Gram-Negative Bacterial Envelope Homeostasis under Oxidative and Nitrosative Stress
Abstract
:1. Oxidative and Nitrosative Stresses: An Omnipresent Challenge
2. The Gram-Negative Bacterial Envelope
2.1. The Outer Membrane (OM)
2.2. The Inner Membrane (IM)
2.3. The Periplasm and the Peptidoglycan (PG)
3. Reactive Oxygen and Nitrogen Species: A Complex Reactions Network
3.1. Oxidative Stress Chemistry
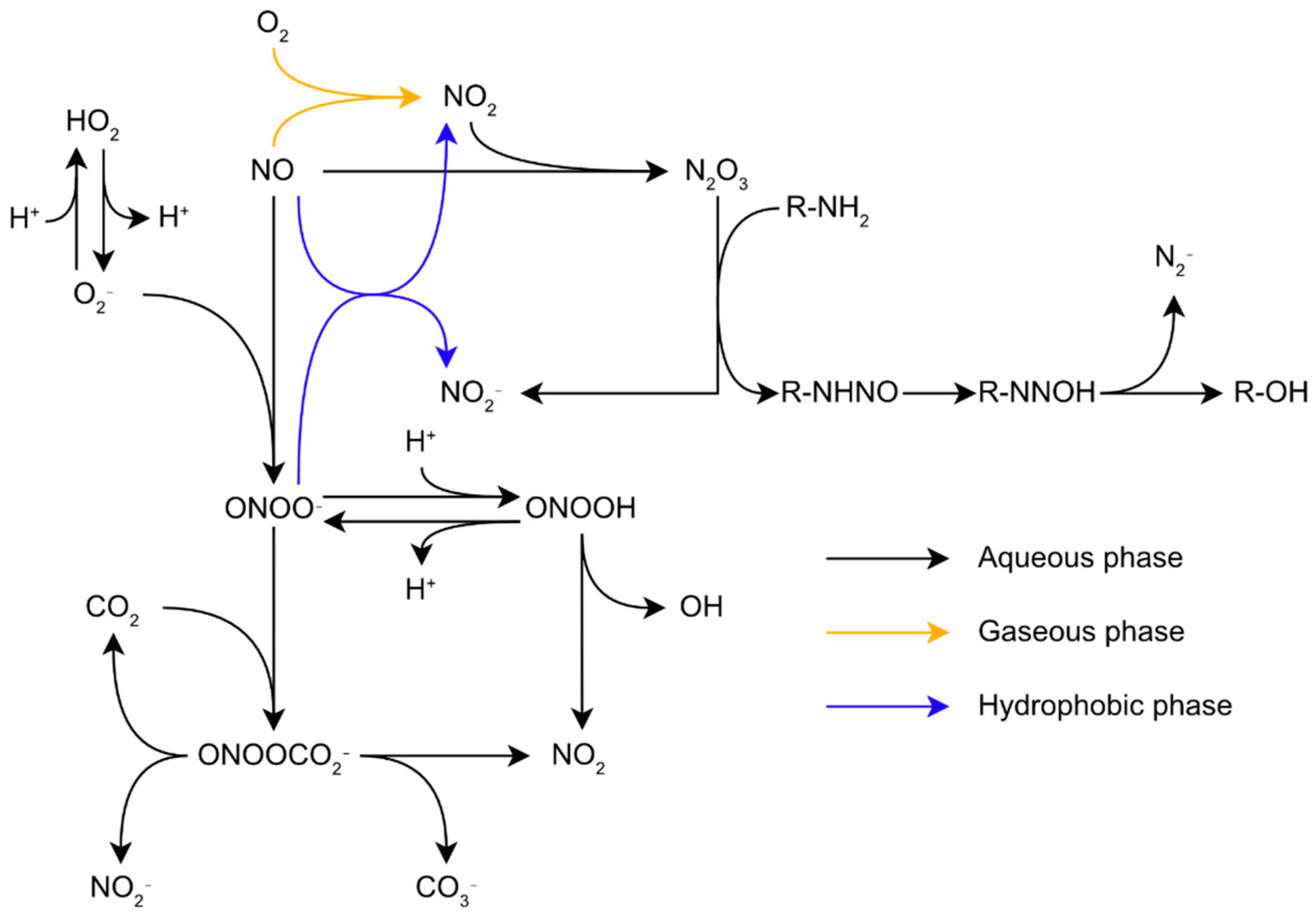
3.2. Nitrosative Stress Chemistry
3.3. Exogenous Sources of Oxidative/Nitrosative Stress
3.3.1. Biotic Stress Sources
3.3.2. Abiotic Stress Sources
3.4. Targets of ROS and RNS
3.4.1. Phospholipids
3.4.2. Peptidoglycan
3.4.3. Envelope Proteins
3.4.4. Protein Carbonylation
3.4.5. Protein S-Nitrosylation
3.4.6. Tyrosine Nitration
4. Defenses against Oxidative and Nitrosative Stresses
4.1. Reducing Systems
4.1.1. Superoxide Dismutases
4.1.2. Catalases/Peroxidases
4.1.3. NO-Reductases
4.1.4. Periplasmic Cytochrome c Nitrite Reductase
4.1.5. Cytochrome bd
4.2. Repair Systems
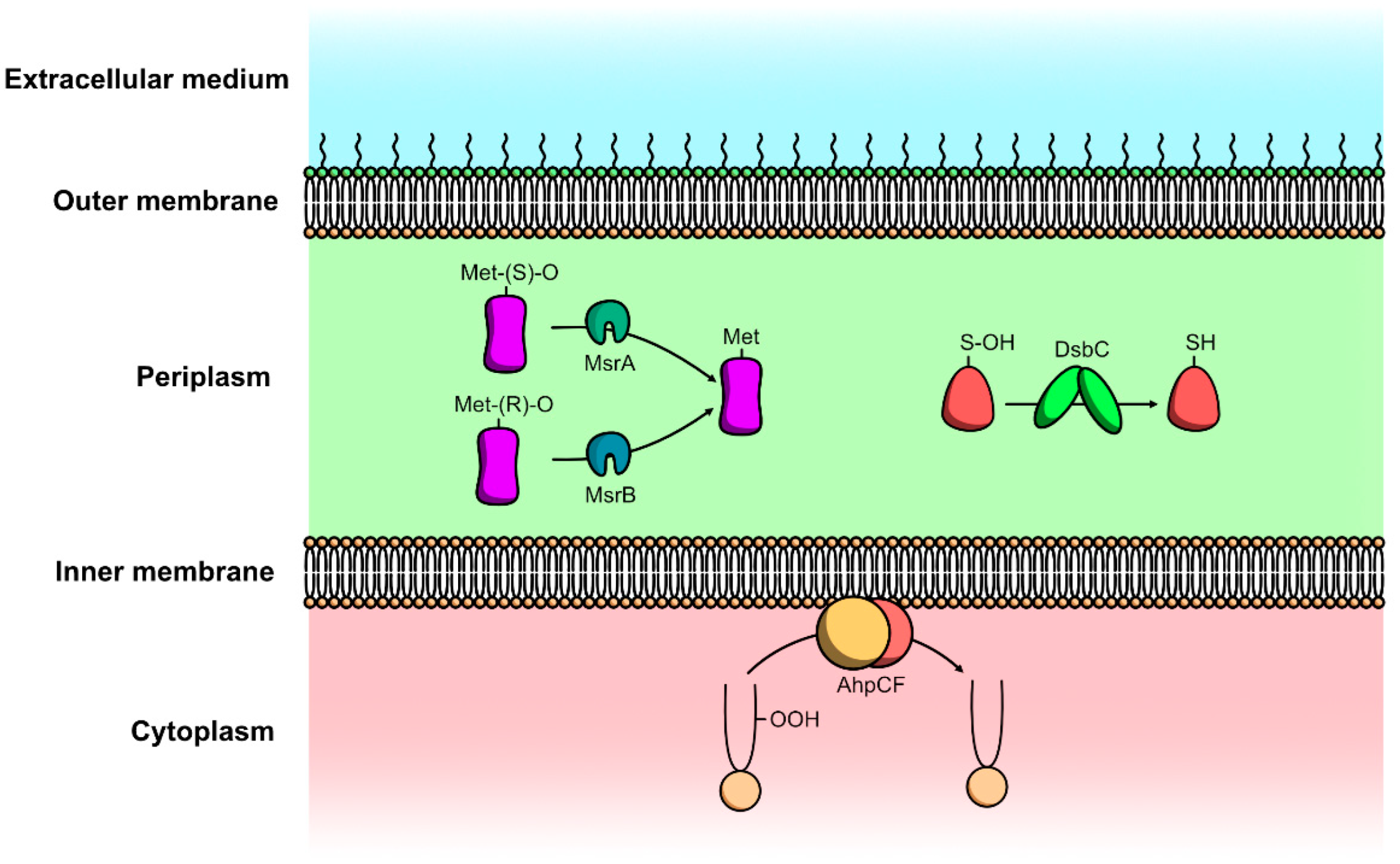
4.2.1. Protein Repair Mechanisms
4.2.2. PG Repair Mechanisms
4.2.3. Membrane Repair Mechanisms
4.3. Envelope Stress Response (ESR)
4.3.1. Cpx Complex in E. coli
4.3.2. σ Factors
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Barraud, N.; Hassett, D.J.; Hwang, S.-H.; Rice, S.A.; Kjelleberg, S.; Webb, J.S. Involvement of Nitric Oxide in Biofilm Dispersal of Pseudomonas aeruginosa. J. Bacteriol. 2006, 188, 7344–7353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ignarro, L.J.; Buga, G.M.; Wood, K.S.; Byrns, R.E.; Chaudhuri, G. Endothelium-derived relaxing factor produced and released from artery and vein is nitric oxide. Proc. Natl. Acad. Sci. USA 1987, 84, 9265–9269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmer, R.M.J.; Ferrige, A.G.; Moncada, S. Nitric oxide release accounts for the biological activity of endothelium-derived relaxing factor. Nature 1987, 327, 524–526. [Google Scholar] [CrossRef] [PubMed]
- Raetz, C.R.H.; Whitfield, C. Lipopolysaccharide Endotoxins. Annu. Rev. Biochem. 2002, 71, 635–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chevalier, S.; Bouffartigues, E.; Bodilis, J.; Maillot, O.; Lesouhaitier, O.; Feuilloley, M.G.J.; Orange, N.; Dufour, A.; Cornelis, P. Structure, function and regulation of Pseudomonas aeruginosa porins. FEMS Microbiol. Rev. 2017, 41, 698–722. [Google Scholar] [CrossRef]
- Vergalli, J.; Bodrenko, I.V.; Masi, M.; Moynié, L.; Acosta-Gutiérrez, S.; Naismith, J.H.; Davin-Regli, A.; Ceccarelli, M.; van den Berg, B.; Winterhalter, M.; et al. Porins and small-molecule translocation across the outer membrane of Gram-negative bacteria. Nat. Rev. Microbiol. 2020, 18, 164–176. [Google Scholar] [CrossRef]
- Sankaran, K.; Wu, H.C. Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J. Biol. Chem. 1994, 269, 19701–19706. [Google Scholar] [CrossRef]
- Cole, G.B.; Bateman, T.J.; Moraes, T.F. The surface lipoproteins of gram-negative bacteria: Protectors and foragers in harsh environments. J. Biol. Chem. 2020, 296, 100147. [Google Scholar] [CrossRef]
- Wilson, M.M.; Bernstein, H.D. Surface-Exposed Lipoproteins: An Emerging Secretion Phenomenon in Gram-Negative Bacteria. Trends Microbiol. 2016, 24, 198–208. [Google Scholar] [CrossRef]
- Kondakova, T.; D’Heygère, F.; Feuilloley, M.J.; Orange, N.; Heipieper, H.J.; Duclairoir Poc, C. Glycerophospholipid synthesis and functions in Pseudomonas. Chem. Phys. Lipids 2015, 190, 27–42. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The Bacterial Cell Envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef] [PubMed]
- Pazos, M.; Peters, K. Peptidoglycan. In Bacterial Cell Walls and Membranes; Kuhn, A., Ed.; Subcellular Biochemistry; Springer International Publishing: Cham, Switzerland, 2019; pp. 127–168. ISBN 978-3-030-18768-2. [Google Scholar]
