Characterization of High-Risk HPV/EBV Co-Presence in Pre-Malignant Cervical Lesions and Squamous Cell Carcinomas
Abstract
:1. Introduction
2. Materials and Methods
2.1. Tissue Samples
2.2. DNA Extraction
2.3. Polymerase Chain Reaction (PCR)
2.4. HPV Genotyping
2.5. In Situ Hybridization for EBV
2.6. Reverse Transcriptase-PCR
2.7. Cell Culture and Transfection
2.8. Phosphoproteomic NF-kB Array
2.9. Cell Migration Assay
2.10. Cell Proliferation Assay
2.11. Anchorage-Independent Growth Assay
2.12. Western Blot
2.13. Statistical Analyses
3. Results
3.1. Patients
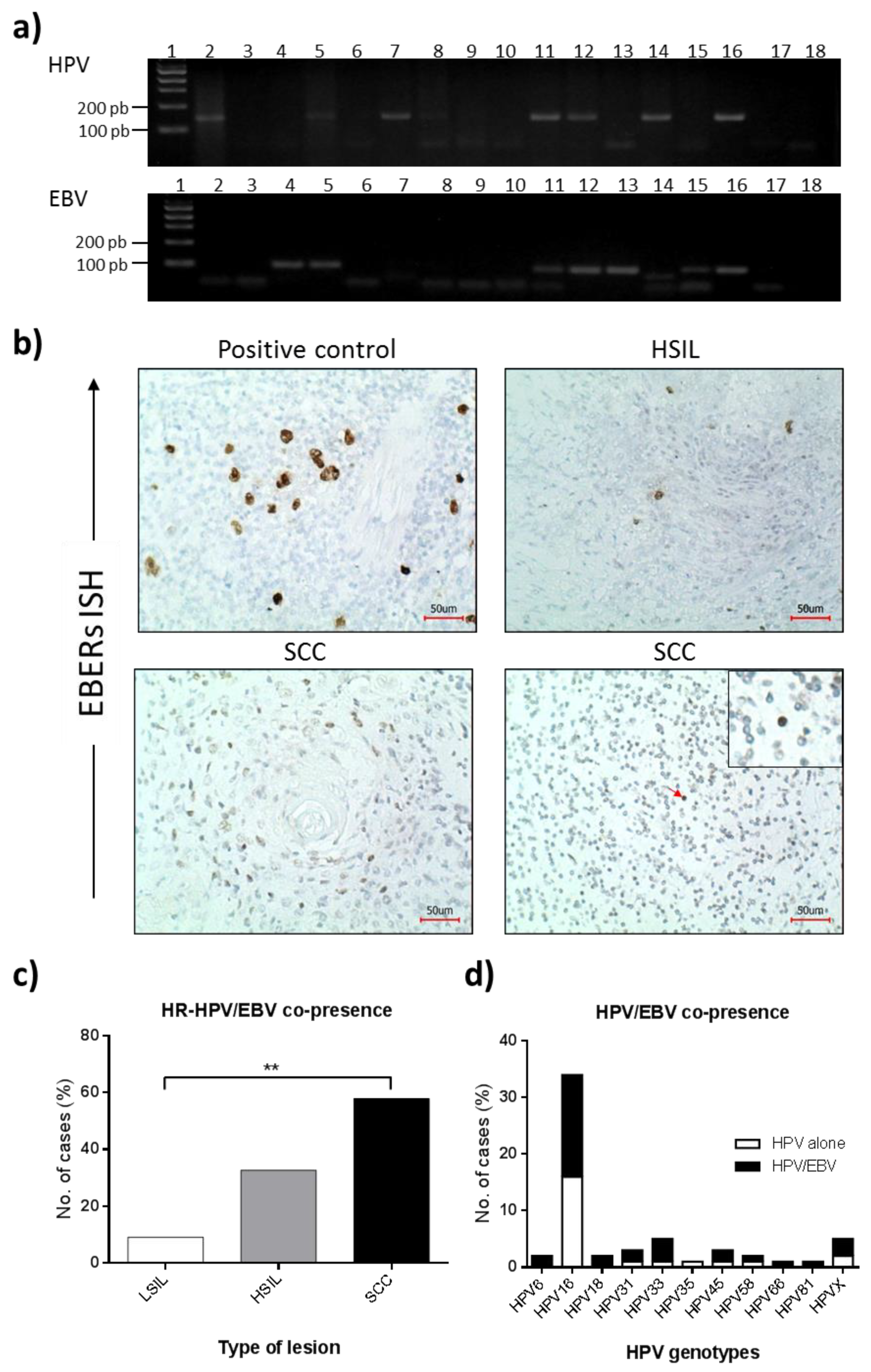
3.2. HPV Presence in Cervical Lesions
3.3. EBV Presence in Cervical Lesions
3.4. BARF1 Transcripts in EBV-Positive Cervical Pre-Malignant Lesions and Squamous Cell Carcinomas
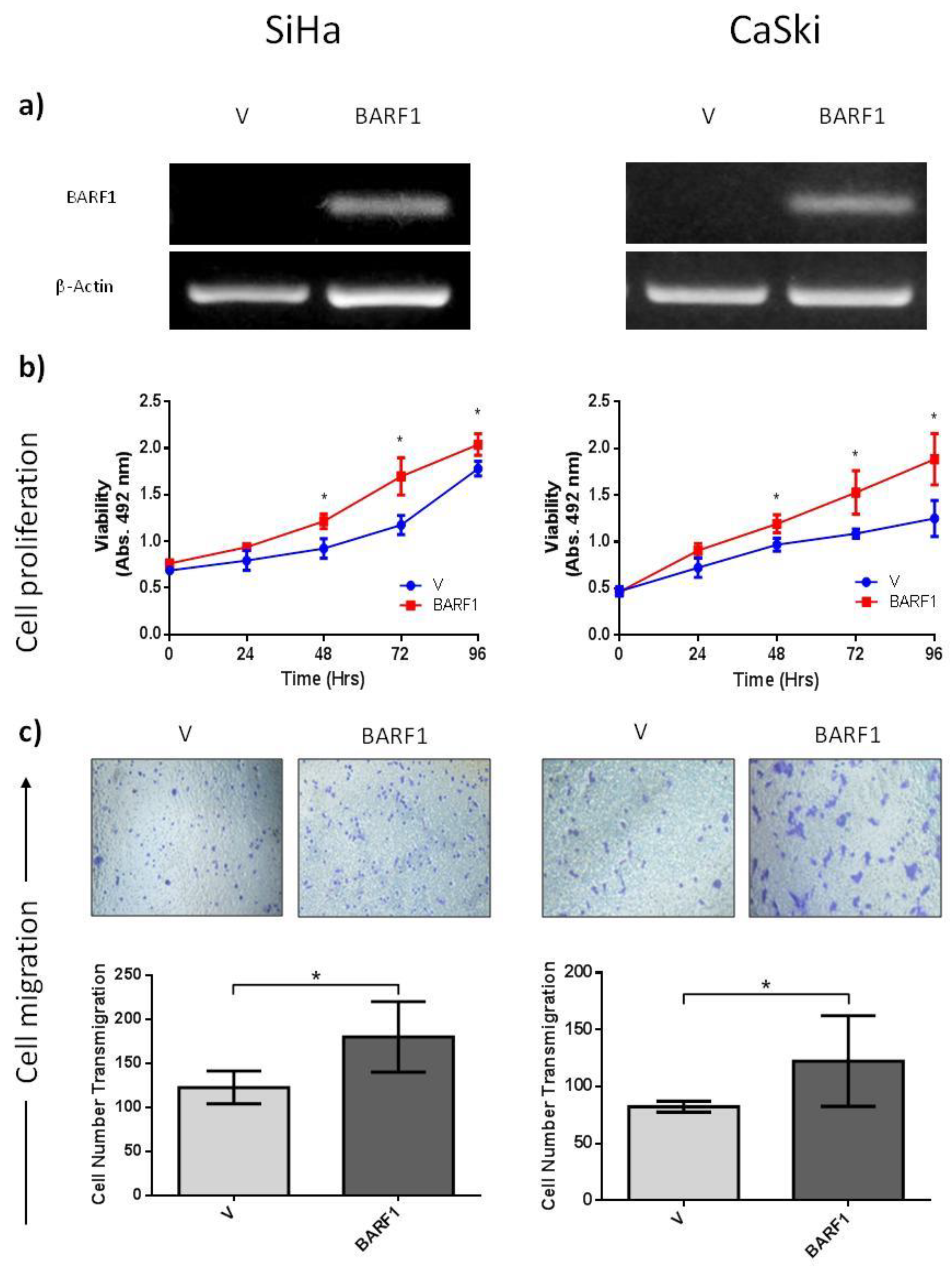
3.5. BARF1 Increases Cervical Cancer Cell Proliferation
3.6. BARF1 Promotes Cervical Cancer Cell Migration
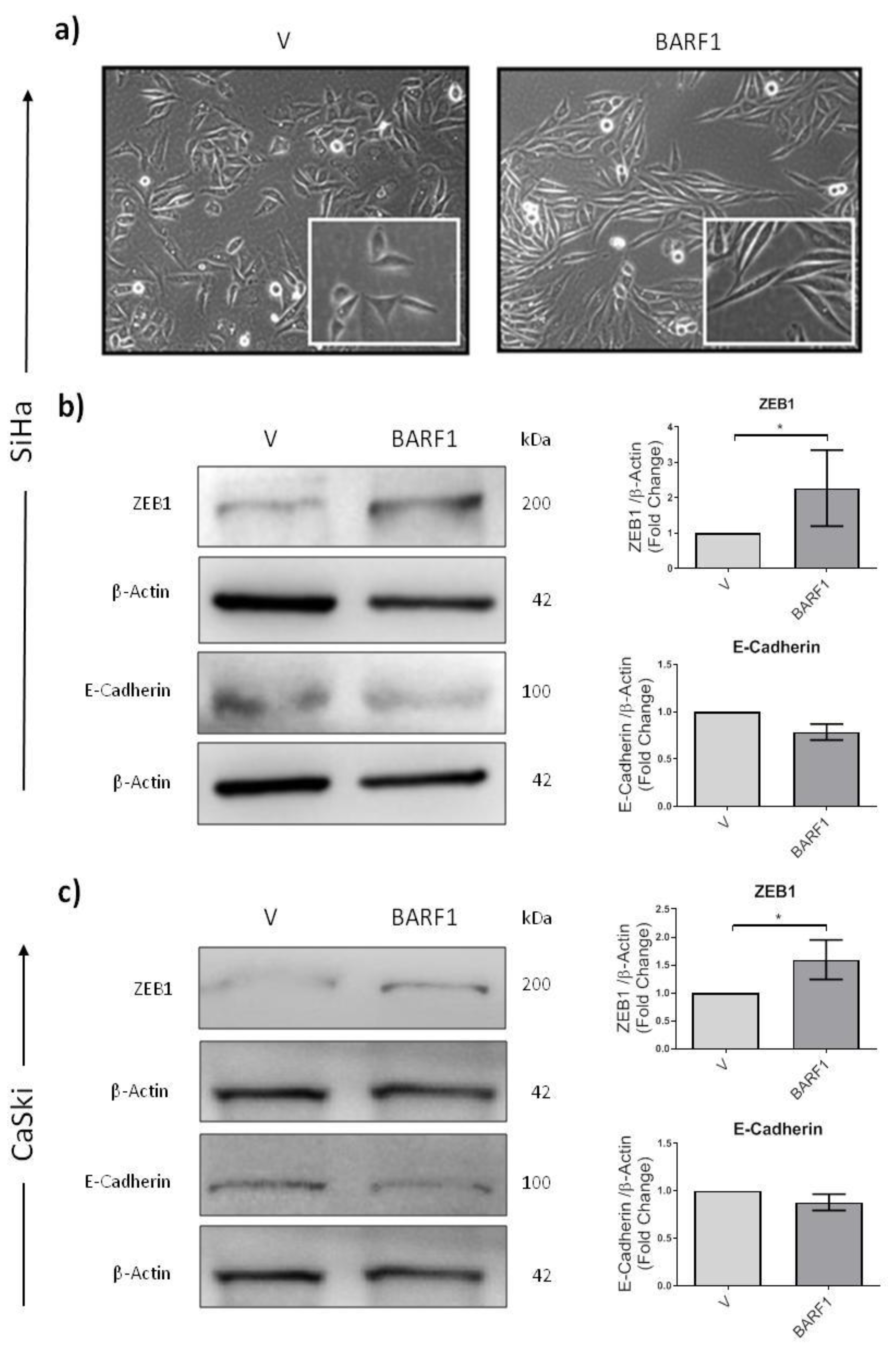
3.7. BARF1 Promotes Epithelial to Mesenchymal Transition in Cervical Cancer Cells
3.8. BARF1 Is Unable to Increase Anchorage-Independent Growth in Cervical Cancer Cells
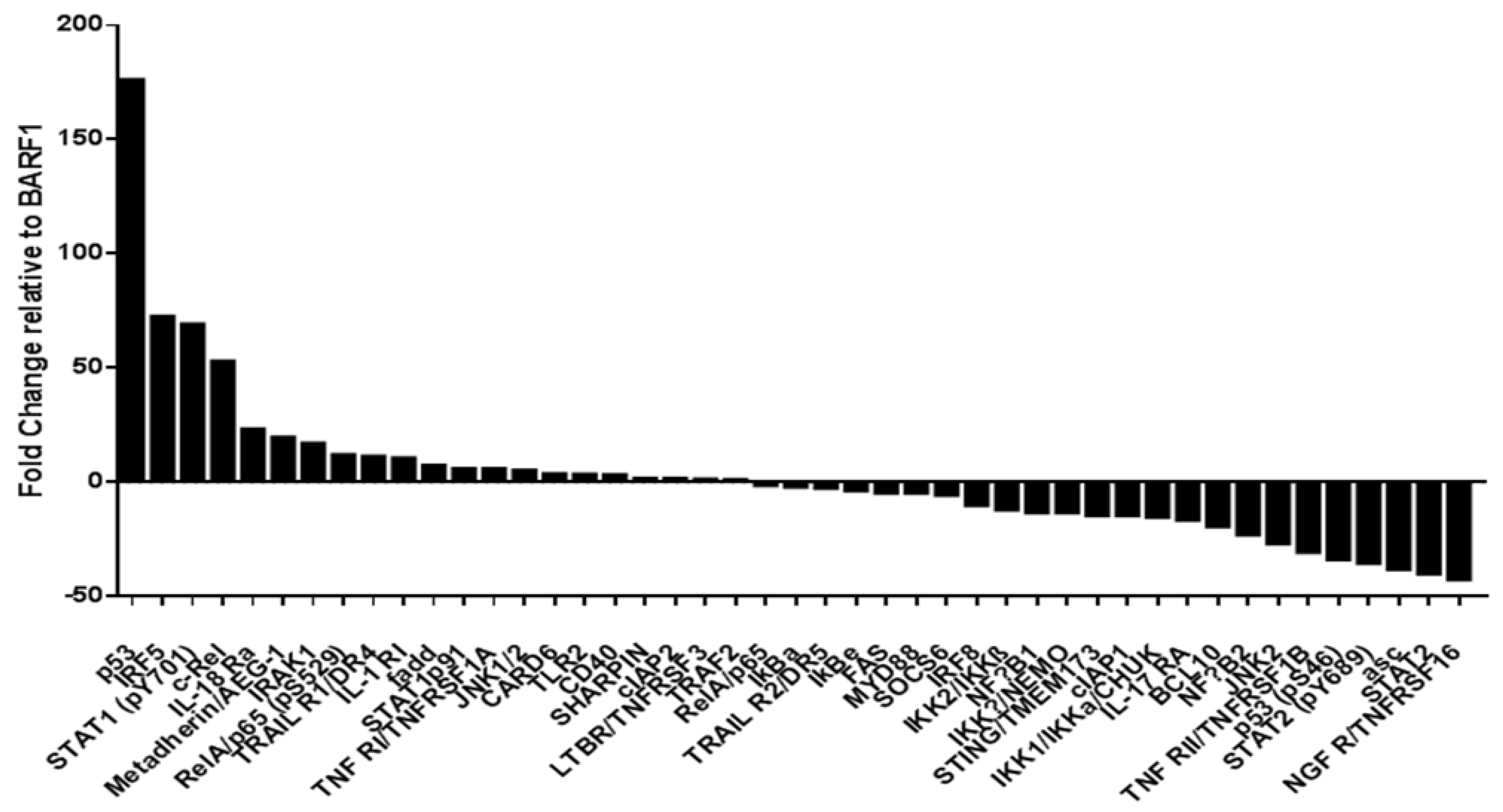
3.9. Phosphoproteomic NF-kB Assay in SiHa Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Arbyn, M.; Weiderpass, E.; Bruni, L.; de Sanjosé, S.; Saraiya, M.; Ferlay, J.; Bray, F. Estimates of incidence and mortality of cervical cancer in 2018: A worldwide analysis. Lancet Glob. Health 2020, 8, e191–e203. [Google Scholar] [CrossRef] [Green Version]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [Green Version]
- Nowakowski, A.; Cybulski, M.; Buda, I.; Janosz, I.; Olszak-Wąsik, K.; Bodzek, P.; Śliwczyński, A.; Teter, Z.; Olejek, A.; Baranowski, W. Cervical Cancer Histology, Staging and Survival before and after Implementation of Organised Cervical Screening Programme in Poland. PLoS ONE 2016, 11, e0155849. [Google Scholar] [CrossRef] [Green Version]
- Chiang, Y.C.; Chen, Y.Y.; Hsieh, S.F.; Chiang, C.J.; You, S.L.; Cheng, W.F.; Lai, M.S.; Chen, C.A.; Taiwan Cervical Cancer Prevention Surveillance Center. Screening frequency and histologic type influence the efficacy of cervical cancer screening: A nationwide cohort study. Taiwan J. Obs. Gynecol 2017, 56, 442–448. [Google Scholar] [CrossRef]
- Walboomers, J.M.; Jacobs, M.V.; Manos, M.M.; Bosch, F.X.; Kummer, J.A.; Shah, K.V.; Snijders, P.J.; Peto, J.; Meijer, C.J.; Muñoz, N. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 1999, 189, 12–19. [Google Scholar] [CrossRef]
