Microbiota and Transcriptomic Effects of an Essential Oil Blend and Its Delivery Route Compared to an Antibiotic Growth Promoter in Broiler Chickens
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Egg Incubation and In Ovo Injection Procedure
2.3. Birds, Housing, and Diets
2.4. Sample Collection
2.5. DNA Extraction, Qualification, Library Preparation, and Sequencing
2.6. Short-Chain Fatty Acid Concentration and Total Bacteria Density
2.7. RNA Extraction, Qualification, Library Preparation, and Sequencing
2.8. Bioinformatics and Statistical Analysis
3. Results
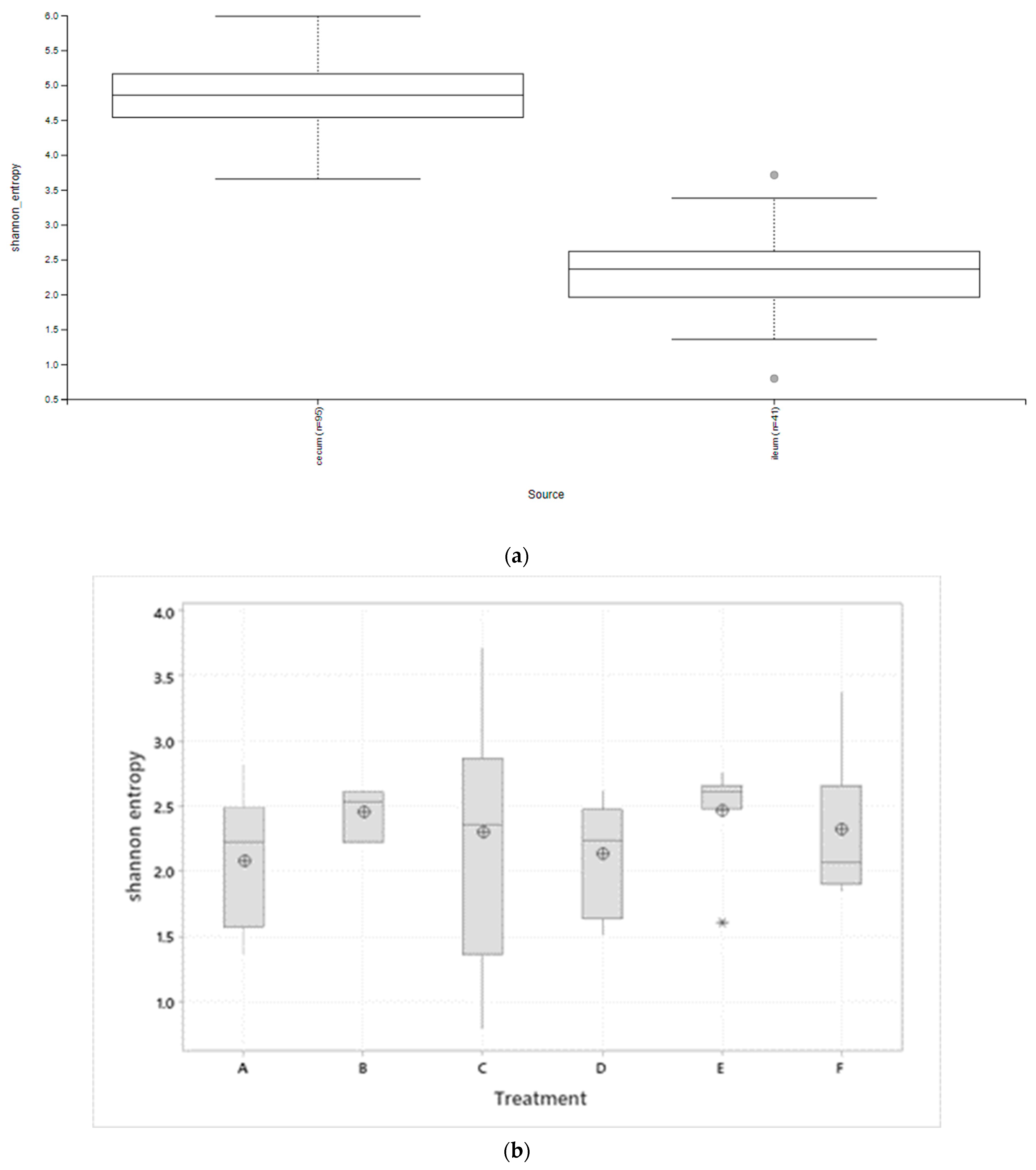
3.1. Microbiota Diversity
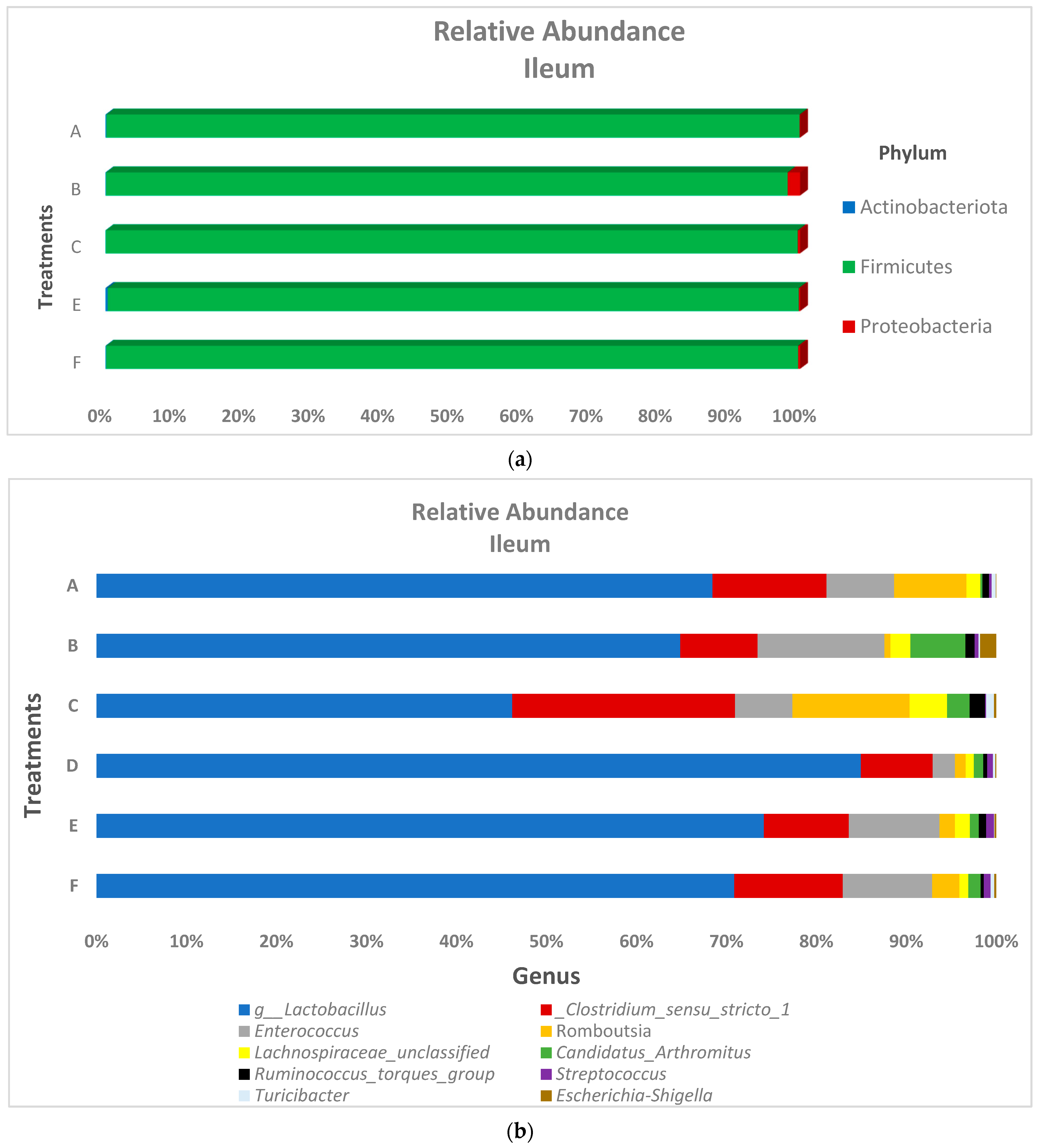
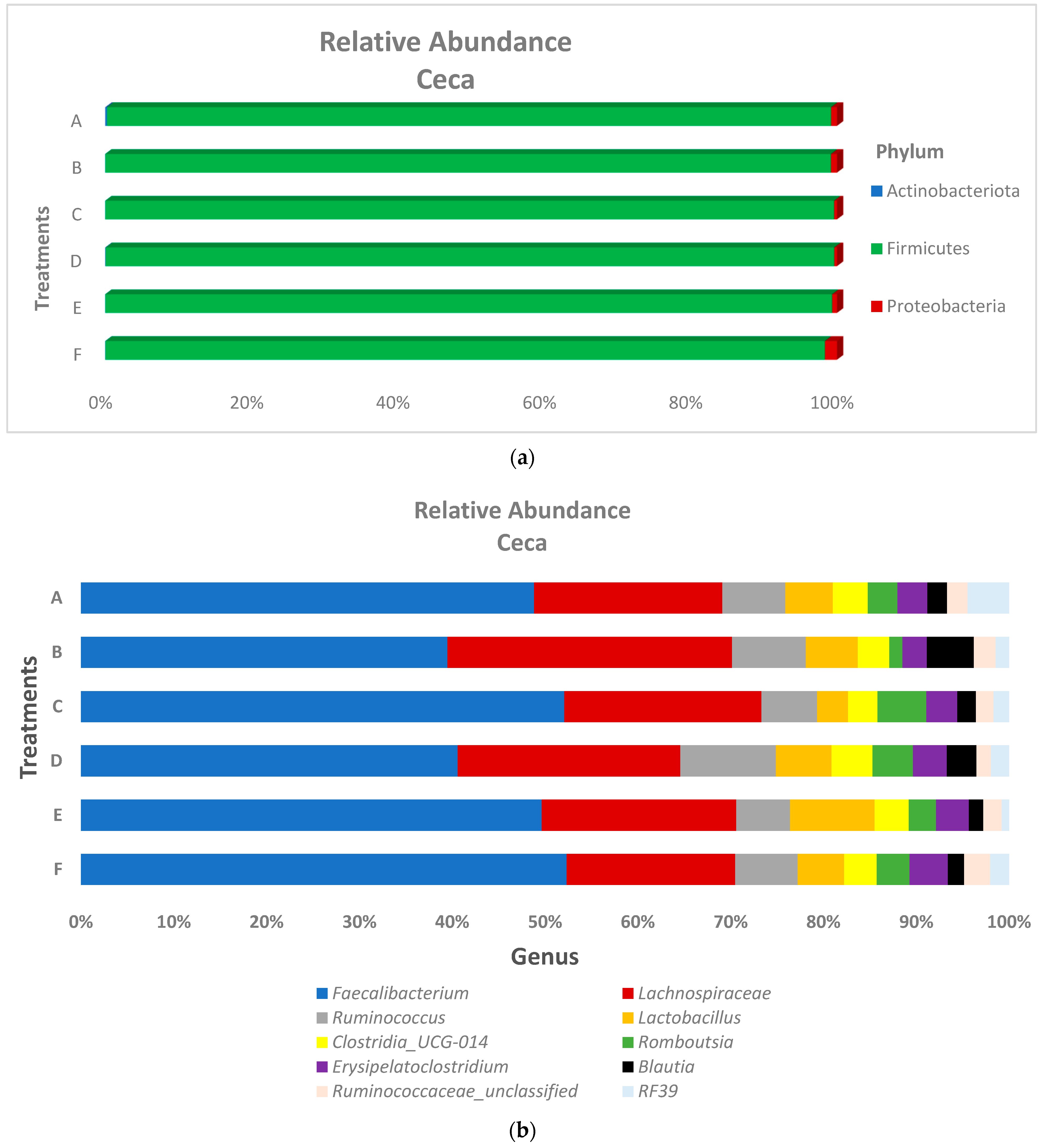
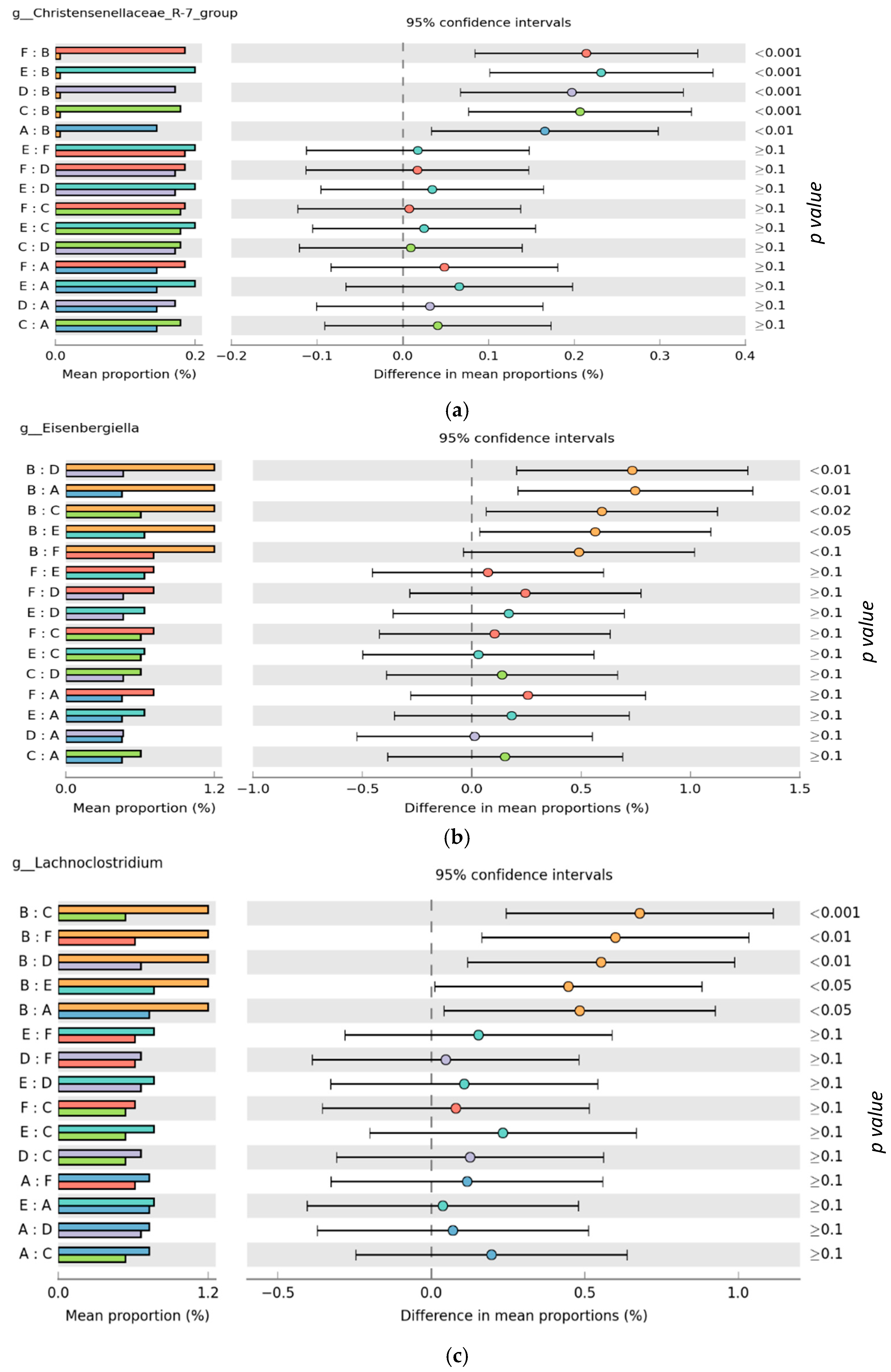
3.2. Microbiota Composition
3.3. Ceca SCFA Concentration
3.4. Transcriptome Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mahmood, T.; Guo, Y. Dietary fiber and chicken microbiome interaction: Where will it lead to? Anim. Nutr. 2020, 6, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Ma, F.; Xu, S.; Tang, Z.; Li, Z.; Zhang, L. Use of antimicrobials in food animals and impact of transmission of antimicrobial resistance on humans. Biosaf. Health 2021, 3, 32–38. [Google Scholar] [CrossRef]
- Thakur, A.; Kumar, A.; Sharma, M.; Kumar, R.; Vanita, B. Strategies to Minimize the Impact of Antibiotic Resistance in Livestock Production System. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 2293–2310. [Google Scholar] [CrossRef]
- Castanon, J.I.R. History of the Use of Antibiotic as Growth Promoters in European Poultry Feeds. Poult. Sci. 2007, 86, 2466–2471. [Google Scholar] [CrossRef] [PubMed]
- FDA. Guidance for Industry #213-New Animal Drugs and New Animal Drug Combination Products Administered in or on Medicated Feed or Drinking Water of Food-Producing Animals: Recommendations for Drug Sponsors for Voluntarily Aligning Product Use Conditions with GFI# 209; U.S. Department of Health and Human Services: Washington, DC, USA, 2013; p. 18.