- Miller, S.I.; Salama, N.R. The gram-negative bacterial periplasm: Size matters. PLoS Biol. 2018, 16, e2004935. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, S.; Bouffartigues, E.; Bazire, A.; Tahrioui, A.; Duchesne, R.; Tortuel, D.; Maillot, O.; Clamens, T.; Orange, N.; Feuilloley, M.G.J.; et al. Extracytoplasmic function sigma factors in Pseudomonas aeruginosa. Biochim. Biophys. Acta BBA Gene Regul. Mech. 2019, 1862, 706–721. [Google Scholar] [CrossRef] [PubMed]
- Arts, I.S.; Gennaris, A.; Collet, J.-F. Reducing systems protecting the bacterial cell envelope from oxidative damage. FEBS Lett. 2015, 589, 1559–1568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrer-Sueta, G.; Campolo, N.; Trujillo, M.; Bartesaghi, S.; Carballal, S.; Romero, N.; Alvarez, B.; Radi, R. Biochemistry of Peroxynitrite and Protein Tyrosine Nitration. Chem. Rev. 2018, 118, 1338–1408. [Google Scholar] [CrossRef]
- Ligeza, A.; Tikhonov, A.N.; Hyde, J.S.; Subczynski, W.K. Oxygen permeability of thylakoid membranes: Electron paramagnetic resonance spin labeling study. Biochim. Biophys. Acta 1998, 1365, 453–463. [Google Scholar] [CrossRef] [Green Version]
- Imlay, J.A. The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat. Rev. Microbiol. 2013, 11, 443–454. [Google Scholar] [CrossRef] [Green Version]
- Korshunov, S.; Imlay, J.A. Two sources of endogenous H2O2 in Escherichia coli. Mol. Microbiol. 2010, 75, 1389–1401. [Google Scholar] [CrossRef] [Green Version]
- Bielski, B.H.J. Reevaluation of the Spectral and Kinetic Properties of Ho2 and O2- Free Radicals. Photochem. Photobiol. 1978, 28, 645–649. [Google Scholar] [CrossRef]
- McCord, J.M.; Fridovich, I. Superoxide Dismutase: An enzymic function for erythrocuprein (hemocuprein). J. Biol. Chem. 1969, 244, 6049–6055. [Google Scholar] [CrossRef]
- Panov, A.V.; Dikalov, S.I. Cardiolipin, Perhydroxyl Radicals, and Lipid Peroxidation in Mitochondrial Dysfunctions and Aging. Oxid. Med. Cell. Longev. 2020, 2020, 1323028. [Google Scholar] [CrossRef] [PubMed]
- Glaeser, J.; Nuss, A.M.; Berghoff, B.A.; Klug, G. Chapter 4—Singlet Oxygen Stress in Microorganisms. In Advances in Microbial Physiology; Poole, R.K., Ed.; Academic Press: Cambridge, MA, USA, 2011; Volume 58, pp. 141–173. [Google Scholar]
- Garcia, X.; Stein, F. Nitric Oxide. Semin. Pediatr. Infect. Dis. 2006, 17, 55–57. [Google Scholar] [CrossRef] [PubMed]
- Fang, F.C.; Frawley, E.R.; Tapscott, T.; Vazquez-Torres, A. Bacterial Stress Responses during Host Infection. Cell Host Microbe 2016, 20, 133–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flint, A.; Stintzi, A.; Saraiva, L.M. Oxidative and nitrosative stress defences of Helicobacter and Campylobacter species that counteract mammalian immunity. FEMS Microbiol. Rev. 2016, 40, 938–960. [Google Scholar] [CrossRef] [Green Version]
- Poole, R.K. Nitric oxide and nitrosative stress tolerance in bacteria. Biochem. Soc. Trans. 2005, 33, 176–180. [Google Scholar] [CrossRef]
- Hughes, M.N. Relationships between nitric oxide, nitroxyl ion, nitrosonium cation and peroxynitrite. Biochim. Biophys. Acta BBA Bioenerg. 1999, 1411, 263–272. [Google Scholar] [CrossRef] [Green Version]
- Koppenol, W.H.; Moreno, J.J.; Pryor, W.A.; Ischiropoulos, H.; Beckman, J.S. Peroxynitrite, a cloaked oxidant formed by nitric oxide and superoxide. Chem. Res. Toxicol. 1992, 5, 834–842. [Google Scholar] [CrossRef]
- Wink, D.A.; Ford, P.C. Nitric Oxide Reactions Important to Biological Systems: A Survey of Some Kinetics Investigations. Methods 1995, 7, 14–20. [Google Scholar] [CrossRef]
- Hughes, M.N. Chapter One—Chemistry of Nitric Oxide and Related Species. In Methods in Enzymology; Poole, R.K., Ed.; Globins and Other Nitric Oxide-Reactive Proteins, Part A; Academic Press: Cambridge, MA, USA, 2008; Volume 436, pp. 3–19. [Google Scholar]
- Kashyap, D.R.; Kuzma, M.; Kowalczyk, D.A.; Gupta, D.; Dziarski, R. Bactericidal peptidoglycan recognition protein induces oxidative stress in Escherichia coli through a block in respiratory chain and increase in central carbon catabolism. Mol. Microbiol. 2017, 105, 755–776. [Google Scholar] [CrossRef] [Green Version]