- Iorga, L.; Dragos Marcu, R.; Cristina Diaconu, C.; Maria Alexandra Stanescu, A.; Pantea Stoian, A.; Liviu Dorel Mischianu, D.; Surcel, M.; Bungau, S.; Constantin, T.; Boda, D.; et al. Penile carcinoma and HPV infection (Review). Exp. Ther. Med. 2020, 20, 91–96. [Google Scholar] [CrossRef] [Green Version]
- Rakislova, N.; Saco, A.; Sierra, A.; Del Pino, M.; Ordi, J. Role of Human Papillomavirus in Vulvar Cancer. Adv. Anat. Pathol. 2017, 24, 201–214. [Google Scholar] [CrossRef]
- Rotondo, J.C.; Oton-Gonzalez, L.; Mazziotta, C.; Lanzillotti, C.; Iaquinta, M.R.; Tognon, M.; Martini, F. Simultaneous Detection and Viral DNA Load Quantification of Different Human Papillomavirus Types in Clinical Specimens by the High Analytical Droplet Digital PCR Method. Front. Microbiol. 2020, 11, 591452. [Google Scholar] [CrossRef]
- Bernard, E.; Pons-Salort, M.; Favre, M.; Heard, I.; Delarocque-Astagneau, E.; Guillemot, D.; Thiébaut, A.C. Comparing human papillomavirus prevalences in women with normal cytology or invasive cervical cancer to rank genotypes according to their oncogenic potential: A meta-analysis of observational studies. BMC Infect Dis. 2013, 13, 373. [Google Scholar] [CrossRef]
- Ciapponi, A.; Bardach, A.; Glujovsky, D.; Gibbons, L.; Picconi, M.A. Type-specific HPV prevalence in cervical cancer and high-grade lesions in Latin America and the Caribbean: Systematic review and meta-analysis. PLoS ONE 2011, 6, e25493. [Google Scholar] [CrossRef] [Green Version]
- Pinidis, P.; Tsikouras, P.; Iatrakis, G.; Zervoudis, S.; Koukouli, Z.; Bothou, A.; Galazios, G.; Vladareanu, S. Human Papilloma Virus’ Life Cycle and Carcinogenesis. Maedica 2016, 11, 48–54. [Google Scholar]
- Vats, A.; Trejo-Cerro, O.; Thomas, M.; Banks, L. Human papillomavirus E6 and E7: What remains? Tumour Virus Res. 2021, 11, 200213. [Google Scholar] [CrossRef]
- Scheffner, M.; Werness, B.A.; Huibregtse, J.M.; Levine, A.J.; Howley, P.M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell 1990, 63, 1129–1136. [Google Scholar] [CrossRef]
- Boyer, S.N.; Wazer, D.E.; Band, V. E7 protein of human papilloma virus-16 induces degradation of retinoblastoma protein through the ubiquitin-proteasome pathway. Cancer Res. 1996, 56, 4620–4624. [Google Scholar]
- Silveira, F.A.; Almeida, G.; Furtado, Y.L.; Cavalcanti, S.; Silva, K.S.; Maldonado, P.; Carvalho, M.G. The association of HPV genotype with the regression, persistence or progression of low-grade squamous intraepithelial lesions. Exp. Mol. Pathol. 2015, 99, 702–706. [Google Scholar] [CrossRef]