- Chicken Farmers of Canada. Antibiotics. Available online: https://www.chickenfarmers.ca/antibiotics/. (accessed on 12 September 2021).
- Paiva, D.; McElroy, A. Necrotic enteritis: Applications for the poultry industry. J. Appl. Poult. Res. 2014, 23, 557–566. [Google Scholar] [CrossRef]
- Rinttilä, T.; Apajalahti, J. Intestinal microbiota and metabolites—Implications for broiler chicken health and performance. J. Appl. Poult. Res. 2013, 22, 647–658. [Google Scholar] [CrossRef]
- Yadav, S.; Jha, R. Strategies to modulate the intestinal microbiota and their effects on nutrient utilization, performance, and health of poultry. J. Anim. Sci. Biotechnol. 2019, 10, 2. [Google Scholar] [CrossRef]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef]
- Burkholder, K.M.; Thompson, K.L.; Einstein, M.E.; Applegate, T.; Patterson, J.A. Influence of Stressors on Normal Intestinal Microbiota, Intestinal Morphology, and Susceptibility to Salmonella Enteritidis Colonization in Broilers. Poult. Sci. 2008, 87, 1734–1741. [Google Scholar] [CrossRef]
- Shi, D.; Bai, L.; Qu, Q.; Zhou, S.; Yang, M.; Guo, S.; Li, Q.; Liu, C. Impact of gut microbiota structure in heat-stressed broilers. Poult. Sci. 2019, 98, 2405–2413. [Google Scholar] [CrossRef]
- Kers, J.G.; Velkers, F.C.; Fischer, E.A.J.; Hermes, G.D.A.; Stegeman, J.A.; Smidt, H. Host and Environmental Factors Affecting the Intestinal Microbiota in Chickens. Front. Microbiol. 2018, 9, 235. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Idris, U.; Harmon, B.; Hofacre, C.; Maurer, J.J.; Lee, M.D. Diversity and Succession of the Intestinal Bacterial Community of the Maturing Broiler Chicken. Appl. Environ. Microbiol. 2003, 69, 6816–6824. [Google Scholar] [CrossRef] [PubMed]
- Oviedo-Rondón, E.O.; Hume, M.E.; Barbosa, N.A.; Sakomura, N.K.; Weber, G.; Wilson, J.W. Ileal and Caecal Microbial Populations in Broilers Given Specific Essential Oil Blends and Probiotics in two Consecutive Grow-Outs. Avian Biol. Res. 2010, 3, 157–169. [Google Scholar] [CrossRef]
- Thompson, K.; Burkholder, K.; Patterson, J.; Applegate, T.J. Microbial Ecology Shifts in the Ileum of Broilers During Feed Withdrawal and Dietary Manipulations. Poult. Sci. 2008, 87, 1624–1632. [Google Scholar] [CrossRef]
- Shang, Y.; Kumar, S.; Oakley, B.; Kim, W.K. Chicken Gut Microbiota: Importance and Detection Technology. Front. Veter. Sci. 2018, 5, 254. [Google Scholar] [CrossRef] [PubMed]
- Bassolé, I.H.N.; Juliani, H.R. Essential Oils in Combination and Their Antimicrobial Properties. Molecules 2012, 17, 3989–4006. [Google Scholar] [CrossRef]
- Gopi, M.; Karthik, K.; Manjunathachar, H.V.; Tamilmahan, P.; Kesavan, M.; Dashprakash, M.; Balaraju, B.L.; Purushothaman, M.R. Essential oils as a feed additive in poultry nutrition. Adv. Anim. Vet. Sci. 2014, 2, 1–7. [Google Scholar] [CrossRef]
- Stevanović, Z.D.; Bošnjak-Neumüller, J.; Pajić-Lijaković, I.; Raj, J.; Vasiljević, M. Essential Oils as Feed Additives—Future Perspectives. Molecules 2018, 23, 1717. [Google Scholar] [CrossRef]
- Swamy, M.K.; Akhtar, M.S.; Sinniah, U.R. Antimicrobial properties of plant essential oils against human pathogens and their mode of action: An updated review. Evid.-Based Complement. Altern. Med. 2016, 2016, 3012462. [Google Scholar] [CrossRef]