- Heesterbeek, D.A.C.; Muts, R.M.; van Hensbergen, V.P.; de Saint Aulaire, P.; Wennekes, T.; Bardoel, B.W.; van Sorge, N.M.; Rooijakkers, S.H.M. Outer membrane permeabilization by the membrane attack complex sensitizes Gram-negative bacteria to antimicrobial proteins in serum and phagocytes. PLOS Pathog. 2021, 17, e1009227. [Google Scholar] [CrossRef]
- Liu, H.; Zheng, F.; Cao, Q.; Ren, B.; Zhu, L.; Striker, G.; Vlassara, H. Amelioration of oxidant stress by the defensin lysozyme. Am. J. Physiol.-Endocrinol. Metab. 2006, 290, E824–E832. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.M.; Tan, A.X.; Vlassara, H. Antibacterial activity of lysozyme and lactoferrin is inhibited by binding of advanced glycation–modified proteins to a conserved motif. Nat. Med. 1995, 1, 1057–1061. [Google Scholar] [CrossRef] [PubMed]
- Ragland, S.A.; Criss, A.K. From bacterial killing to immune modulation: Recent insights into the functions of lysozyme. PLoS Pathog. 2017, 13, e1006512. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Iida, K.; Saito, M.; Nakayama, H.; Yoshida, S. Hydrogen peroxide production in Streptococcus pyogenes: Involvement of lactate oxidase and coupling with aerobic utilization of lactate. J. Bacteriol. 2004, 186, 2046–2051. [Google Scholar] [CrossRef] [Green Version]
- Spellerberg, B.; Cundell, D.R.; Sandros, J.; Pearce, B.J.; Idanpaan-Heikkila, I.; Rosenow, C.; Masure, H.R. Pyruvate oxidase, as a determinant of virulence in Streptococcus pneumoniae. Mol. Microbiol. 1996, 19, 803–813. [Google Scholar] [CrossRef]
- Liu, X.; Ramsey, M.M.; Chen, X.; Koley, D.; Whiteley, M.; Bard, A.J. Real-time mapping of a hydrogen peroxide concentration profile across a polymicrobial bacterial biofilm using scanning electrochemical microscopy. Proc. Natl. Acad. Sci. USA 2011, 108, 2668–2673. [Google Scholar] [CrossRef] [Green Version]
- Tong, H.; Chen, W.; Merritt, J.; Qi, F.; Shi, W.; Dong, X. Streptococcus oligofermentans inhibits Streptococcus mutans through conversion of lactic acid into inhibitory H2O2: A possible counteroffensive strategy for interspecies competition. Mol. Microbiol. 2007, 63, 872–880. [Google Scholar] [CrossRef]
- Baron, S.S.; Terranova, G.; Rowe, J.J. Molecular mechanism of the antimicrobial action of pyocyanin. Curr. Microbiol. 1989, 18, 223–230. [Google Scholar] [CrossRef]
- Hassan, H.M.; Fridovich, I. Mechanism of the antibiotic action pyocyanine. J. Bacteriol. 1980, 141, 156–163. [Google Scholar] [CrossRef] [Green Version]
- Mourenza, Á.; Gil, J.A.; Mateos, L.M.; Letek, M. Oxidative Stress-Generating Antimicrobials, a Novel Strategy to Overcome Antibacterial Resistance. Antioxidants 2020, 9, 361. [Google Scholar] [CrossRef]
- Keren, I.; Wu, Y.; Inocencio, J.; Mulcahy, L.R.; Lewis, K. Killing by bactericidal antibiotics does not depend on reactive oxygen species. Science 2013, 339, 1213–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahoney, T.F.; Silhavy, T.J. The Cpx Stress Response Confers Resistance to Some, but Not All, Bactericidal Antibiotics. J. Bacteriol. 2013, 195, 1869–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albesa, I.; Becerra, M.C.; Battán, P.C.; Páez, P.L. Oxidative stress involved in the antibacterial action of different antibiotics. Biochem. Biophys. Res. Commun. 2004, 317, 605–609. [Google Scholar] [CrossRef] [PubMed]
- Goswami, M.; Mangoli, S.H.; Jawali, N. Involvement of Reactive Oxygen Species in the Action of Ciprofloxacin against Escherichia coli. Antimicrob. Agents Chemother. 2006, 50, 949–954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ladjouzi, R.; Bizzini, A.; Lebreton, F.; Sauvageot, N.; Rincé, A.; Benachour, A.; Hartke, A. Analysis of the tolerance of pathogenic Enterococci and Staphylococcus aureus to cell wall active antibiotics. J. Antimicrob. Chemother. 2013, 68, 2083–2091. [Google Scholar] [CrossRef]
