Abstract
Escherichia coli K1 is the most common Gram-negative bacterium that causes neonatal meningitis; thus, a better understanding of its pathogenic molecular mechanisms is critical. However, the mechanisms by which E. coli K1 senses the signals of the host and expresses toxins for survival are poorly understood. As an extracytoplasmic function sigma factor, RpoE controls a wide range of pathogenesis-associated pathways in response to environmental stress. We found that the ΔrpoE mutant strain reduced the binding and invasion rate in human brain microvascular endothelial cells (HBMECs) in vitro, level of bacteremia, and percentage of meningitis in vivo. To confirm the direct targets of RpoE in vivo, we performed qRT-PCR and ChIP-qPCR on known toxic genes. RpoE was found to regulate pathogenic target genes, namely, ompA, cnf1, fimB, ibeA, kpsM, and kpsF directly and fimA, aslA, and traJ indirectly. The expression of these genes was upregulated when E. coli K1 was cultured with antibacterial peptides, whereas remained unchanged in the presence of the ΔrpoE mutant strain. Moreover, RpoE reduced IL-6 and IL-8 levels in E. coli K1-infected HBMECs. Altogether, these findings demonstrate that RpoE mediates the host adaptation capacity of E. coli K1 via a regulatory mechanism on virulence factors.
1. Introduction
During the first few postnatal months of life, Escherichia coli K1 is one of the primary causative agents of neonatal bacterial meningitis, which is associated with high mortality and morbidity worldwide every year [1,2,3]. Clinical data show that E. coli has emerged as a common cause of early onset sepsis and meningitis among very low birth weight infants, which can cause serious complications, with a morbidity rate of 1.04% and mortality rate of 35.3% [4]. Empiric treatment often incorporates multiple antibiotics, such as ampicillin and cefotaxime, or similar third-generation cephalosporins, while anti-inflammatory agents may be added to prevent neurologic sequelae, such as hearing loss [5,6,7]. Despite timely treatment, many patients experience long-term neurological complications. To date, our incomplete understanding of the pathogenesis of this disease has hindered our ability to decrease its mortality and morbidity rates.
The pathology and clinical symptoms of E. coli K1 infection are sepsis and meningitis [8,9]. When E. coli K1 crosses the blood–brain barrier in vivo, it requires a threshold level of bacteremia to invade the brain microvascular endothelial cells (HBMECs) and arrive at the niche cerebrospinal fluid (CSF) [10]. Previous studies have shown that high-degree bacteremia (e.g., >105 CFU/mL of neonatal animal blood [10]) significantly promotes the development of meningitis compared to low-degree bacteremia (e.g., <104 CFU/mL of neonatal animal blood [10]) in laboratory animals by allowing E. coli K1 to escape host defenses and cause meningitis. E. coli K1 crosses the blood–brain barrier (BBB) through a transcytosis process without disrupting the tight junction in the BBB. Binding to and invasion of HBMECs by E. coli K1 are the prerequisites for successful penetration into the brain parenchyma [11]. During infection, many bacterial factor toxins, adhesins (S fimbriae, Type I fimbriae, and nonfimbrial adhesins) [12,13,14,15], protein secretion systems (T6SSs) [16], invasins (IbeA and CNF1) [17,18,19,20,21], and K1 capsules (KpsMT and NeuDB) [10,22,23,24] help E. coli K1 survive the host innate immunity, thereby facilitating the binding, invasion and crossing of brain microvascular endothelial cells.
RpoE, whose molecular weight is 24 kDa, is an alternative sigma factor that regulates the transcription of various genes in E. coli in response to cell envelope stress and plays essential roles in the pathogenesis of numerous bacterial pathogens, including E. coli [25,26], Salmonella [27,28,29], Vibrio cholerae [30], Vibrio alginolyticus [31,32], and Treponema pallidum [33]. In E. coli, rpoE is a component of a five-gene operon (rseD-rpoE-rseA-rseB-rseC), which also includes genes encoding the anti-sigma factor RseA, the negative regulator RseB, a positive modulator RseC, and the leader peptide RseD. Further, RpoE controls the expression of conserved genes that are functionally related to membrane integrity (e.g., ompA, ompC, and ompF) [34]; thus, it is essential for bacterial survival.
However, the specific role of RpoE in mediating E. coli K1 virulence has not been elucidated. In this study, we investigated the involvement of RpoE in the virulence of E. coli K1. The deletion of rpoE reduced the expression of many virulence factors and the virulence of E. coli K1 in vitro and in vivo. Moreover, we found that RpoE is induced by antibacterial peptides present in the blood and that it directly regulates many virulence genes. This is the first report to describe the regulation mechanism of RpoE in E. coli K1.
2. Materials and Methods
2.1. Ethics Statement
All animal experiments were conducted in accordance with the criteria specified in the Guide for the Nursing and Use of Laboratory Animals. Animal research procedures were approved by the Institutional Animal Care Committee at Nankai University and Tianjin Institute of Pharmaceutical Research New Drug Evaluation (IACUC 2016032102, 21 March 2016), Tianjin, China. Every effort was made to minimize animal suffering and reduce the number of animals used.
2.2. Bacterial Strains, Plasmids and Growth Conditions
The bacterial strains and plasmids used in the present study are listed in Supplementary Table S1. The oligonucleotide primers used in this study are listed in Supplementary Table S2. E. coli K1 RS218 was used as the WT strain. Mutant strains were generated by the λ Red recombinase system of pSim6 using primers carrying 50-bp homologous regions flanking the start and stop codons of the gene to be deleted, as previously described [35]. All strains were grown at 37 °C in brain-heart infusion (BHI) medium.