- Cho, J.; Kim, H.; Kim, I. Effects of phytogenic feed additive on growth performance, digestibility, blood metabolites, intestinal microbiota, meat color and relative organ weight after oral challenge with Clostridium perfringens in broilers. Livest. Sci. 2014, 160, 82–88. [Google Scholar] [CrossRef]
- Hashemipour, H.; Khaksar, V.; Rubio, L.; Veldkamp, T.; van Krimpen, M. Effect of feed supplementation with a thymol plus carvacrol mixture, in combination or not with an NSP-degrading enzyme, on productive and physiological parameters of broilers fed on wheat-based diets. Anim. Feed Sci. Technol. 2016, 211, 117–131. [Google Scholar] [CrossRef]
- Pathak, M.; Mandal, G.P.; Patra, A.K.; Samanta, I.; Pradhan, S.; Haldar, S. Effects of dietary supplementation of cinnamaldehyde and formic acid on growth performance, intestinal microbiota and immune response in broiler chickens. Anim. Prod. Sci. 2017, 57, 821–827. [Google Scholar] [CrossRef]
- Mitsch, P.; Zitterl-Eglseer, K.; Köhler, B.; Gabler, C.; Losa, R.; Zimpernik, I. The effect of two different blends of essential oil components on the proliferation of Clostridium perfringens in the intestines of broiler chickens. Poult. Sci. 2004, 83, 669–675. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.-C.; Steiner, T.; Aufy, A.; Lien, T.-F. Effects of supplemental essential oil on growth performance, lipid metabolites and immunity, intestinal characteristics, microbiota and carcass traits in broilers. Livest. Sci. 2012, 144, 253–262. [Google Scholar] [CrossRef]
- Paraskeuas, V.; Mountzouris, K.C. Broiler gut microbiota and expressions of gut barrier genes affected by cereal type and phytogenic inclusion. Anim. Nutr. 2019, 5, 22–31. [Google Scholar] [CrossRef]
- Bilia, A.R.; Guccione, C.; Isacchi, B.; Righeschi, C.; Firenzuoli, F.; Bergonzi, M.C. Essential Oils Loaded in Nanosystems: A Developing Strategy for a Successful Therapeutic Approach. Evid.-Based Complement. Altern. Med. 2014, 2014, 651593. [Google Scholar] [CrossRef]
- Heydarian, M.; Ebrahimnezhad, Y.; Meimandipour, A.; Hosseini, S.A.; Banabazi, M.H. Effects of Dietary Inclusion of the Encapsulated Thyme and Oregano Essential Oils Mixture and Probiotic on Growth Performance, Immune Response and Intestinal Morphology of Broiler Chickens. Poult. Sci. J. 2020, 8, 17–25. [Google Scholar]
- Maenner, K.; Vahjen, W.; Simon, O. Studies on the effects of essential-oil-based feed additives on performance, ileal nutrient digestibility, and selected bacterial groups in the gastrointestinal tract of piglets1. J. Anim. Sci. 2011, 89, 2106–2112. [Google Scholar] [CrossRef]
- Malayoğlu, H.B.; Baysal, Ş.; Misirlioğlu, Z.; Polat, M.; Yilmaz, H.; Turan, N. Effects of oregano essential oil with or without feed enzymes on growth performance, digestive enzyme, nutrient digestibility, lipid metabolism and immune response of broilers fed on wheat–soybean meal diets. Br. Poult. Sci. 2010, 51, 67–80. [Google Scholar] [CrossRef]
- Mountzouris, K.; Paraskevas, V.; Tsirtsikos, P.; Palamidi, I.; Steiner, T.; Schatzmayr, G.; Fegeros, K. Assessment of a phytogenic feed additive effect on broiler growth performance, nutrient digestibility and caecal microflora composition. Anim. Feed Sci. Technol. 2011, 168, 223–231. [Google Scholar] [CrossRef]
