta, A.; Garancini, M.; Nespoli, L.; Alfieri, S.; et al. Multi-Center Randomized Controlled Trial on the Effect of Triclosan-Coated Sutures on Surgical Site Infection after Colorectal Surgery. Surg. Infect. 2015, 16, 226–235. [Google Scholar] [CrossRef]
- Ruiz-Tovar, J.; Alonso, N.; Morales, V.; Llavero, C. Association between Triclosan-Coated Sutures for Abdominal Wall Closure and Incisional Surgical Site Infection after Open Surgery in Patients Presenting with Fecal Peritonitis: A Randomized Clinical Trial. Surg. Infect. 2015, 16, 588–594. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, T.; Kashimura, N.; Noji, T.; Suzuki, O.; Ambo, Y.; Nakamura, F.; Kishida, A. Triclosan-coated sutures reduce the incidence of wound infections and the costs after colorectal surgery: A randomized controlled trial. Surgery 2013, 153, 576–583. [Google Scholar] [CrossRef] [PubMed]
- Justinger, C.; Slotta, J.E.; Ningel, S.; Gräber, S.; Kollmar, O.; Schilling, M.K. Surgical-site infection after abdominal wall closure with triclosan-impregnated polydioxanone sutures: Results of a randomized clinical pathway facilitated trial (NCT00998907). Surgery 2013, 154, 589–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thimour-Bergström, L.; Roman-Emanuel, C.; Scherstén, H.; Friberg, Ö.; Gudbjartsson, T.; Jeppsson, A. Triclosan-coated sutures reduce surgical site infection after open vein harvesting in coronary artery bypass grafting patients: A randomised controlled trial. Eur. J. Cardio-Thorac. Surg. 2013, 44, 931–938. [Google Scholar] [CrossRef] [PubMed]
- Isik, I.; Selimen, D.; Senay, S.; Alhan, C. Efficiency of Antibacterial Suture Material in Cardiac Surgery: A Double-Blind Randomized Prospective Study. Heart Surg. Forum 2012, 15, E40–E45. [Google Scholar] [CrossRef] [PubMed]
- Mingmalairak, C.; Ungbhakorn, P.; Paocharoen, V. Efficacy of antimicrobial coating suture coated polyglactin 910 with tricosan (Vicryl plus) compared with polyglactin 910 (Vicryl) in reduced surgical site infection of appendicitis, double blind randomized control trial, preliminary safety report. J. Med. Assoc. Thail. 2009, 92, 770–775. [Google Scholar]
- Rozzelle, C.J.; Leonardo, J.; Li, V. Antimicrobial suture wound closure for cerebrospinal fluid shunt surgery: A prospective, double-blinded, randomized controlled trial. J. Neurosurg. Pediatr. 2008, 2, 111–117. [Google Scholar] [CrossRef] [Green Version]
- Chuanchuen, R.; Karkhoff-Schweizer, R.R.; Schweizer, H.P. High-level triclosan resistance in Pseudomonas aeruginosa is solely a result of efflux. Am. J. Infect. Control 2003, 31, 124–127. [Google Scholar] [CrossRef]