- Schlecht, N.F.; Platt, R.W.; Duarte-Franco, E.; Costa, M.C.; Sobrinho, J.P.; Prado, J.C.; Ferenczy, A.; Rohan, T.E.; Villa, L.L.; Franco, E.L. Human papillomavirus infection and time to progression and regression of cervical intraepithelial neoplasia. J. Natl. Cancer Inst. 2003, 95, 1336–1343. [Google Scholar] [CrossRef] [Green Version]
- Imajoh, M.; Hashida, Y.; Nemoto, Y.; Oguri, H.; Maeda, N.; Furihata, M.; Fukaya, T.; Daibata, M. Detection of Merkel cell polyomavirus in cervical squamous cell carcinomas and adenocarcinomas from Japanese patients. Virol. J. 2012, 9, 154. [Google Scholar] [CrossRef] [Green Version]
- Al-Thawadi, H.; Ghabreau, L.; Aboulkassim, T.; Yasmeen, A.; Vranic, S.; Batist, G.; Al Moustafa, A.E. Co-Incidence of Epstein-Barr Virus and High-Risk Human Papillomaviruses in Cervical Cancer of Syrian Women. Front Oncol. 2018, 8, 250. [Google Scholar] [CrossRef] [Green Version]
- Farrell, P.J. Epstein-Barr Virus and Cancer. Annu. Rev. Pathol. 2019, 14, 29–53. [Google Scholar] [CrossRef]
- Suntornlohanakul, R.; Wanlapakorn, N.; Vongpunsawad, S.; Thongmee, T.; Chansaenroj, J.; Poovorawan, Y. Seroprevalence of Anti-EBV IgG among Various Age Groups from Khon Kaen Province, Thailand. Asian Pac. J. Cancer Prev. 2015, 16, 7583–7587. [Google Scholar] [CrossRef]
- Smatti, M.K.; Yassine, H.M.; AbuOdeh, R.; AlMarawani, A.; Taleb, S.A.; Althani, A.A.; Nasrallah, G.K. Prevalence and molecular profiling of Epstein Barr virus (EBV) among healthy blood donors from different nationalities in Qatar. PLoS ONE 2017, 12, e0189033. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Wang, J.; Yu, D.; Liu, Y.; Xue, K.; Zhao, X. Role of Epstein-Barr Virus in the Development of Nasopharyngeal Carcinoma. Open Med. 2017, 12, 171–176. [Google Scholar] [CrossRef]
- Mozaffari, H.R.; Ramezani, M.; Janbakhsh, A.; Sadeghi, M. Malignant Salivary Gland Tumors and Epstein-Barr Virus (EBV) Infection: A Systematic Review and Meta-Analysis. Asian Pac. J. Cancer Prev. 2017, 18, 1201–1206. [Google Scholar] [CrossRef]
- Naseem, M.; Barzi, A.; Brezden-Masley, C.; Puccini, A.; Berger, M.D.; Tokunaga, R.; Battaglin, F.; Soni, S.; McSkane, M.; Zhang, W.; et al. Outlooks on Epstein-Barr virus associated gastric cancer. Cancer Treat Rev. 2018, 66, 15–22. [Google Scholar] [CrossRef]
- Shannon-Lowe, C.; Rickinson, A.B.; Bell, A.I. Epstein-Barr virus-associated lymphomas. Philos. Trans. R. Soc. London. Ser. B Biol. Sci. 2017, 372. [Google Scholar] [CrossRef]
- de Lima, M.A.P.; Neto, P.J.N.; Lima, L.P.M.; Goncalves Junior, J.; Teixeira Junior, A.G.; Teodoro, I.P.P.; Facundo, H.T.; da Silva, C.G.L.; Lima, M.V.A. Association between Epstein-Barr virus (EBV) and cervical carcinoma: A meta-analysis. Gynecol. Oncol. 2018, 148, 317–328. [Google Scholar] [CrossRef]