- Oladokun, S.; Adewole, D.I. In ovo delivery of bioactive substances: An alternative to the use of antibiotic growth promoters in poultry production—A review. J. Appl. Poult. Res. 2020, 29, 744–763. [Google Scholar] [CrossRef]
- Slawinska, A.; Plowiec, A.; Siwek, M.; Jaroszewski, M.; Bednarczyk, M. Long-Term Transcriptomic Effects of Prebiotics and Synbiotics Delivered In Ovo in Broiler Chickens. PLoS ONE 2016, 11, e0168899. [Google Scholar] [CrossRef] [PubMed]
- Roto, S.M.; Kwon, Y.M.; Ricke, S.C. Applications of In Ovo Technique for the Optimal Development of the Gastrointestinal Tract and the Potential Influence on the Establishment of Its Microbiome in Poultry. Front. Veter. Sci. 2016, 3, 63. [Google Scholar] [CrossRef]
- Glendinning, L.; Watson, K.A.; Watson, M. Development of the duodenal, ileal, jejunal and caecal microbiota in chickens. Anim. Microbiome 2019, 1, 17. [Google Scholar] [CrossRef]
- Gong, J.; Si, W.; Forster, R.J.; Huang, R.; Yu, H.; Yin, Y.; Yang, C.; Han, Y. 16S rRNA gene-based analysis of mucosa-associated bacterial community and phylogeny in the chicken gastrointestinal tracts: From crops to ceca. FEMS Microbiol. Ecol. 2007, 59, 147–157. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Chambers, J.R.; Wheatcroft, R.; Chen, S.; Forster, R.J.; Yu, H.; Sabour, P.M. Molecular analysis of bacterial populations in the ileum of broiler chickens and comparison with bacteria in the cecum. FEMS Microbiol. Ecol. 2002, 41, 171–179. [Google Scholar] [CrossRef]
- Sabino, M.; Capomaccio, S.; Cappelli, K.; Verini-Supplizi, A.; Bomba, L.; Marsan, P.A.; Cobellis, G.; Olivieri, O.; Pieramati, C.; Trabalza-Marinucci, M. Oregano dietary supplementation modifies the liver transcriptome profile in broilers: RNASeq analysis. Res. Veter. Sci. 2018, 117, 85–91. [Google Scholar] [CrossRef]
- Li, W.; He, Z.-Q.; Zhang, X.-Y.; Chen, Y.-J.; Zuo, J.-J.; Cao, Y. Proteome and Transcriptome Analysis of the Antioxidant Mechanism in Chicken Regulated by Eucalyptus Leaf Polyphenols Extract. Oxidative Med. Cell. Longev. 2020, 2020, 1384907. [Google Scholar] [CrossRef]
- Bastos, M.S.; Del Vesco, A.P.; Santana, T.P.; Santos, T.S.; De Oliveira Junior, G.M.; Fernandes, R.P.M.; Barbosa, L.T.; Gasparino, E. The role of cinnamon as a modulator of the expression of genes related to antioxidant activity and lipid metabolism of laying quails. PLoS ONE 2017, 12, e0189619. [Google Scholar] [CrossRef]
- Li, H.; Wang, T.; Xu, C.; Wang, D.; Ren, J.; Li, Y.; Tian, Y.; Wang, Y.; Jiao, Y.; Kang, X.; et al. Transcriptome profile of liver at different physiological stages reveals potential mode for lipid metabolism in laying hens. BMC Genom. 2015, 16, 763. [Google Scholar] [CrossRef]
- Akbarian, A.; Golian, A.; Gilani, A.; Kermanshahi, H.; Zhaleh, S.; Akhavan, A.; De Smet, S.; Michiels, J. Effect of feeding citrus peel extracts on growth performance, serum components, and intestinal morphology of broilers exposed to high ambient temperature during the finisher phase. Livest. Sci. 2013, 157, 490–497. [Google Scholar] [CrossRef]
- Lillehoj, H.S.; Kim, D.K.; Bravo, D.M.; Lee, S.H. Effects of dietary plant-derived phytonutrients on the genome-wide profiles and coccidiosis resistance in the broiler chickens. BMC Proc. 2011, 5, S34. [Google Scholar] [CrossRef] [PubMed]