2.3. E. coli Binding and Invasion Assay in HBMECs
HBMECs were cultured in RPMI 1640 medium (Thermo Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS, Hyclone, Marlborough, MA, USA), 10% Nu-serum (BD Biosciences, Franklin Lakes, NJ, USA), 2 mM glutamine, 1% MEM nonessential amino acids (Wako, Osaka, Japan), 1 × MEM vitamin (Sigma, Irvine, UK), 100 U/mL penicillin, 100 μg/mL streptomycin, and 1 mM sodium pyruvate (Thermo Scientific). E. coli K1 strains were resuscitated from −80 °C, cultured overnight, and then subcultured in fresh BHI medium at a dilution of 1:100 until strains were grown to the exponential phase at an OD600 of 0.6. Bacteria were collected by centrifugation and resuspended in RPMI 1640 medium containing 10% FBS. HBMECs infected at a multiplicity of infection (MOI) of 100 were incubated for 1.5 h in a cell incubator. The HBMECs were then washed with phosphate-buffered saline (PBS) to remove unbound bacteria and incubated with new medium containing gentamicin (100 mg/mL) for 1 h to kill extracellular E. coli. HBMECs were washed, lysed with 0.5% triton X-100, and plated on Luria broth (LB) agar plates for the determination of CFUs. A binding assay was performed similarly to the invasion assay except that the gentamicin treatment step was omitted. All experiments were conducted in duplicate and performed at least in triplicates.
2.4. Animal Model of E. coli Bacteremia and Hematogenous Meningitis
E. coli bacteremia was induced in 14-day-old BalB/C mice (Beijing Vital River Laboratory Animal Technology Co., Beijing, China). All mice were injected with 1 × 107 CFU of E. coli in the exponential phase at an OD600 of 0.6 via the tail vein. At 4 h after bacterial inoculation, blood specimens were obtained for quantitative cultures and RNA extraction. For the determination of CFUs, bacteria in the samples from the blood were assessed by plating on LB agar plates. For RNA extraction, blood samples were processed using TRIzol reagent (Invitrogen, Waltham, MA, USA). For hematogenous meningitis, all 14-day-old BalB/C mice were injected with 1 × 106 CFU of E. coli in the exponential phase at an OD600 of 0.6 via the tail vein. At 4 h after bacterial inoculation, CSF specimens were obtained to enumerate CFUs.
2.5. qRT–PCR
For the antibacterial peptide assay, the bacteria were cultured in M9 medium with or without LL-37 (Sigma) and incubated at 37 °C for 1–2 h. Blood or bacteria were pelleted by centrifugation. RNA samples were isolated using TRIzol (Invitrogen), reverse transcribed using the PrimeScriptTM RT reagent Kit (TaKaRa, Kusatsu, Japan), and processed for qRT–PCR. Each qRT–PCR was reacted in Power SYBR Green PCR Master Mix (Applied Biosystems, Waltham, MA, USA). The fold-change in the target gene relative to the housekeeping gene dnaE was determined using the 2−ΔΔCt method. At least three biological replicates were used for each qRT–PCR analysis.
2.6. ChIP-qPCR
RS218 WT containing pBAD24-RpoE/FLAG was cultured in RPMI 1640 medium supplemented with 0.1% arabinose until the exponential phase and then treated with 1% formaldehyde for Cross-linking, then stopped by adding 125 mM glycine. The bacterial pellets were washed with PBS, resuspended in immunoprecipitation buffer (IP; 50 mM pH 7.5 HEPES–KOH, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, and protease inhibitor cocktail (MCE)), and then subjected to sonication. The program was set to 30 cycles of a 5 s sonication followed by 10 s pauses (30% amplitude, 7.5 min total). Insoluble cellular debris was removed by centrifugation, and the supernatant was used as the input sample in the IP experiments. Both mock and IP samples were incubated with mouse IgG1 isotype control antibodies and anti-FLAG antibodies (Sigma), respectively, and then incubated with protein A beads (Life Technologies, Carlsbad, CA, USA) in IP buffer. Washing, crosslink reversal, and purification of the ChIP DNA were performed.
ChIP-qPCR was performed as described previously [36]. To measure the enrichment of potential RpoE-binding targets in the immunoprecipitated DNA samples, percent input and fold enrichment were performed using SYBR green mix. The relative target levels were calculated using the ΔCt method. lacZ was used as a negative control, with specific details referring to the ChIP Analysis [36]. The results were reported as the average enrichment of three biological replicates.
2.7. ELISA Analyses
HBMECs (1 × 106) were grown in 6-well plates and incubated with 50 g/mL Hcp family proteins per well for 6 h at 37 °C in a 5% CO2 incubator. The supernatants were collected and centrifuged to remove cell debris. The cytokine levels in the supernatant were determined using the Human IL-6 and IL-8 DuoSet® ELISA detection kits (R&D Systems, Minneapolis, MN, USA).
2.8. Fluorescent Actin Staining
Fluorescent actin staining (FAS) was performed as described previously [37]. Briefly, overnight bacteria were subcultured in BHI and incubated until an OD600 of 0.6 was obtained. Then, the bacteria were diluted on coverslips to infect HBMECs at MOI = 100 in the exponential phase at an OD600 of 0.6. After incubation, the coverslips were washed and fixed with formaldehyde; the cells were permeabilized with 0.1% Triton-X and stained with AF647-labeled phalloidin to visualize the actin filaments. E. coli was stained with FITC labeled anti-E. coli K and O antigen antibody (Abcam, Cambridge, UK). HBMEC nuclei were stained with 6-diamidino-2-phenylindol (DAPI). Binding and invasion assays on each cell line were performed, with three slides per experiment.
2.9. Brain Slice Immunofluorescence
Fourteen-day-old BALB/c mice were injected with 1 × 107 CFU of WT or ΔrpoE via the tail vein. Brain specimens were prepared on a Leica Cm1950 cryostat platform (Leica Microsystems, Buffalo Grove, IL, USA). The slices were fixed, and blocked. E. coli was stained with FITC-conjugated Anti-E. coli O and K antigen antibody (Abcam). DAPI was stained for nucleus visualization. Slices were then observed using a Zeiss LSM800 confocal microscope (Zeiss, Oberkochen, Germany). Each experiment was repeated at least three times.