- McCormick, T.M.; Canedo, N.H.; Furtado, Y.L.; Silveira, F.A.; de Lima, R.J.; Rosman, A.D.; Almeida Filho, G.L.; Carvalho, M.D.A.G. Association between human papillomavirus and Epstein—Barr virus DNA and gene promoter methylation of RB1 and CDH1 in the cervical lesions: A transversal study. Diagn. Pathol. 2015, 10, 59. [Google Scholar] [CrossRef] [Green Version]
- Munz, C. The Role of Lytic Infection for Lymphomagenesis of Human gamma-Herpesviruses. Front. Cell. Infect. Microbiol. 2021, 11, 605258. [Google Scholar] [CrossRef]
- Morales-Sánchez, A.; Fuentes-Panana, E.M. The Immunomodulatory Capacity of an Epstein-Barr Virus Abortive Lytic Cycle: Potential Contribution to Viral Tumorigenesis. Cancers 2018, 10, 98. [Google Scholar] [CrossRef] [Green Version]
- Hayes, D.P.; Brink, A.A.; Vervoort, M.B.; Middeldorp, J.M.; Meijer, C.J.; van den Brule, A.J. Expression of Epstein-Barr virus (EBV) transcripts encoding homologues to important human proteins in diverse EBV associated diseases. Mol. Pathol. 1999, 52, 97–103. [Google Scholar] [CrossRef] [Green Version]
- Jiang, R.; Cabras, G.; Sheng, W.; Zeng, Y.; Ooka, T. Synergism of BARF1 with Ras induces malignant transformation in primary primate epithelial cells and human nasopharyngeal epithelial cells. Neoplasia 2009, 11, 964–973. [Google Scholar] [CrossRef] [Green Version]
- Khenchouche, A.; Sadouki, N.; Boudriche, A.; Houali, K.; Graba, A.; Ooka, T.; Bouguermouh, A. Human papillomavirus and Epstein-Barr virus co-infection in cervical carcinoma in Algerian women. Virol. J. 2013, 10, 340. [Google Scholar] [CrossRef] [Green Version]
- Muñoz, J.P.; Blanco, R.; Osorio, J.C.; Oliva, C.; Diaz, M.J.; Carrillo-Beltrán, D.; Aguayo, R.; Castillo, A.; Tapia, J.C.; Calaf, G.M.; et al. Merkel cell polyomavirus detected in head and neck carcinomas from Chile. Infect Agent Cancer 2020, 15, 4. [Google Scholar] [CrossRef]
- Carrillo-Beltran, D.; Munoz, J.P.; Guerrero-Vasquez, N.; Blanco, R.; Leon, O.; de Souza Lino, V.; Tapia, J.C.; Maldonado, E.; Dubois-Camacho, K.; Hermoso, M.A.; et al. Human Papillomavirus 16 E7 Promotes EGFR/PI3K/AKT1/NRF2 Signaling Pathway Contributing to PIR/NF-kappaB Activation in Oral Cancer Cells. Cancers 2020, 12, 1904. [Google Scholar] [CrossRef]
- Munoz, J.P.; Carrillo-Beltran, D.; Aedo-Aguilera, V.; Calaf, G.M.; Leon, O.; Maldonado, E.; Tapia, J.C.; Boccardo, E.; Ozbun, M.A.; Aguayo, F. Tobacco Exposure Enhances Human Papillomavirus 16 Oncogene Expression via EGFR/PI3K/Akt/c-Jun Signaling Pathway in Cervical Cancer Cells. Front. Microbiol. 2018, 9, 3022. [Google Scholar] [CrossRef]