- Assadian, O.; Wehse, K.; Hübner, N.-O.; Koburger, T.; Bagel, S.; Jethon, F.; Kramer, A. Minimum inhibitory (MIC) and minimum microbicidal concentration (MMC) of polihexanide and triclosan against antibiotic sensitive and resistant Staphylococcus aureus and Escherichia coli strains. GMS Krankenhhyg Interdiszip 2011, 6, Doc06. [Google Scholar] [CrossRef]
- Sanchez, P.; Moreno, E.; Martinez, J.L. The Biocide Triclosan Selects Stenotrophomonas maltophilia Mutants That Overproduce the SmeDEF Multidrug Efflux Pump. Antimicrob. Agents Chemother. 2005, 49, 781–782. [Google Scholar] [CrossRef] [Green Version]
- Rose, H.; Baldwin, A.; Dowson, C.; Mahenthiralingam, E. Biocide susceptibility of the Burkholderia cepacia complex. J. Antimicrob. Chemother. 2009, 63, 502–510. [Google Scholar] [CrossRef] [PubMed]
- Anegundi, R.T.; Gowda, J.; Tavarageri, A.; Kulkarni, R.; Janardhan, A.; A Bhat, M. Comparative Assessment of the Antimicrobial Efficacy of Triclosan, Amoxicillin and Eugenol against Enterococcus faecalis. Int. J. Clin. Pediatr. Dent. 2021, 14, 59–62. [Google Scholar] [CrossRef] [PubMed]
- Edmiston, C.E.; Krepel, C.J.; Marks, R.M.; Rossi, P.J.; Sanger, J.; Goldblatt, M.; Graham, M.B.; Rothenburger, S.; Collier, J.; Seabrook, G.R. Microbiology of Explanted Suture Segments from Infected and Noninfected Surgical Patients. J. Clin. Microbiol. 2013, 51, 417–421. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Item | Specification |
|---|---|
| Patients | Surgically operated patients |
| Intervention | Surgical wound closure with any TS |
| Comparator | Surgical wound closure with any NTS |
| Outcome | Count of each microorganism isolated in ccSSIs |
| Study | Patients TS, NTS | Enrollment | Type of Surgery | Sutures TS/NTS | Diagnostic Criteria and Follow-Up | ccSSIs/Microorganisms TS, NTS |
|---|---|---|---|---|---|---|
| Ruiz-Tovar 2020 [58] | 45 and 50 BTS), 47 | 4 centers, Spain, 2018–2019 | Midline laparotomy, acute abdomen | PDS+ and Stratafix), PDS II | CDC + culture, 30 days | 4/4, 11/22 |
| Arslan 2018 [59] | 86, 91 | 1 center, Turkey, 2011–2013 | Excision of pilonidal disease | Vicryl+ and PDS+, Vicryl & polypropylene | CDC + culture, 30 days | 9/11, 19/22 |
| Ichida 2018 [60] | 508, 505 | 1 center, Japan, 2009–2011 | Digestive tract surgery | Vicryl+ and PDS+, Vicryl & PDS II | CDC + culture, 30 days | 22/72, 19/59 |
| Lin 2018 [61] | 51, 51 | 1 center, ROC, 2011–2012 | Total knee arthroplasty | Vicryl+, Vicryl | Own rules + cultures, 6 months | 0/0, 1/1 |
| Mattavelli 2015 [62] | 140, 141 | 4 centers, Italy, 2010–2013 | Elective colorectal resection | Vicryl+ and PDS+, Vicryl and PDS II | CDC + culture, 30 days | 11/18, 8/13 |
| Ruiz-Tovar 2015 [63] | 50, 51 | 2 centers, Spain, 2007–2013 | Fecal peritonitis | Vicryl+, Vicryl | CDC + culture, 60 days | 5/5, 18/35 |
| Nakamura 2013 [64] | 206, 204 | 1 center, Japan, 2009–2011 | Elective colorectal | Vicryl+, Vicryl | CDC + culture, 30 days | 7/12, 13/17 |