2.10. Capsule Staining
Capsular staining [38] was performed using a capsular staining kit (Solarbio, Beijing, China). Staining was performed according to the manufacturer’s instructions. The capsule and colony morphology were observed via optical microscopy (Leica DM2500).
2.11. Growth Assay
To determine the growth curve of each strain, overnight cultures were diluted to a 1:100 ratio in a flask containing 200 mL of BHI broth without antibiotics and incubated at 37 °C with shaking at 180 rpm. A 200 μL aliquot was added to a 96-well flat bottom microplate and incubated at 37 °C with shaking at high speed for 24 h. The absorbance at 600 nm was recorded. Experiments were independently performed three times.
2.12. Statistical Analysis
Statistical analyses were conducted using the GraphPad Prism software (v8.3.0). GraphPad Software, San Diego, CA, USA; https://www.graphpad.com/scientific-software/prism/ (accessed on 20 April 2022). The mean ± standard deviation values of three independent experiments are shown in the figures. Differences between two mean values were evaluated using a two-tailed Student’s t-test. Statistical significance was assessed using the two-sided Fisher exact test in the mouse experiments. Statistical significance was set at p < 0.05.
3. Results
3.1. rpoE Promotes E. coli K1 Binding and Invasion of HBMECs
To determine the role of rpoE in the invasion of HBMEC by E. coli K1, an isogenic in-frame deletion mutant of rpoE was constructed. The regulatory gene rpoE is required for the binding and invasion of HBMECs by E. coli K1 strain RS218, which causes neonatal meningitis. To examine the virulence phenotype of the rpoE deletion mutant, we performed a comparative study of the binding and invasion capability of RS218 WT (parent strain) and ΔrpoE. The binding and invasion rates of rpoE deletion in HBMECs reduced by 20.2% and 99.28%, respectively, compared with those in the WT strain (Figure 1A). The binding and invasion rates of the complemented strain (crpoE) were comparable to that of the WT strain (Figure 1A). Immunofluorescence microscopy confirmed the effect of rpoE deletion on E. coli K1 binding and HBMEC invasion. Consistent with the findings of the in vitro assay, FAS shows less binding by the ΔrpoE mutant and invasion of HBMECs, compared to RS218 WT and the rpoE-overexpression strain (rpoEOE) induces higher binding and invasion rates than that of the WT strain during HBMECs infection (Figure 1C,D). The growth curve of the ΔrpoE mutant is similar to that of the RS218 WT strain (Figure 1B), indicating that the decrease in binding and invasion is not due to the observed growth rates.
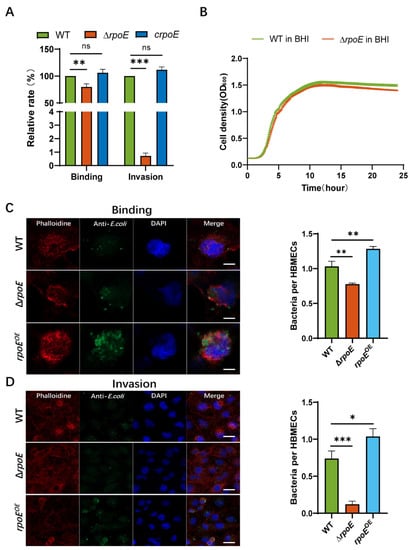
Figure 1.
Deletion of rpoE reduces Escherichia coli K1 binding to and invasion of human brain microvascular endothelial cells (HBMECs). (A) The binding and invasion abilities of ΔrpoE and crpoE strains were calculated relative to those of the wild-type (WT) strain. (B) Growth of RS218 WT and ΔrpoE strains in brain-heart infusion (BHI) medium. (C) Number of bacterial cells per each HBMEC determined in random fields of fluorescent actin staining (FAS) in a binding assay. Average of at least 50 cells is shown (left). Scale bar, 5 μm. (D) Number of bacterial cells per HBMEC determined in random fields of FAS in an invasion assay. Average of at least 50 cells is shown (left). Actin cytoskeleton (red) and nuclei (blue) of the HBMECs, and bacterium (green) are shown. Scale bar, 20 μm. * p < 0.05, ** p < 0.01, *** p < 0.001 by Student’s t-test. ns, not significant.
3.2. RpoE Is Involved in the Survival of E. coli K1 in Blood and Meningitis In Vivo
To investigate the role of rpoE in E. coli K1 infection, we examined the ability of the RS218 WT and ΔrpoE strains to survive in the blood of BALB/c mice. Each animal received 1 × 107 CFU of RS218 WT and ΔrpoE strains via tail vein injection to induce meningitis. Then, blood specimens were collected and cultured for divergent levels of bacteremia (WT, 8.14 ± 0.10 mean log CFU/mL of blood; ΔrpoE, 5.39 ± 0.40 mean log CFU/mL of blood, p < 0.001, Figure 2A), suggesting that rpoE expression plays a role in promoting E. coli K1 proliferation in mouse blood.
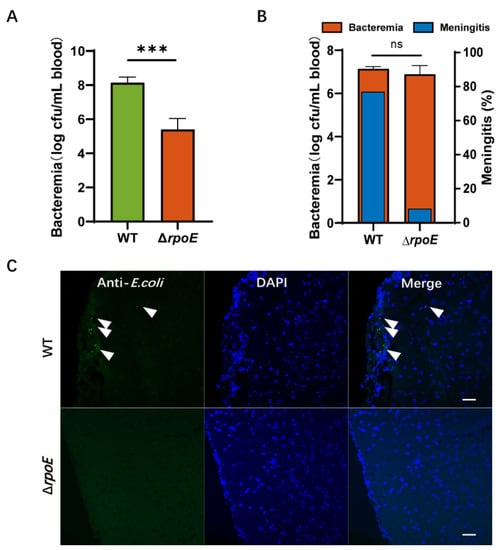
Figure 2.