- Tsang, C.M.; Yip, Y.L.; Lo, K.W.; Deng, W.; To, K.F.; Hau, P.M.; Lau, V.M.; Takada, K.; Lui, V.W.; Lung, M.L.; et al. Cyclin D1 overexpression supports stable EBV infection in nasopharyngeal epithelial cells. Proc. Natl. Acad. Sci. USA 2012, 109, E3473–E3482. [Google Scholar] [CrossRef] [Green Version]
- Yip, Y.L.; Tsang, C.M.; Deng, W.; Cheung, P.Y.; Jin, Y.; Cheung, A.L.; Lung, M.L.; Tsao, S.W. Expression of Epstein-Barr virus-encoded LMP1 and hTERT extends the life span and immortalizes primary cultures of nasopharyngeal epithelial cells. J. Med. Virol. 2010, 82, 1711–1723. [Google Scholar] [CrossRef]
- Abudoukadeer, A.; Niyazi, M.; Aikula, A.; Kamilijian, M.; Sulaiman, X.; Mutailipu, A.; Abudula, A. Association of EBV and HPV co-infection with the development of cervical cancer in ethnic Uyghur women. Eur. J. Gynaecol. Oncol. 2015, 36, 546–550. [Google Scholar]
- Zehbe, I.; Rylander, E.; Edlund, K.; Wadell, G.; Wilander, E. Detection of human papillomavirus in cervical intra-epithelial neoplasia, using in situ hybridization and various polymerase chain reaction techniques. Virchows Arch. 1996, 428, 151–157. [Google Scholar] [CrossRef]
- Ogembo, R.K.; Gona, P.N.; Seymour, A.J.; Park, H.S.; Bain, P.A.; Maranda, L.; Ogembo, J.G. Prevalence of human papillomavirus genotypes among African women with normal cervical cytology and neoplasia: A systematic review and meta-analysis. PLoS ONE 2015, 10, e0122488. [Google Scholar] [CrossRef]
- Park, E.; Kim, J.Y.; Choi, S.; Kim, D.S.; Oh, Y.L. Carcinogenic risk of human papillomavirus (HPV) genotypes and potential effects of HPV vaccines in Korea. Sci. Rep. 2019, 9, 12556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sasagawa, T.; Shimakage, M.; Nakamura, M.; Sakaike, J.; Ishikawa, H.; Inoue, M. Epstein-Barr virus (EBV) genes expression in cervical intraepithelial neoplasia and invasive cervical cancer: A comparative study with human papillomavirus (HPV) infection. Hum. Pathol. 2000, 31, 318–326. [Google Scholar] [CrossRef]
- Landers, R.J.; O’Leary, J.J.; Crowley, M.; Healy, I.; Annis, P.; Burke, L.; O’Brien, D.; Hogan, J.; Kealy, W.F.; Lewis, F.A.; et al. Epstein-Barr virus in normal, pre-malignant, and malignant lesions of the uterine cervix. J. Clin. Pathol. 1993, 46, 931–935. [Google Scholar] [CrossRef] [Green Version]
- Baena-Del Valle, J.A.; Zheng, Q.; Hicks, J.L.; Fedor, H.; Trock, B.J.; Morrissey, C.; Corey, E.; Cornish, T.C.; Sfanos, K.S.; De Marzo, A.M. Rapid Loss of RNA Detection by In Situ Hybridization in Stored Tissue Blocks and Preservation by Cold Storage of Unstained Slides. Am. J. Clin. Pathol. 2017, 148, 398–415. [Google Scholar] [CrossRef]