| Jüstinger 2013 [65] | 485, 371 | 1 center, Germany, 2009–2011 | Laparotomy for various causes | PDS+, PDS II | CDC + culture, 30 days | 28/28, 30/30 |
| Thimour-Bergström 2013 [66] | 184, 190 | 1 center, Sweden, 2009–2012 | Saphenous vein harvesting, CABG | Vicryl+ and Monocryl+, Vicryl and Monocryl | CDC + culture, 60 days | 14/22, 23/29 |
| Isik 2012 [67] | 170, 340 | 1 center, Turkey, 2008–2009 | Sternal and saphenous vein harvesting, CABG | Vicryl+, Vicryl | CDC + culture, 30 days | 5/5, 9/9 |
| Mingmalairak 2009 [68] | 50, 50 | 1 center, Thailand, 2006–2007 | Appendectomy | Vicryl+, Vicryl | Criteria not reported + culture, 30 days | 1/1, 1/1 |
| Rozelle 2008 [69] | 46, 38 | 1 center, USA, 2005–2006 | CSF shunt in children | Vicryl+, Vicryl | Criteria not reported + culture, 6 months | 2/2, 8/8 |
| Microbial Designations | TS n | TS % | NTS n | NTS % | Total n | Total % |
|---|---|---|---|---|---|---|
| Staphylococcus aureus | 10 | 5.6% | 26 | 10.6% | 36 | 8.5% |
| MRSA | 1 | 0.6% | 2 | 0.8% | 3 | 0.7% |
| Coagulase-negative Staphylococcus | 4 | 2.2% | 7 | 2.8% | 11 | 2.6% |
| Staphylococcus epidermidis | 5 | 2.8% | 5 | 2.0% | 10 | 2.3% |
| Staphylococcus spp. | 25 | 13.9% | 29 | 11.8% | 54 | 12.7% |
| Escherichia coli | 22 | 12.2% | 52 | 21.1% | 74 | 17.4% |
| Enterococcus spp. | 18 | 10.0% | 16 | 6.5% | 34 | 8.0% |
| Enterococcus fecalis | 8 | 4.4% | 12 | 4.9% | 20 | 4.7% |
| Enterococcus fecium | 0 | 0.0% | 2 | 0.8% | 2 | 0.5% |
| Enterococcus avium | 1 | 0.6% | 0 | 0.0% | 1 | 0.2% |
| Klebsiella pneumoniae | 13 | 7.2% | 17 | 6.9% | 30 | 7.0% |
| Klebsiella spp. | 4 | 2.2% | 11 | 4.5% | 15 | 3.5% |
| Koxytoca | 1 | 0.6% | 0 | 0.0% | 1 | 0.2% |
| Pseudomonas aeruginosa | 7 | 3.9% | 17 | 6.9% | 24 | 5.6% |
| Pseudomonas spp. | 6 | 3.3% | 3 | 1.2% | 9 | 2.1% |
| Enterobacter spp. | 5 | 2.8% | 7 | 2.8% | 12 | 2.8% |
| Enterobacter cloacae | 4 | 2.2% | 5 | 2.0% | 9 | 2.1% |
| Streptococcus mutans | 2 | 1.1% | 7 | 2.8% | 9 | 2.1% |
| Streptococcus spp. | 3 | 1.7% | 2 | 0.8% | 5 | 1.2% |
| Streptococcus anginosus | 1 | 0.6% | 0 | 0.0% | 1 | 0.2% |
| Bacteroides fragilis | 4 | 2.2% | 6 | 2.4% | 10 | 2.3% |
| Bacteroides spp. | 2 | 1.1% | 1 | 0.4% | 3 | 0.7% |
| Bacteroides ovatus | 0 | 0.0% | 1 | 0.4% | 1 | 0.2% |
| Bacteroides thetaiotaomicron | 0 | 0.0% | 1 | 0.4% | 1 | 0.2% |
| Proteus mirabilis | 2 | 1.1% | 0 | 0.0% | 2 | 0.5% |
| Proteus vulgaris | 2 | 1.1% | 0 | 0.0% | 2 | 0.5% |
| Citrobacter freundii | 0 | 0.0% | 1 | 0.4% | 1 | 0.2% |
| Citrobacter koseri | 1 | 0.6% | 0 | 0.0% | 1 | 0.2% |
| Morganella morganii | 1 | 0.6% | 1 | 0.4% | 2 | 0.5% |
| Peptostreptococcus magnus (*) | 1 | 0.6% | 0 | 0.0% | 1 | 0.2% |
| Corynebacterium ssp. | 0 | 0.0% | 1 | 0.4% | 1 | 0.2% |
| Moraxella catarrhalis | 1 | 0.6% | 0 | 0.0% | 1 | 0.2% |
| Serratia marcescens | 0 | 0.0% | 1 | 0.4% | 1 | 0.2% |