Deletion of rpoE reduces RS218 survival in the blood and penetration of the blood–brain barrier (BBB). (A) Magnitude of bacteremia in BALB/c mice. Bacterial counts in the blood (log CFU/mL) were determined. (B) Blood samples were collected for bacteremia measurement, and cerebrospinal fluid (CSF) was collected and cultured to assess the passage of bacteria through the BBB. (C) The brains were collected after the mice were intravenously inoculated with bacteria and were sectioned for immunofluorescent staining. E. coli K1 (green) and nuclei (blue) are shown. Arrows indicate bacteria. Scale bar, 20 μm. The data represent the mean ± standard deviation from three independent experiments performed in triplicate. *** p < 0.001 by Student’s t-test, ns, not significant.
CSF was collected and cultured to illustrate the onset of bacterial infection in the central nervous system (CNS). The rate of meningitis occurrence, defined as a positive CSF culture with meningeal inflammation, is reduced to 8.33% by the ΔrpoE strain (n = 12), which is significantly (p = 0.001 by two-sided Fisher exact test) lower than the 76.92% by the parent strain RS218 WT (n = 13, Figure 2B). In the mice with E. coli meningitis, we found the presence of WT strains in the brain’s parenchyma (5.0 ± 1.4 bacteria/slice, Figure 2C), but ΔrpoE was not found. These data suggest that the rpoE gene contributes to penetration of the blood–brain barrier in vivo.
3.3. Identification of the Virulence Factors That Are Regulated by RpoE
To investigate the regulatory network of RpoE, qRT-PCR analysis was performed. Bacterial RNA was extracted from the blood of mice infected with RS218 WT and rpoE mutant strains via tail vein injection and used for qRT–PCR. E. coli K1 replication in blood and invasion of HBMECs depends on various outer membrane proteins, fimbriae, and effectors, such as K1 capsule (kpsM, kpsF), cnf1, fimA, fimB, ibeA, ompA, aslA, and traJ, the levels of which decreased by 0.064-, 0.101-, 0.371-, 0.442-, 0.261-, 0.154-, 0.156-, 0.346-, and 0.302-fold (Figure 3A), respectively, in rpoE mutant. We speculate RpoE can directly upregulate the yield of the K1 capsule by activating the expression kpsM and kpsF, which are the first gene of region 2–3 and region 1 transcriptional units, respectively [39]. Capsule staining shows K1 capsule presence in the WT strain but not in the ΔneuDB and ΔrpoE mutant strains (Figure 3B).

Figure 3.
RpoE is a positive virulence regulator of E. coli K1. (A) Fold changes in genes transcription in the rpoE mutant strain relative to their expression in the WT strain are shown. (B) Capsule morphology Scale bar, 3 μm. Capsule staining of WT, ΔneuDB, and ΔrpoE strains * p < 0.05; ** p < 0.01; *** p < 0.001.
ChIP-qPCR was performed to identify which toxic genes could be directly regulated by RpoE. RpoE binds to a highly conserved ‘AAC’ motif and a clustering of ‘CGT’ trinucleotides in the 35 region and 10 region, respectively [40], and activates its own promoter in response to cytoplasmic stress [34]. Therefore, we used rpoE as a positive control and a (GC)-rich gene lacZ as a negative control for the ChIP assay. ChIP-qPCR analysis revealed that PrpoE, PkpsM, PkpsF, Pcnf1, PfimB, PibeA, and PompA, were enriched 8.25-, 9.77-, 14.19-, 8.05-, 6.8-, 8.66-, and 13.55-fold, respectively, in IP-ChIP samples compared to those of mock ChIP samples (Figure 4A,B), confirming the binding of virulence genes in vivo. In contrast, the fold enrichment of lacZ, PfimA, PaslA, and PtraJ does not differ between the IP-ChIP and mock-ChIP samples (Figure 4A,B). These results demonstrate that RpoE influences the expression of many virulence genes through direct or indirect regulation in vivo.
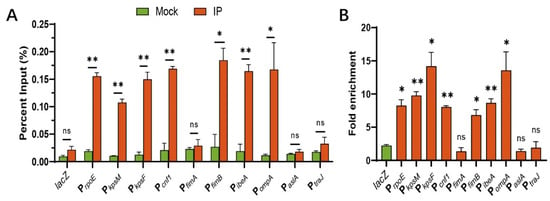
Figure 4.
Virulence factors are directly regulated by RpoE in vivo. (A) Percent input of the promoter region of virulence factors in RpoE-ChIP samples. (B) Fold enrichment of the promoter region of virulence factors in RpoE-ChIP samples. * p < 0.05; ** p < 0.01; ns, not significant by Student’s t-test.
3.4. The Expression of rpoE Is Upregulated after the Infection of E. coli K1 In Vitro and In Vivo
The transcriptional levels of rpoE vary during the diffident infectious phase. The expression of the rpoE gene increases the binding and invasion 2.141- and 2.418-fold, respectively (Figure 5), compared with that cultured in BHI. These data suggest that rpoE needs more expression to enhance the virulence in E. coli K1 binding and invasion of HBMECs. As the WT strain was able to form high-level bacteremia, we tested the expression of rpoE in blood compared to that in BHI (Figure 5). The expression level of rpoE was 3.385-fold higher in blood than that in the BHI medium. The above results illustrate that some signals in serum of PMI 1640 medium or blood activate the expression of rpoE.