- Nuovo, A.J.; Garofalo, M.; Mikhail, A.; Nicol, A.F.; Vianna-Andrade, C.; Nuovo, G.J. The effect of aging of formalin-fixed paraffin-embedded tissues on the in situ hybridization and immunohistochemistry signals in cervical lesions. Diagn. Mol. Pathol. 2013, 22, 164–173. [Google Scholar] [CrossRef] [PubMed]
- Joharinia, N.; Faghihinejad, S.; Seyedi, K.; Farhadi, A.; Hosseini, S.Y.; Safaei, A.; Bahrampour, H.; Sarvari, J. Co-existing of HSV1/2 or EBV Infection with the Presence of High-Risk HPV DNA in Cervical Lesions in the Southwest of Iran. Asian Pac. J. Cancer Prev. 2020, 21, 1459–1464. [Google Scholar] [CrossRef]
- Szostek, S.; Zawilinska, B.; Kopec, J.; Kosz-Vnenchak, M. Herpesviruses as possible cofactors in HPV-16-related oncogenesis. Acta Biochim. Pol. 2009, 56, 337–342. [Google Scholar] [CrossRef] [Green Version]
- Kahla, S.; Oueslati, S.; Achour, M.; Kochbati, L.; Chanoufi, M.B.; Maalej, M.; Oueslati, R. Correlation between ebv co-infection and HPV16 genome integrity in Tunisian cervical cancer patients. Braz. J. Microbiol. 2012, 43, 744–753. [Google Scholar] [CrossRef] [Green Version]
- Jiang, R.; Ekshyyan, O.; Moore-Medlin, T.; Rong, X.; Nathan, S.; Gu, X.; Abreo, F.; Rosenthal, E.L.; Shi, M.; Guidry, J.T.; et al. Association between human papilloma virus/Epstein-Barr virus coinfection and oral carcinogenesis. Am. Acad. Oral Pathol. 2015, 44, 28–36. [Google Scholar] [CrossRef]
- Dona, M.G.; Spriano, G.; Pichi, B.; Rollo, F.; Laquintana, V.; Covello, R.; Pellini, R.; Giuliani, M.; Pescarmona, E.; Benevolo, M. Human papillomavirus infection and p16 overexpression in oropharyngeal squamous cell carcinoma: A case series from 2010 to 2014. Future Microbiol. 2015, 10, 1283–1291. [Google Scholar] [CrossRef]
- Tung, Y.C.; Lin, K.H.; Chu, P.Y.; Hsu, C.C.; Kuo, W.R. Detection of human papilloma virus and Epstein-Barr virus DNA in nasopharyngeal carcinoma by polymerase chain reaction. Kaohsiung J. Med. Sci. 1999, 15, 256–262. [Google Scholar]
- Al-Thawadi, H.; Gupta, I.; Jabeen, A.; Skenderi, F.; Aboulkassim, T.; Yasmeen, A.; Malki, M.I.; Batist, G.; Vranic, S.; Al Moustafa, A.E. Co-presence of human papillomaviruses and Epstein-Barr virus is linked with advanced tumor stage: A tissue microarray study in head and neck cancer patients. Cancer Cell Int. 2020, 20, 361. [Google Scholar] [CrossRef]
- Blanco, R.; Aguayo, F. Role of BamHI-A Rightward Frame 1 in Epstein-Barr Virus-Associated Epithelial Malignancies. Biology 2020, 9, 461. [Google Scholar] [CrossRef]