- Dam, S.; Pagès, J.-M.; Masi, M. Stress responses, outer membrane permeability control and antimicrobial resistance in Enterobacteriaceae. Microbiol. Read. Engl. 2018, 164, 260–267. [Google Scholar] [CrossRef]
- Lesser, M.P. Oxidative stress in marine environments: Biochemistry and Physiological Ecology. Annu. Rev. Physiol. 2006, 68, 253–278. [Google Scholar] [CrossRef] [Green Version]
- Meslé, M.M.; Beam, J.P.; Jay, Z.J.; Bodle, B.; Bogenschutz, E.; Inskeep, W.P. Hydrogen Peroxide Cycling in High-Temperature Acidic Geothermal Springs and Potential Implications for Oxidative Stress Response. Front. Mar. Sci. 2017, 4, 130. [Google Scholar] [CrossRef] [Green Version]
- Santos, A.L.; Gomes, N.C.M.; Henriques, I.; Almeida, A.; Correia, A.; Cunha, Â. Growth conditions influence UVB sensitivity and oxidative damage in an estuarine bacterial isolate. Photochem. Photobiol. Sci. 2013, 12, 974–986. [Google Scholar] [CrossRef]
- Depayras, S.; Kondakova, T.; Heipieper, H.J.; Feuilloley, M.G.; Orange, N.; Duclairoir-Poc, C. The Hidden Face of Nitrogen Oxides Species: From Toxic Effects to Potential Cure? IntechOpen: London, UK, 2018; ISBN 978-1-78923-385-8. [Google Scholar]
- Skalska, K.; Miller, J.S.; Ledakowicz, S. Trends in NOx abatement: A review. Sci. Total Environ. 2010, 408, 3976–3989. [Google Scholar] [CrossRef]
- Chautrand, T.; Souak, D.; Kondakova, T.; Depayras, S.; Machour, N.; Heipieper, H.; Feuilloley, M.; Orange, N.; Poc, C. Air pollution and other environmental stresses: Gaseous NO2 exposure leads to specific alterations of Pseudomonas fluorescens. WIT Trans. Ecol. Environ. 2020, 244, 53–63. [Google Scholar]
- Depayras, S.; Kondakova, T.; Merlet-Machour, N.; Heipieper, H.J.; Barreau, M.; Catovic, C.; Feuilloley, M.; Orange, N.; Poc, C.D. Impact of gaseous NO2 on P. fluorescens strain in the membrane adaptation and virulence. Int. J. Environ. Impacts 2018, 1, 183–192. [Google Scholar] [CrossRef]
- Blee, J.A.; Roberts, I.S.; Waigh, T.A. Membrane potentials, oxidative stress and the dispersal response of bacterial biofilms to 405 nm light. Phys. Biol. 2020, 17, 036001. [Google Scholar] [CrossRef] [PubMed]
- Strahl, H.; Errington, J. Bacterial Membranes: Structure, Domains, and Function. Annu. Rev. Microbiol. 2017, 71, 519–538. [Google Scholar] [CrossRef]
- Dailey, F.E.; McGraw, J.E.; Jensen, B.J.; Bishop, S.S.; Lokken, J.P.; Dorff, K.J.; Ripley, M.P.; Munro, J.B. The Microbiota of Freshwater Fish and Freshwater Niches Contain Omega-3 Fatty Acid-Producing Shewanella Species. Appl. Environ. Microbiol. 2015, 82, 218–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moravec, A.R.; Siv, A.W.; Hobby, C.R.; Lindsay, E.N.; Norbash, L.V.; Shults, D.J.; Symes, S.J.K.; Giles, D.K. Exogenous Polyunsaturated Fatty Acids Impact Membrane Remodeling and Affect Virulence Phenotypes among Pathogenic Vibrio Species. Appl. Environ. Microbiol. 2017, 83, e01415-17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, J.-H.; Hassan, K.A.; Begg, S.L.; Rupasinghe, T.W.T.; Naidu, V.; Pederick, V.G.; Khorvash, M.; Whittall, J.J.; Paton, J.C.; Paulsen, I.T.; et al. Identification of Novel Acinetobacter baumannii Host Fatty Acid Stress Adaptation Strategies. mBio 2019, 10, e02056-18. [Google Scholar] [CrossRef] [Green Version]
- Shin, S.Y.; Bajpai, V.K.; Kim, H.R.; Kang, S.C. Antibacterial activity of bioconverted eicosapentaenoic (EPA) and docosahexaenoic acid (DHA) against foodborne pathogenic bacteria. Int. J. Food Microbiol. 2007, 113, 233–236. [Google Scholar] [CrossRef]
- Desbois, A.P.; Lawlor, K.C. Antibacterial Activity of Long-Chain Polyunsaturated Fatty Acids against Propionibacterium acnes and Staphylococcus aureus. Mar. Drugs 2013, 11, 4544–4557. [Google Scholar] [CrossRef] [Green Version]
- Eijkelkamp, B.A.; Begg, S.L.; Pederick, V.G.; Trapetti, C.; Gregory, M.K.; Whittall, J.J.; Paton, J.C.; McDevitt, C.A. Arachidonic Acid Stress Impacts Pneumococcal Fatty Acid Homeostasis. Front. Microbiol. 2018, 9, 813. [Google Scholar] [CrossRef]