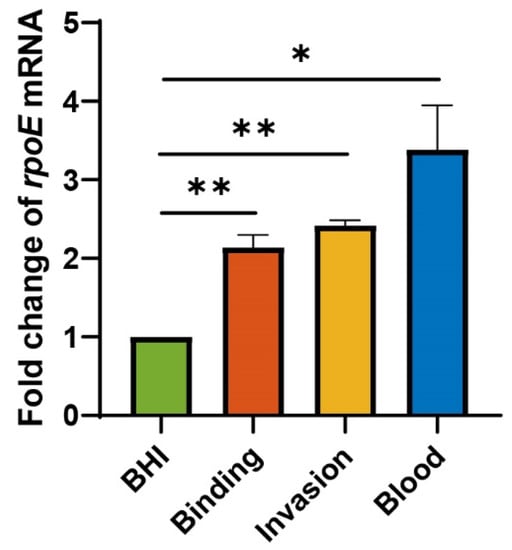
Figure 5.
rpoE transcriptional levels are upregulated upon E. coli K1 infection in vitro and in vivo. * p < 0.05; ** p < 0.01 by Student’s t-test. Data are presented as the mean ± standard deviation; n = 3.
3.5. RpoE Is Essential for the Cationic Antimicrobial Peptide-Dependent Expression of the Virulence Factors
The serum concentrations of LL-37 in patients infected with Enterobacteriaceae were up to 86.12 ng/mL [41], but the concentration of LL-37 in a healthy infant was 0.3 ng/mL [42]. RpoE plays a necessary role in responding to cell envelope stresses such as a cationic antimicrobial peptide (AMP) [43]. The serum mean concentrations of LL-37 in mice with meningitis was 35 ng/mL (data is not published). To further investigate the molecular mechanism underlying RpoE-mediation, the regulation of virulence gene expression was measured using qRT–PCR with the cationic antimicrobial peptide LL-37 (35 ng/mL). Total RNA obtained from the WT or ΔrpoE cells was cultured in the M9 medium with or without LL-37. The expression of the rpoE, kpsM, kpsF, cnf1, fimA, fimB, ibeA, ompA, aslA, and traJ genes showed a 2.338-, 1.727-, 1.902-, 3.532-, 3.953-, 3.131-, 5.237-, 3.149-, 3.261-, and 3.320-fold increase with LL-37, respectively, compared to that in cultures without LL-37 (Figure 6A). Gene induction by LL-37 was abolished in the rpoE mutant strain (Figure 6B). The above results show that toxin induction by LL-37 requires RpoE.

Figure 6.
RpoE enhances the expression of virulence factors induced by LL-37. (A) Fold change in the transcriptional levels of toxic genes in the WT strain cultured with LL-37. (B) Fold change in the transcriptional levels of toxic genes in the ΔrpoE mutant strain cultured with LL-37. * p < 0.05; ** p < 0.01; *** p < 0.001; ns, not significant by Student’s t-test.
3.6. RpoE Attenuates Inflammation of HBMECs Caused by E. coli K1
Since increased levels of cytokines occur in bacterial meningitis [16] and rpoE mutation attenuates the ability of E. coli K1 to cross the BBB, we investigated the role of RpoE in cytokine and chemokine secretion in HBMECs. Compared to those of the WT strain, significantly higher levels of IL-6 and IL-8 are released after the binding of the ΔrpoE strain to HBMECs (Figure 7A). After E. coli K1 invasion in HBMECs, the ΔrpoE strain caused significant increases in cytokine and chemokine release in HBMECs compared to those caused by the WT strain (Figure 7B). These results illustrate that RpoE attributes to attenuating inflammation of HBMECs caused by E. coli K1.
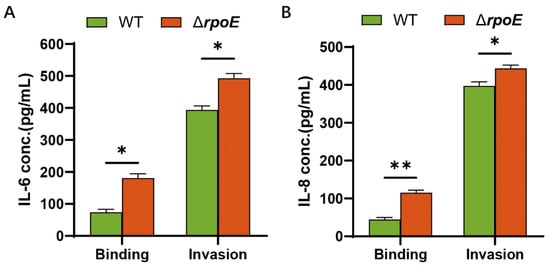
Figure 7.
Cytokine release by HBMECs incubated with E. coli K1. (A) The levels of IL-6 in the culture supernatants of HBMECs that infected WT and ΔrpoE. (B) The levels of IL-8 in the culture supernatants of HBMECs that infected WT and ΔrpoE. Data were obtained from three independent experiments and analyzed using Student’s t-test. * p < 0.05, ** p < 0.01.
4. Discussion
The results of the present study revealed that RpoE regulates additional biological pathways, including those related to the capsule assembly, fimbria expression, and cell necrosis factor biogenesis, which were compared to the main biological pathways, including those for lipopolysaccharide biogenesis and outer membrane porin assembly [34,44]. There were significant differences associated with targets of RpoE between the results of our study based on E. coli K1 and those previously validated in E. coli K12. In this study, we established that RpoE promotes high-level bacteremia and meningitis associated with E. coli K1. RpoE upregulated the expression of toxic genes such as cnf1, fimB, ibeA, ompA, kpsM, and kpsF directly. Except for ompA, the rest were verified to be new target genes of RpoE. Meanwhile, RpoE indirectly upregulated the expression of toxic genes such as fimA, aslA, and traJ.
In response to environmental stress in Salmonella-containing vacuoles, RpoE upregulates the expression of several genes, including htrA, which is required for survival in Salmonella typhimurium intramacrophage [28]. The rpoE-mutant of Salmonella strain had reduced survival and infirmity to oxidative stress [28]. In contrast to the parental Vibrio cholerae in an infant mouse model, the lethality of the rpoE mutant was attenuated significantly, with a decreasing colonization ability to the intestine [30]. The increased expression level of RpoE was observed in the WT E. coli LF82 strain at high osmolarity, and the overexpression of RpoE in the ΔompR isogenic mutant restored the phenotype of the WT strain [26]. The rpoE mutant Vibrio alginolyticus was incapacitated to adapt to environmental and host stresses, indicating that rpoE is critical for the activation of virulence genes in response to temperature [31]. To investigate whether RpoE is temperature-dependent with respect to the regulation of virulence in E. coli K1, qRT-PCR analysis was performed. However, the expression of kpsM and kpsF was not different between WT and rpoE mutant strains at various temperatures (Supplementary Figure S1), indicating that RpoE cannot regulate virulence in response to temperature in E. coli K1.