- Sakka, E.; Zur Hausen, A.; Houali, K.; Liu, H.; Fiorini, S.; Ooka, T. Cellular localization of BARF1 oncoprotein and its cell stimulating activity in human epithelial cell. Virus Res. 2013, 174, 8–17. [Google Scholar] [CrossRef]
- Chang, M.S.; Kim, D.H.; Roh, J.K.; Middeldorp, J.M.; Kim, Y.S.; Kim, S.; Han, S.; Kim, C.W.; Lee, B.L.; Kim, W.H.; et al. Epstein-Barr virus-encoded BARF1 promotes proliferation of gastric carcinoma cells through regulation of NF-κB. J. Virol. 2013, 87, 10515–10523. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.H.; Chang, M.S.; Yoon, C.J.; Middeldorp, J.M.; Martinez, O.M.; Byeon, S.J.; Rha, S.Y.; Kim, S.H.; Kim, Y.S.; Woo, J.H. Epstein-Barr virus BARF1-induced NFκB/miR-146a/SMAD4 alterations in stomach cancer cells. Oncotarget 2016, 7, 82213–82227. [Google Scholar] [CrossRef]
- Hunter, J.E.; Leslie, J.; Perkins, N.D. c-Rel and its many roles in cancer: An old story with new twists. Br. J. Cancer 2016, 114, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Hoebe, E.K.; Le Large, T.Y.; Greijer, A.E.; Middeldorp, J.M. BamHI-A rightward frame 1, an Epstein-Barr virus-encoded oncogene and immune modulator. Rev. Med. Virol. 2013, 23, 367–383. [Google Scholar] [CrossRef] [Green Version]
- Laurson, J.; Khan, S.; Chung, R.; Cross, K.; Raj, K. Epigenetic repression of E-cadherin by human papillomavirus 16 E7 protein. Carcinogenesis 2010, 31, 918–926. [Google Scholar] [CrossRef] [Green Version]
- Seto, E.; Ooka, T.; Middeldorp, J.; Takada, K. Reconstitution of nasopharyngeal carcinoma-type EBV infection induces tumorigenicity. Cancer Res. 2008, 68, 1030–1036. [Google Scholar] [CrossRef] [Green Version]
- Wiech, T.; Nikolopoulos, E.; Lassman, S.; Heidt, T.; Schopflin, A.; Sarbia, M.; Werner, M.; Shimizu, Y.; Sakka, E.; Ooka, T.; et al. Cyclin D1 expression is induced by viral BARF1 and is overexpressed in EBV-associated gastric cancer. Virchows Arch. Int. J. Pathol. 2008, 452, 621–627. [Google Scholar] [CrossRef] [PubMed]
| Type of Lesion | Age at Diagnosis | |||
|---|---|---|---|---|
| Total | ≤38 | >38 | p-Value | |
| No. Cases | No. (%) | No. (%) | ||
| LSIL | 22 | 16 (72.7) | 6 (27.3) | 0.001 a |
| HSIL | 52 | 28 (53.8) | 24 (46.2) | |
| SCC | 19 | 3 (18.8) | 16 (84.2) | |
| Type of Lesion | Generic HPV | HPV16 | ||||||
|---|---|---|---|---|---|---|---|---|
| Total | HPV− | HPV+ | p-Value | Total | HPV16− | HPV16+ | p-Value | |
| No. Cases | No. (%) | No. (%) | No. Cases | No. (%) | No. (%) | |||
| LSIL | 22 | 16 (72.7) | 6 (27.3) | <0.001 a | 22 | 22 (100) | 0 (0) | <0.001 b |
| HSIL | 52 | 14 (26.9) | 38 (73.1) | 52 | 29 (55.8) | 23 (44.2) | ||