- Krute, C.N.; Ridder, M.J.; Seawell, N.A.; Bose, J.L.Y. Inactivation of the exogenous fatty acid utilization pathway leads to increased resistance to unsaturated fatty acids in Staphylococcus aureus. Microbiology 2021, 165, 197–207. [Google Scholar] [CrossRef] [PubMed]
- Beavers, W.N.; Monteith, A.J.; Amarnath, V.; Mernaugh, R.L.; Roberts, L.J.; Chazin, W.J.; Davies, S.S.; Skaar, E.P. Arachidonic Acid Kills Staphylococcus aureus through a Lipid Peroxidation Mechanism. mBio 2019, 10, e01333-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thibessard, A.; Fernandez, A.; Gintz, B.; Leblond-Bourget, N.; Decaris, B. Effects of rodA and pbp2b disruption on cell morphology and oxidative stress response of Streptococcus thermophilus CNRZ368. J. Bacteriol. 2002, 184, 2821–2826. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, G.; Olczak, A.; Forsberg, L.S.; Maier, R.J. Oxidative Stress-induced Peptidoglycan Deacetylase in Helicobacter pylori. J. Biol. Chem. 2009, 284, 6790–6800. [Google Scholar] [CrossRef] [Green Version]
- Davis, M.M.; Brock, A.M.; DeHart, T.G.; Boribong, B.P.; Lee, K.; McClune, M.E.; Chang, Y.; Cramer, N.; Liu, J.; Jones, C.N.; et al. The peptidoglycan-associated protein NapA plays an important role in the envelope integrity and in the pathogenesis of the lyme disease spirochete. PLoS Pathog. 2021, 17, e1009546. [Google Scholar] [CrossRef]
- Chautrand, T.; Depayras, S.; Souak, D.; Kondakova, T.; Barreau, M.; Kentache, T.; Hardouin, J.; Tahrioui, A.; Thoumire, O.; Konto-Ghiorghi, Y.; et al. Gaseous NO2 induces various envelope alterations in Pseudomonas fluorescens MFAF76a. Sci. Rep. 2022; in press. [Google Scholar]
- Giacomucci, S.; Alvarez, L.; Rodrigues, C.D.A.; Cava, F.; Paradis-Bleau, C. Hydroxyl Radical Overproduction in the Envelope: An Achilles’ Heel in Peptidoglycan Synthesis. Microbiol. Spectr. 2022, 10, e01203-21. [Google Scholar] [CrossRef]
- Ezraty, B.; Gennaris, A.; Barras, F.; Collet, J.-F. Oxidative stress, protein damage and repair in bacteria. Nat. Rev. Microbiol. 2017, 15, 385–396. [Google Scholar] [CrossRef]
- Lee, B.C.; Gladyshev, V.N. The biological significance of methionine sulfoxide stereochemistry. Free Radic. Biol. Med. 2011, 50, 221–227. [Google Scholar] [CrossRef] [Green Version]
- Davies, M.J. The oxidative environment and protein damage. Biochim. Biophys. Acta 2005, 1703, 93–109. [Google Scholar] [CrossRef]
- Roos, G.; Messens, J. Protein sulfenic acid formation: From cellular damage to redox regulation. Free Radic. Biol. Med. 2011, 51, 314–326. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, Y.J.; Carini, M.; Butterfield, D.A. Protein Carbonylation. Antioxid. Redox Signal. 2010, 12, 323–325. [Google Scholar] [CrossRef] [PubMed]
- Stadtman, E.R.; Levine, R.L. Protein oxidation. Ann. N. Y. Acad. Sci. 2000, 899, 191–208. [Google Scholar] [CrossRef] [PubMed]
- Grimsrud, P.A.; Xie, H.; Griffin, T.J.; Bernlohr, D.A. Oxidative Stress and Covalent Modification of Protein with Bioactive Aldehydes. J. Biol. Chem. 2008, 283, 21837–21841. [Google Scholar] [CrossRef] [Green Version]
- Sayre, L.M.; Lin, D.; Yuan, Q.; Zhu, X.; Tang, X. Protein adducts generated from products of lipid oxidation: Focus on HNE and one. Drug Metab. Rev. 2006, 38, 651–675. [Google Scholar] [CrossRef]
- Dalle-Donne, I.; Giustarini, D.; Colombo, R.; Rossi, R.; Milzani, A. Protein carbonylation in human diseases. Trends Mol. Med. 2003, 9, 169–176. [Google Scholar] [CrossRef]
- Ballesteros, M.; Fredriksson, Å.; Henriksson, J.; Nyström, T. Bacterial senescence: Protein oxidation in non-proliferating cells is dictated by the accuracy of the ribosomes. EMBO J. 2001, 20, 5280–5289. [Google Scholar] [CrossRef] [Green Version]
- Seth, D.; Hausladen, A.; Stamler, J.S. Anaerobic Transcription by OxyR: A Novel Paradigm for Nitrosative Stress. Antioxid. Redox Signal. 2020, 32, 803–816. [Google Scholar] [CrossRef]
- Bartesaghi, S.; Wenzel, J.; Trujillo, M.; López, M.; Joseph, J.; Kalyanaraman, B.; Radi, R. Lipid peroxyl radicals mediate tyrosine dimerization and nitration in membranes. Chem. Res. Toxicol. 2010, 23, 821–835. [Google Scholar] [CrossRef] [Green Version]