Vibrio cholerae El Tor was reported to resist the cationic antimicrobial peptide (AMP) of the intestinal tract, but the rpoE mutant of V. cholerae El Tor was more sensitive than the WT was to the cationic AMP (polymyxin B) [43,45,46]. Before E. coli K1 invades the BBB, it needs to replicate in the blood to induce high levels of bacteremia. Therefore, E. coli K1 can sense the cationic antimicrobial peptide of the blood via RpoE to upregulate the expression of many toxic genes to evade host immunity and cross the BBB finally.
The ΔrpoE strain stimulated HBMECs to release more IL-6 and IL-8 than the WT strain did, which are crucial inflammatory mediators that elicit leukocyte infiltration in bacterial meningitis [16]. Recruitment of neutrophils is not conducive to the survival of bacteria. Meanwhile, the released cytokines disrupt the tight junctions of the BBB, causing a marked inflammatory response in the CNS [11,47]. This phenomenon may explain why E. coli K1 crossing the BBB is a key point in meningitis and central nervous system damage. RpoE-regulated target genes, such as K1 capsule, contribute to the evasion of host immune recognition by E. coli K1 [48,49].
In conclusion, our data demonstrate that the extracytoplasmic function sigma factor RpoE contributes to the virulence of E. coli K1 by influencing the expression of known virulence factors (Figure 8). This regulatory mechanism is critical for the pathogenicity of E. coli K1 as the rpoE mutant results in significantly reduced virulence. The response to cationic AMP results in the activation of toxic genes via RpoE. Thus, our findings deepen our understanding of how E. coli K1 senses environmental signals to facilitate the overall adaptability of the pathogen.
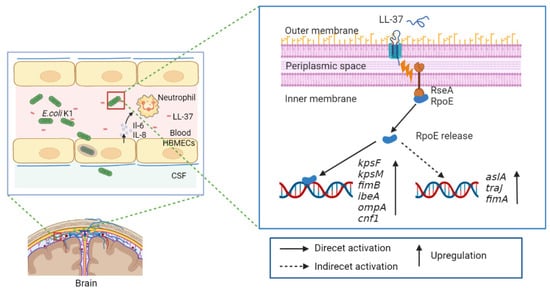
Figure 8.
Model of the regulatory pathway of RpoE-mediated cationic antimicrobial peptide-dependent signaling in E. coli K1.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/microorganisms10050879/s1, Figure S1. RpoE cannot respond to temperature to regulate the expression of kpsF and kpsM in E. coli K1; Table S1: Bacterial strains and plasmids used; Table S2: Primers used.
Author Contributions
Conceptualization, B.Y. and D.X.; methodology, Y.F. and J.B.; software, Y.F.; data curation, B.Y.; writing—original draft preparation, Y.F; writing—review and editing, B.Y.; visualization, Y.F.; supervision, B.Y.; project administration, Y.F.; funding acquisition, B.Y. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Natural Science Foundation of China (Grant No. 31800125), the Natural Science Foundation of Tianjin (Grant No. 20JCQNJC01970), and the Fundamental Research Funds for the Central Universities, Nankai University (Grant No. 980/63211149).
Institutional Review Board Statement
All mice experiments in this study were conducted according to protocols approved by the Institutional Animal Care Committee at Nankai University (Tianjin, China; protocol code 20190325.02, approval date 21 March 2016).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available in the Supplementary Material.
Acknowledgments
Human brain microvascular endothelial cells (HBMECs) were a generous gift from K. S. Kim (Johns Hopkins University, Baltimore, MD, USA).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Teng, C.-H.; Cai, M.; Shin, S.; Xie, Y.; Kim, K.-J.; Khan, N.A.; Di Cello, F.; Kim, K.S. Escherichia coli K1 RS218 interacts with human brain microvascular endothelial cells via type 1 fimbria bacteria in the fimbriated state. Infect. Immun. 2005, 73, 2923–2931. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffiths, M.J.; Guhadasan, R.; Carrol, E.D. Acute Bacterial Meningitis. In Hunter’s Tropical Medicine and Emerging Infectious Diseases; Elsevier: Amsterdam, The Netherlands, 2020; pp. 541–547. [Google Scholar]
- Pick, A.M.; Sweet, D.C.; Begley, K.J. A review of pediatric bacterial meningitis. US Pharm 2016, 41, 41–45. [Google Scholar]
- Mendoza-Palomar, N.; Balasch-Carulla, M.; González-Di Lauro, S.; Céspedes, M.C.; Andreu, A.; Frick, M.A.; Linde, M.; Soler-Palacin, P. Escherichia coli early-onset sepsis: Trends over two decades. Eur. J. Pediatr. 2017, 176, 1227–1234. [Google Scholar] [CrossRef] [PubMed]
- Klugman, K.P.; Dagan, R. Randomized comparison of meropenem with cefotaxime for treatment of bacterial meningitis. Meropenem Meningitis Study Group. Antimicrob. Agents Chemother. 1995, 39, 1140–1146. [Google Scholar] [CrossRef] [Green Version]
- Belohradsky, B.; Geiss, D.; Marget, W.; Bruch, K.; Kafetzis, D.; Peters, G. Intravenous cefotaxime in children with bacterial meningitis. Lancet 1980, 315, 61–63. [Google Scholar] [CrossRef]
- Shin, S.H.; Kim, K.S. Treatment of bacterial meningitis: An update. Expert Opin. Pharmacother. 2012, 13, 2189–2206. [Google Scholar] [CrossRef] [PubMed]
- Deshmukh, H.S.; Liu, Y.; Menkiti, O.R.; Mei, J.; Dai, N.; O’leary, C.E.; Oliver, P.M.; Kolls, J.K.; Weiser, J.N.; Worthen, G.S. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat. Med. 2014, 20, 524–530. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.S. Investigating bacterial penetration of the blood-brain barrier for the pathogenesis, prevention, and therapy of bacterial meningitis. ACS Infect. Dis. 2020, 6, 34–42. [Google Scholar] [CrossRef]