| SCC | 19 | 4 (21.1) | 15 (78.9) | 19 | 8 (42.1) | 11 (57.9) | ||
| Type of Lesion | EBV/PCR | EBV/ISH(Epithelial Cells) | |||||
|---|---|---|---|---|---|---|---|
| Total | EBV− | EBV+ | p-Value | EBV− | EBV+ | p-Value | |
| No. Cases | No. (%) | No. (%) | No. (%) | No. (%) | |||
| LSIL | 22 | 6 (27.3) | 16 (72.7) | 0.177 a | 16 (100) | 0 (0) | 0.539 b |
| HSIL | 52 | 25 (48.1) | 27 (51.9) | 26 (98.1) | 1 (1.9) | ||
| SCC | 19 | 6 (31.6) | 13 (68.4) | 12 (94.7) | 1 (5.3) | ||
| HPV16 Detection | EBV Detected by PCR | |||
|---|---|---|---|---|
| Total | EBV− | EBV+ | p-Value | |
| No. Cases | No. (%) | No. (%) | ||
| HPV16+ | 34 | 16 (47.1) | 18 (52.9) | 0.277 a |
| HPV16− | 59 | 21 (35.6) | 38 (64.4) | |
| Type of Lesion | BARF1 Positivity | BARF1 Expression Level | ||||||
|---|---|---|---|---|---|---|---|---|
| Total | BARF1− | BARF1+ | p-Value | Total | BARF1 Low | BARF1 High | p-Value | |
| No. Cases | No. (%) | No. (%) | No. Cases | No. (%) | No. (%) | |||
| LSIL | 16 | 4 (25.0) | 12 (75.00) | 0.655 a | 12 | 4 (33.3) | 8 (66.7) | 0.360 b |
| HSIL | 26 | 10 (38.5) | 16 (61.5) | 15 * | 9 (60.0) | 6 (40.0) | ||
| SCC | 13 | 4 (30.8) | 9 (69.2) | 9 | 5 (55.6) | 4 (44.4) | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blanco, R.; Carrillo-Beltrán, D.; Muñoz, J.P.; Osorio, J.C.; Tapia, J.C.; Burzio, V.A.; Gallegos, I.; Calaf, G.M.; Chabay, P.; Aguayo, F. Characterization of High-Risk HPV/EBV Co-Presence in Pre-Malignant Cervical Lesions and Squamous Cell Carcinomas. Microorganisms 2022, 10, 888. https://doi.org/10.3390/microorganisms10050888
Blanco R, Carrillo-Beltrán D, Muñoz JP, Osorio JC, Tapia JC, Burzio VA, Gallegos I, Calaf GM, Chabay P, Aguayo F. Characterization of High-Risk HPV/EBV Co-Presence in Pre-Malignant Cervical Lesions and Squamous Cell Carcinomas. Microorganisms. 2022; 10(5):888. https://doi.org/10.3390/microorganisms10050888
Chicago/Turabian StyleBlanco, Rancés, Diego Carrillo-Beltrán, Juan P. Muñoz, Julio C. Osorio, Julio C. Tapia, Verónica A. Burzio, Iván Gallegos, Gloria M. Calaf, Paola Chabay, and Francisco Aguayo. 2022. "Characterization of High-Risk HPV/EBV Co-Presence in Pre-Malignant Cervical Lesions and Squamous Cell Carcinomas" Microorganisms 10, no. 5: 888. https://doi.org/10.3390/microorganisms10050888
APA StyleBlanco, R., Carrillo-Beltrán, D., Muñoz, J. P., Osorio, J. C., Tapia, J. C., Burzio, V. A., Gallegos, I., Calaf, G. M., Chabay, P., & Aguayo, F. (2022). Characterization of High-Risk HPV/EBV Co-Presence in Pre-Malignant Cervical Lesions and Squamous Cell Carcinomas. Microorganisms, 10(5), 888. https://doi.org/10.3390/microorganisms10050888