- Folkes, L.K.; Bartesaghi, S.; Trujillo, M.; Radi, R.; Wardman, P. Kinetics of oxidation of tyrosine by a model alkoxyl radical. Free Radic. Res. 2012, 46, 1150–1156. [Google Scholar] [CrossRef]
- Raivio, T.L. MicroReview: Envelope stress responses and Gram-negative bacterial pathogenesis. Mol. Microbiol. 2005, 56, 1119–1128. [Google Scholar] [CrossRef] [PubMed]
- Korshunov, S.S.; Imlay, J.A. A potential role for periplasmic superoxide dismutase in blocking the penetration of external superoxide into the cytosol of Gram-negative bacteria. Mol. Microbiol. 2002, 43, 95–106. [Google Scholar] [CrossRef] [PubMed]
- Yost, F.J.; Fridovich, I. An Iron-containing Superoxide Dismutase from Escherichia coli. J. Biol. Chem. 1973, 248, 4905–4908. [Google Scholar] [CrossRef]
- Keele, B.B.; McCord, J.M.; Fridovich, I. Superoxide Dismutase from Escherichia coli B: A new manganese-containing enzyme. J. Biol. Chem. 1970, 245, 6176–6181. [Google Scholar] [CrossRef]
- Benov, L.T.; Fridovich, I. Escherichia coli expresses a copper- and zinc-containing superoxide dismutase. J. Biol. Chem. 1994, 269, 25310–25314. [Google Scholar] [CrossRef]
- De Groote, M.A.; Ochsner, U.A.; Shiloh, M.U.; Nathan, C.; McCord, J.M.; Dinauer, M.C.; Libby, S.J.; Vazquez-Torres, A.; Xu, Y.; Fang, F.C. Periplasmic superoxide dismutase protects Salmonella from products of phagocyte NADPH-oxidase and nitric oxide synthase. Proc. Natl. Acad. Sci. USA 1997, 94, 13997–14001. [Google Scholar] [CrossRef] [Green Version]
- Cho, S.-H.; Parsonage, D.; Thurston, C.; Dutton, R.J.; Poole, L.B.; Collet, J.-F.; Beckwith, J. A new family of membrane electron transporters and its substrates, including a new cell envelope peroxiredoxin, reveal a broadened reductive capacity of the oxidative bacterial cell envelope. mBio 2012, 3, e00291-11. [Google Scholar] [CrossRef] [Green Version]
- Shapleigh, J.P. The Denitrifying Prokaryotes. In The Prokaryotes: Volume 2: Ecophysiology and Biochemistry; Dworkin, M., Falkow, S., Rosenberg, E., Schleifer, K.-H., Stackebrandt, E., Eds.; Springer: New York, NY, USA, 2006; pp. 769–792. ISBN 978-0-387-30742-8. [Google Scholar]
- Shiro, Y. Structure and function of bacterial nitric oxide reductases: Nitric oxide reductase, anaerobic enzymes. Biochim. Biophys. Acta BBA Bioenerg. 2012, 1817, 1907–1913. [Google Scholar] [CrossRef] [Green Version]
- Vázquez-Torres, A.; Bäumler, A. Nitrate, nitrite and nitric oxide reductases: From the last universal common ancestor to modern bacterial pathogens. Curr. Opin. Microbiol. 2016, 29, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Poock, S.R.; Leach, E.R.; Moir, J.W.B.; Cole, J.A.; Richardson, D.J. Respiratory Detoxification of Nitric Oxide by the Cytochromec Nitrite Reductase of Escherichia coli. J. Biol. Chem. 2002, 277, 23664–23669. [Google Scholar] [CrossRef] [Green Version]
- Kern, M.; Volz, J.; Simon, J. The oxidative and nitrosative stress defence network of Wolinella succinogenes: Cytochrome c nitrite reductase mediates the stress response to nitrite, nitric oxide, hydroxylamine and hydrogen peroxide. Environ. Microbiol. 2011, 13, 2478–2494. [Google Scholar] [CrossRef] [PubMed]
- Pittman, M.S.; Elvers, K.T.; Lee, L.; Jones, M.A.; Poole, R.K.; Park, S.F.; Kelly, D.J. Growth of Campylobacter jejuni on nitrate and nitrite: Electron transport to NapA and NrfA via NrfH and distinct roles for NrfA and the globin Cgb in protection against nitrosative stress. Mol. Microbiol. 2007, 63, 575–590. [Google Scholar] [CrossRef] [PubMed]
- Runkel, S.; Wells, H.C.; Rowley, G. Chapter Three—Living with Stress: A Lesson from the Enteric Pathogen Salmonella enterica. In Advances in Applied Microbiology; Sariaslani, S., Gadd, G.M., Eds.; Academic Press: Cambridge, MA, USA, 2013; Volume 83, pp. 87–144. [Google Scholar]