- Kim, K.S.; Itabashi, H.; Gemski, P.; Sadoff, J.; Warren, R.L.; Cross, A.S. The K1 capsule is the critical determinant in the development of Escherichia coli meningitis in the rat. J. Clin. Investig. 1992, 90, 897–905. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.S. Pathogenesis of bacterial meningitis: From bacteraemia to neuronal injury. Nat. Rev. Neurosci. 2003, 4, 376–385. [Google Scholar] [CrossRef]
- Prasadarao, N.V.; Wass, C.A.; Hacker, J.; Jann, K.; Kim, K.S. Adhesion of S-fimbriated Escherichia coli to brain glycolipids mediated by sfaA gene-encoded protein of S-fimbriae. J. Biol. Chem. 1993, 268, 10356–10363. [Google Scholar] [CrossRef]
- Prasadarao, N.V.; Wass, C.A.; Kim, K.S. Identification and characterization of S fimbria-binding sialoglycoproteins on brain microvascular endothelial cells. Infect. Immun. 1997, 65, 2852–2860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Wen, Z.G.; Kim, K.S. Role of S fimbriae in Escherichia coli K1 binding to brain microvascular endothelial cells in vitro and penetration into the central nervous system in vivo. Microb. Pathog. 2004, 37, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.A.; Kim, Y.; Shin, S.; Kim, K.S. FimH-mediated Escherichia coli K1 invasion of human brain microvascular endothelial cells. Cell Microbiol. 2007, 9, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Tao, J.; Yu, H.; Ni, J.; Zeng, L.; Teng, Q.; Kim, K.S.; Zhao, G.P.; Guo, X.; Yao, Y. Hcp family proteins secreted via the type VI secretion system coordinately regulate Escherichia coli K1 interaction with human brain microvascular endothelial cells. Infect. Immun. 2012, 80, 1243–1251. [Google Scholar] [CrossRef] [Green Version]
- Khan, N.A.; Wang, Y.; Kim, K.J.; Chung, J.W.; Wass, C.A.; Kim, K.S. Cytotoxic necrotizing factor-1 contributes to Escherichia coli K1 invasion of the central nervous system. J. Biol. Chem. 2002, 277, 15607–15612. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.H.; Kim, K.S. Cytotoxic necrotizing factor 1 contributes to Escherichia coli meningitis. Toxins 2013, 5, 2270–2280. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Pearce, D.; Kim, K.S. Prevention of Escherichia coli K1 penetration of the blood-brain barrier by counteracting the host cell receptor and signaling molecule involved in E. coli invasion of human brain microvascular endothelial cells. Infect. Immun. 2010, 78, 3554–3559. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.H.; Chen, Y.H.; Fu, Q.; Stins, M.; Wang, Y.; Wass, C.; Kim, K.S. Identification and characterization of an Escherichia coli invasion gene locus, ibeB, required for penetration of brain microvascular endothelial cells. Infect. Immun. 1999, 67, 2103–2109. [Google Scholar] [CrossRef] [Green Version]
- Prasadarao, N.V.; Wass, C.A.; Huang, S.H.; Kim, K.S. Identification and characterization of a novel Ibe10 binding protein that contributes to Escherichia coli invasion of brain microvascular endothelial cells. Infect. Immun. 1999, 67, 1131–1138. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.J.; Elliott, S.J.; Di Cello, F.; Stins, M.F.; Kim, K.S. The K1 capsule modulates trafficking of E. coli-containing vacuoles and enhances intracellular bacterial survival in human brain microvascular endothelial cells. Cell Microbiol. 2003, 5, 245–252. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, J.A.; Wass, C.; Stins, M.F.; Kim, K.S. The capsule supports survival but not traversal of Escherichia coli K1 across the blood-brain barrier. Infect. Immun. 1999, 67, 3566–3570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badger, J.L.; Wass, C.A.; Kim, K.S. Identification of Escherichia coli K1 genes contributing to human brain microvascular endothelial cell invasion by differential fluorescence induction. Mol. Microbiol. 2000, 36, 174–182. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulesus, R.R.; Diaz-Perez, K.; Slechta, E.S.; Eto, D.S.; Mulvey, M.A. Impact of the RNA chaperone Hfq on the fitness and virulence potential of uropathogenic Escherichia coli. Infect. Immun. 2008, 76, 3019–3026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rolhion, N.; Carvalho, F.A.; Darfeuille-Michaud, A. OmpC and the sigma(E) regulatory pathway are involved in adhesion and invasion of the Crohn’s disease-associated Escherichia coli strain LF82. Mol. Microbiol. 2007, 63, 1684–1700. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Overall, C.C.; Nakayasu, E.S.; Kidwai, A.S.; Jones, M.B.; Johnson, R.C.; Nguyen, N.T.; McDermott, J.E.; Ansong, C.; Heffron, F.; et al. Analysis of the Salmonella regulatory network suggests involvement of SsrB and H-NS in sigma(E)-regulated SPI-2 gene expression. Front. Microbiol. 2015, 6, 27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Overall, C.C.; Johnson, R.C.; Jones, M.B.; McDermott, J.E.; Heffron, F.; Adkins, J.N.; Cambronne, E.D. ChIP-Seq analysis of the sigmae regulon of Salmonella enterica serovar Typhimurium reveals new genes implicated in heat shock and oxidative stress response. PLoS ONE 2015, 10, e0138466. [Google Scholar]