| Other bacteria | 14 | 7.8% | 11 | 4.5% | 25 | 5.9% |
| Polymicrobial | 12 | 6.7% | 0 | 0.0% | 12 | 2.8% |
| Fungus: C. Albicans | 0 | 0.0% | 2 | 0.8% | 2 | 0.5% |
| TOTAL microorganism count | 180 | 100% | 246 | 100% | 426 | 100% |
| Culture-confirmed SSIs | 124 | 198 | 322 | |||
| Patients included by authors | 2021 | 2079 | 4100 |
| Genus, n (%) | TS | NTS | Total |
|---|---|---|---|
| Staphylococcus | 45 (39.47) | 69 (60.53) | 114 (30.40) |
| Escherichia | 22 (29.73) | 52 (70.27) | 74 (19.73) |
| Enterococcus | 27 (47.37) | 30 (52.63) | 57 (15.20) |
| Klebsiella | 18 (39.13) | 28 (60.87) | 46 (12.27) |
| Pseudomonas | 13 (39.39) | 20 (60.61) | 33 (8.80) |
| Enterobacter | 9 (42.86) | 12 (57.14) | 21 (5.60) |
| Streptococcus | 6 (40.00) | 9 (60.00) | 15 (4.00) |
| Bacteroides | 6 (40.00) | 9 (60.00) | 15 (4.00) |
| Total | 146 (38.93) | 229 (61.07) | 375 (100) |
| Certainty Assessment | Summary of Findings | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Risk of Bias | Inconsistency | Indirectness | Imprecision | Publication Bias | Overall Certainty of Evidence | Study Event Rates (%) | Relative Effect (95% CI) | Anticipated Absolute Effects | |||
| With Sutures without Triclosan | With Sutures with Triclosan | Risk with Sutures without Triclosan | Risk Difference with Sutures with Triclosan | ||||||||
| New outcome (follow up: range 30 days to 365 days; assessed with: clinically and positive culture) | |||||||||||
| 4100 (12 RCTs) | Serious a | Not serious b | Not serious c | Serious d | None observed | ⨁⨁⨁◯ Moderate | 198/2079 (9.5%) | 124/2021 (6.1%) | RR 0.62 [0.47; 0.82] | 95 per 1000 | 36 fewer per 1000 (s50 to 17 fewer) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daoud, F.C.; Coppry, M.; Moore, N.; Rogues, A.-M. Do Triclosan Sutures Modify the Microbial Diversity of Surgical Site Infections? A Systematic Review and Meta-Analysis. Microorganisms 2022, 10, 927. https://doi.org/10.3390/microorganisms10050927
Daoud FC, Coppry M, Moore N, Rogues A-M. Do Triclosan Sutures Modify the Microbial Diversity of Surgical Site Infections? A Systematic Review and Meta-Analysis. Microorganisms. 2022; 10(5):927. https://doi.org/10.3390/microorganisms10050927
Chicago/Turabian StyleDaoud, Frederic C., Maïder Coppry, Nicholas Moore, and Anne-Marie Rogues. 2022. "Do Triclosan Sutures Modify the Microbial Diversity of Surgical Site Infections? A Systematic Review and Meta-Analysis" Microorganisms 10, no. 5: 927. https://doi.org/10.3390/microorganisms10050927
APA StyleDaoud, F. C., Coppry, M., Moore, N., & Rogues, A.-M. (2022). Do Triclosan Sutures Modify the Microbial Diversity of Surgical Site Infections? A Systematic Review and Meta-Analysis. Microorganisms, 10(5), 927. https://doi.org/10.3390/microorganisms10050927