- Borisov, V.B.; Verkhovsky, M.I. Oxygen as Acceptor. EcoSal Plus 2009, 3, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Borisov, V.B.; Forte, E.; Siletsky, S.A.; Arese, M.; Davletshin, A.I.; Sarti, P.; Giuffrè, A. Cytochrome bd protects bacteria against oxidative and nitrosative stress: A potential target for next-generation antimicrobial agents. Biochemistry 2015, 80, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Way, S.S.; Sallustio, S.; Magliozzo, R.S.; Goldberg, M.B. Impact of either Elevated or Decreased Levels of Cytochrome bd Expression on Shigella flexneri Virulence. J. Bacteriol. 1999, 181, 1229–1237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endley, S.; McMurray, D.; Ficht, T.A. Interruption of the cydB Locus inBrucella abortus Attenuates Intracellular Survival and Virulence in the Mouse Model of Infection. J. Bacteriol. 2001, 183, 2454–2462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones-Carson, J.; Husain, M.; Liu, L.; Orlicky, D.J.; Vázquez-Torres, A. Cytochrome bd-Dependent Bioenergetics and Antinitrosative Defenses in Salmonella Pathogenesis. mBio 2016, 7, e02052-16. [Google Scholar] [CrossRef] [Green Version]
- Depuydt, M.; Messens, J.; Collet, J.-F. How Proteins Form Disulfide Bonds. Antioxid. Redox Signal. 2011, 15, 49–66. [Google Scholar] [CrossRef]
- Dutton, R.J.; Boyd, D.; Berkmen, M.; Beckwith, J. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc. Natl. Acad. Sci. USA 2008, 105, 11933–11938. [Google Scholar] [CrossRef] [Green Version]
- Mainardi, J.-L.; Hugonnet, J.-E.; Rusconi, F.; Fourgeaud, M.; Dubost, L.; Moumi, A.N.; Delfosse, V.; Mayer, C.; Gutmann, L.; Rice, L.B.; et al. Unexpected Inhibition of Peptidoglycan LD-Transpeptidase from Enterococcus faecium by the β-Lactam Imipenem. J. Biol. Chem. 2007, 282, 30414–30422. [Google Scholar] [CrossRef] [Green Version]
- Sasindran, S.J.; Saikolappan, S.; Dhandayuthapani, S. Methionine sulfoxide reductases and virulence of bacterial pathogens. Future Microbiol. 2007, 2, 619–630. [Google Scholar] [CrossRef] [PubMed]
- Ezraty, B.; Aussel, L.; Barras, F. Methionine sulfoxide reductases in prokaryotes. Biochim. Biophys. Acta BBA Proteins Proteom. 2005, 1703, 221–229. [Google Scholar] [CrossRef] [PubMed]
- Alamuri, P.; Maier, R.J. Methionine sulphoxide reductase is an important antioxidant enzyme in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 2004, 53, 1397–1406. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.; McClain, M.S.; Forsyth, M.H.; Cover, T.L. Extracellular Release of Antigenic Proteins by Helicobacter pylori. Infect. Immun. 1998, 66, 2984–2986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mintz, K.P.; Moskovitz, J.; Wu, H.; Fives-Taylor, P.M.Y. Peptide methionine sulfoxide reductase (MsrA) is not a major virulence determinant for the oral pathogen Actinobacillus actinomycetemcomitansaa. Microbiology 2002, 148, 3695–3703. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamburro, A.; Robuffo, I.; Heipieper, H.J.; Allocati, N.; Rotilio, D.; Di Ilio, C.; Favaloro, B. Expression of glutathione S-transferase and peptide methionine sulphoxide reductase in Ochrobactrum anthropi is correlated to the production of reactive oxygen species caused by aromatic substrates. FEMS Microbiol. Lett. 2004, 241, 151–156. [Google Scholar] [CrossRef] [Green Version]
- Artymiuk, P.J.; Green, J. The Double Life of Aconitase. Structure 2006, 14, 2–4. [Google Scholar] [CrossRef]
- Goss, D.J.; Theil, E.C. Iron Responsive mRNAs: A Family of Fe2+ Sensitive Riboregulators. Acc. Chem. Res. 2011, 44, 1320–1328. [Google Scholar] [CrossRef] [Green Version]
- Soum, E.; Drapier, J.-C. Nitric oxide and peroxynitrite promote complete disruption of the [4Fe-4S] cluster of recombinant human iron regulatory protein 1. JBIC J. Biol. Inorg. Chem. 2003, 8, 226–232. [Google Scholar] [CrossRef]
- Austin, C.M.; Maier, R.J. Aconitase-Mediated Posttranscriptional Regulation of Helicobacter pylori Peptidoglycan Deacetylase. J. Bacteriol. 2013, 195, 5316–5322. [Google Scholar] [CrossRef] [Green Version]