- Zhang, H.; Jia, Y.; Xie, X.; Wang, M.; Zheng, Y.; Xu, S.; Zhang, W.; Wang, Q.; Huang, X.; Du, H. RpoE promotes invasion and intracellular survival by regulating SPI-1 and SPI-2 in Salmonella enterica serovar Typhi. Future Microbiol. 2016, 11, 1011–1024. [Google Scholar] [CrossRef]
- Kovacikova, G.; Skorupski, K. The alternative sigma factor sigma(E) plays an important role in intestinal survival and virulence in Vibrio cholerae. Infect. Immun. 2002, 70, 5355–5362. [Google Scholar] [CrossRef] [Green Version]
- Gu, D.; Guo, M.; Yang, M.; Zhang, Y.; Zhou, X.; Wang, Q. A sigmaE-mediated temperature gauge controls a switch from LuxR-mediated virulence gene expression to thermal stress adaptation in Vibrio alginolyticus. PLoS Pathog. 2016, 12, e1005645. [Google Scholar] [CrossRef] [Green Version]
- Gu, D.; Zhang, J.; Hao, Y.; Xu, R.; Zhang, Y.; Ma, Y.; Wang, Q. Alternative sigma factor RpoX is a part of the RpoE regulon and plays distinct roles in stress responses, motility, biofilm formation, and hemolytic activities in the marine pathogen Vibrio alginolyticus. Appl. Environ. Microbiol. 2019, 85, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giacani, L.; Denisenko, O.; Tompa, M.; Centurion-Lara, A. Identification of the Treponema pallidum subsp. pallidum TP0092 (RpoE) regulon and its implications for pathogen persistence in the host and syphilis pathogenesis. J. Bacteriol. 2013, 195, 896–907. [Google Scholar] [PubMed] [Green Version]
- Rhodius, V.A.; Suh, W.C.; Nonaka, G.; West, J.; Gross, C.A. Conserved and variable functions of the sigmaE stress response in related genomes. PLoS Biol. 2006, 4, e2. [Google Scholar]
- Juhas, M.; Ajioka, J.W. Lambda Red recombinase-mediated integration of the high molecular weight DNA into the Escherichia coli chromosome. Microb. Cell Factories 2016, 15, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhatia, S.; Matthews, J.; Wells, P.G. Characterization of epigenetic histone activation/repression marks in sequences of genes by chromatin immunoprecipitation-quantitative polymerase chain reaction (ChIP-qPCR). Methods Mol. Biol. 2019, 1965, 389–403. [Google Scholar] [PubMed]
- Liu, Y.; Liu, B.; Yang, P.; Wang, T.; Chang, Z.; Wang, J.; Wang, Q.; Li, W.; Wu, J.; Huang, D.; et al. LysR-type transcriptional regulator OvrB encoded in O island 9 drives enterohemorrhagic Escherichia coli O157:H7 virulence. Virulence 2019, 10, 783–792. [Google Scholar] [CrossRef] [Green Version]
- Gong, Q.; Wang, X.; Huang, H.; Sun, Y.; Qian, X.; Xue, F.; Ren, J.; Dai, J.; Tang, F. Novel host recognition mechanism of the K1 capsule-specific phage of Escherichia coli: Capsular polysaccharide as the first receptor and lipopolysaccharide as the secondary receptor. J. Virol. 2021, 95, e0092021. [Google Scholar] [CrossRef]
- Roberts, I.S. The biochemistry and genetics of capsular polysaccharide production in bacteria. Annu. Rev. Microbiol. 1996, 50, 285–315. [Google Scholar] [CrossRef]
- Staroń, A.; Sofia, H.J.; Dietrich, S.; Ulrich, L.E.; Liesegang, H.; Mascher, T. The third pillar of bacterial signal transduction: Classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol. Microbiol. 2009, 74, 557–581. [Google Scholar] [CrossRef]
- Majewski, K.; Kozłowska, E.; Żelechowska, P.; Brzezińska-Błaszczyk, E. Serum concentrations of antimicrobial peptide cathelicidin LL-37 in patients with bacterial lung infections. Cent. Eur. J. Immunol. 2018, 43, 453–457. [Google Scholar] [CrossRef]
- Thursfield, R.M.; Naderi, K.; Leaver, N.; Rosenthal, M.; Alton, E.; Bush, A.; Davies, J.C. Children with cystic fibrosis demonstrate no respiratory immunological, infective or physiological, consequences of vitamin D deficiency. J. Cyst. Fibros. 2018, 17, 657–665. [Google Scholar] [CrossRef] [PubMed]
- Mathur, J.; Davis, B.M.; Waldor, M.K. Antimicrobial peptides activate the Vibrio cholerae sigmaE regulon through an OmpU-dependent signalling pathway. Mol. Microbiol. 2010, 63, 848–858. [Google Scholar]
- Dartigalongue, C.; Missiakas, D.; Raina, S. Characterization of the Escherichia coli sigma E regulon. J. Biol. Chem. 2001, 276, 20866–20875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haines-Menges, B.; Whitaker, W.B.; Boyd, E.F. Alternative sigma factor RpoE is important for Vibrio parahaemolyticus cell envelope stress response and intestinal colonization. Infect. Immun. 2014, 82, 3667–3677. [Google Scholar] [CrossRef] [Green Version]
- Davis, B.M.; Waldor, M.K. High-throughput sequencing reveals suppressors of Vibrio cholerae rpoE mutations: One fewer porin is enough. Nucleic Acids Res. 2009, 37, 5757–5767. [Google Scholar] [CrossRef] [Green Version]
- Furth, A.; Roord, J.J.; Furth, R.V. Roles of proinflammatory and anti-inflammatory cytokines in pathophysiology of bacterial meningitis and effect of adjunctive therapy. Infect. Immun. 1996, 64, 4883–4890. [Google Scholar] [CrossRef] [Green Version]
- Cross, A.S.; Kim, K.S.; Wright, D.C.; Sadoff, J.C.; Gemski, P. Role of lipopolysaccharide and capsule in the serum resistance of bacteremic strains of Escherichia coli. J. Infect. Dis. 1986, 154, 497–503. [Google Scholar] [CrossRef]
- Jann, K.; Jann, B. Polysaccharide antigens of Escherichia coli. Rev. Infect. Dis. 1987, 9 (Suppl. S5), S517–S526. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).