- Dziarski, R. Recognition of bacterial peptidoglycan by the innate immune system. Cell. Mol. Life Sci. CMLS 2003, 60, 1793–1804. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Maier, S.E.; Lo, L.F.; Maier, G.; Dosi, S.; Maier, R.J. Peptidoglycan Deacetylation in Helicobacter pylori Contributes to Bacterial Survival by Mitigating Host Immune Responses. Infect. Immun. 2010, 78, 4660–4666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dolin, M.I. DPNH peroxidase: Effector activities of DPN+. Biochem. Biophys. Res. Commun. 1977, 78, 393–400. [Google Scholar] [CrossRef] [Green Version]
- Farr, S.B.; Kogoma, T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol. Rev. 1991, 55, 561–585. [Google Scholar] [CrossRef] [PubMed]
- Hews, C.L.; Cho, T.; Rowley, G.; Raivio, T.L. Maintaining Integrity Under Stress: Envelope Stress Response Regulation of Pathogenesis in Gram-Negative Bacteria. Front. Cell. Infect. Microbiol. 2019, 9, 313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, A.M.; Silhavy, T.J. Envelope stress responses: Balancing damage repair and toxicity. Nat. Rev. Microbiol. 2019, 17, 417–428. [Google Scholar] [CrossRef]
- Raivio, T.L. Everything old is new again: An update on current research on the Cpx envelope stress response. Biochim. Biophys. Acta BBA Mol. Cell Res. 2014, 1843, 1529–1541. [Google Scholar] [CrossRef] [Green Version]
- Guest, R.L.; Wang, J.; Wong, J.L.; Raivio, T.L. A Bacterial Stress Response Regulates Respiratory Protein Complexes To Control Envelope Stress Adaptation. J. Bacteriol. 2017, 199, e00153-17. [Google Scholar] [CrossRef] [Green Version]
- Erickson, J.W.; Gross, C.A. Identification of the sigma E subunit of Escherichia coli RNA polymerase: A second alternate sigma factor involved in high-temperature gene expression. Genes Dev. 1989, 3, 1462–1471. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.Y. Two stress sensor proteins for the expression of sigmaE regulon: DegS and RseB. J. Microbiol. 2015, 53, 306–310. [Google Scholar] [CrossRef]
- Nuss, A.M.; Glaeser, J.; Klug, G. RpoHII Activates Oxidative-Stress Defense Systems and Is Controlled by RpoE in the Singlet Oxygen-Dependent Response in Rhodobacter sphaeroides. J. Bacteriol. 2009, 191, 220–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alvarez-Martinez, C.E.; Lourenço, R.F.; Baldini, R.L.; Laub, M.T.; Gomes, S.L. The ECF sigma factor σT is involved in osmotic and oxidative stress responses in Caulobacter crescentus. Mol. Microbiol. 2007, 66, 1240–1255. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Li, Y.; Zhan, W.; Wood, T.K.; Wang, X. Resistance to oxidative stress by inner membrane protein ElaB is regulated by OxyR and RpoS. Microb. Biotechnol. 2019, 12, 392–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sulaiman, J.E.; Lam, H. Proteomic Study of the Survival and Resuscitation Mechanisms of Filamentous Persisters in an Evolved Escherichia coli Population from Cyclic Ampicillin Treatment. mSystems 2020, 5, e00462-20. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chautrand, T.; Souak, D.; Chevalier, S.; Duclairoir-Poc, C. Gram-Negative Bacterial Envelope Homeostasis under Oxidative and Nitrosative Stress. Microorganisms 2022, 10, 924. https://doi.org/10.3390/microorganisms10050924
Chautrand T, Souak D, Chevalier S, Duclairoir-Poc C. Gram-Negative Bacterial Envelope Homeostasis under Oxidative and Nitrosative Stress. Microorganisms. 2022; 10(5):924. https://doi.org/10.3390/microorganisms10050924
Chicago/Turabian StyleChautrand, Thibault, Djouhar Souak, Sylvie Chevalier, and Cécile Duclairoir-Poc. 2022. "Gram-Negative Bacterial Envelope Homeostasis under Oxidative and Nitrosative Stress" Microorganisms 10, no. 5: 924. https://doi.org/10.3390/microorganisms10050924
APA StyleChautrand, T., Souak, D., Chevalier, S., & Duclairoir-Poc, C. (2022). Gram-Negative Bacterial Envelope Homeostasis under Oxidative and Nitrosative Stress. Microorganisms, 10(5), 924. https://doi.org/10.3390/microorganisms10050924