- Rowsell, H.C. Canadian Council on Animal Care: Its Role. Available online: https://www.ccac.ca/en/CCAC_Programs/Guidelines_Policies/GUIDES/ENGLISH/toc_v1.htm (accessed on 17 October 2021). [CrossRef][Green Version]
- Oladokun, S.; Koehler, A.; MacIsaac, J.; Ibeagha-Awemu, E.M.; Adewole, D.I. Bacillus subtilis delivery route: Effect on growth performance, intestinal morphology, cecal short-chain fatty acid concentration, and cecal microbiota in broiler chickens. Poult. Sci. 2021, 100, 100809. [Google Scholar] [CrossRef] [PubMed]
- Oladokun, S.; MacIsaac, J.; Rathgeber, B.; Adewole, D. Essential Oil Delivery Route: Effect on Broiler Chicken’s Growth Performance, Blood Biochemistry, Intestinal Morphology, Immune, and Antioxidant Status. Animals 2021, 11, 3386. [Google Scholar] [CrossRef] [PubMed]
- National Research Council. Nutrient Requirements of Poultry; The National Academies Press: Washington, DC, USA, 1994. [Google Scholar]
- Comeau, A.M.; Douglas, G.M.; Langille, M.G.I. Microbiome Helper: A Custom and Streamlined Workflow for Microbiome Research. mSystems 2017, 2, e00127-16. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef]
- Amir, A.; McDonald, D.; Navas-Molina, J.A.; Kopylova, E.; Morton, J.T.; Zech Xu, Z.; Kightley, E.P.; Thompson, L.R.; Hyde, E.R.; Gonzalez, A.; et al. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems 2017, 2, e00191-16. [Google Scholar] [CrossRef]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.-P.; Mushayamaha, T.; Thomas, P.D. PANTHER version 16: A revised family classification, tree-based classification tool, enhancer regions and extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Kim, D.K.; Lillehoj, H.S.; Lee, S.H.; Jang, S.I.; Bravo, D. High-throughput gene expression analysis of intestinal intraepithelial lymphocytes after oral feeding of carvacrol, cinnamaldehyde, or Capsicum oleoresin. Poult. Sci. 2010, 89, 68–81. [Google Scholar] [CrossRef]
- Huyghebaert, G.; Ducatelle, R.; Van Immerseel, F. An update on alternatives to antimicrobial growth promoters for broilers. Veter. J. 2011, 187, 182–188. [Google Scholar] [CrossRef]
- Wang, J.; Fan, H.; Han, Y.; Wei, J.; Zhao, J.; Zhou, Z. Pyrosequencing of the broiler chicken gastrointestinal tract reveals the regional similarity and dissimilarity of microbial community. Can. J. Anim. Sci. 2016, 97, 302–313. [Google Scholar] [CrossRef]
- Oakley, B.B.; Lillehoj, H.S.; Kogut, M.H.; Kim, W.K.; Maurer, J.J.; Pedroso, A.; Lee, M.D.; Collett, S.R.; Johnson, T.; Cox, N.A. The chicken gastrointestinal microbiome. FEMS Microbiol. Lett. 2014, 360, 100–112. [Google Scholar] [CrossRef]
- Choi, J.H.; Kim, G.B.; Cha, C.J. Spatial heterogeneity and stability of bacterial community in the gastrointestinal tracts of broiler chickens. Poult. Sci. 2014, 93, 1942–1950. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Yu, H.; Liu, T.; Gill, J.; Chambers, J.; Wheatcroft, R.; Sabour, P. Effects of zinc bacitracin, bird age and access to range on bacterial microbiota in the ileum and caeca of broiler chickens. J. Appl. Microbiol. 2008, 104, 1372–1382. [Google Scholar] [CrossRef] [PubMed]
- Owens, B.; Tucker, L.; Collins, M.A.; McCracken, K.J. Effects of different feed additives alone or in combination on broiler performance, gut microflora and ileal histology. Br. Poult. Sci. 2008, 49, 202–212. [Google Scholar] [CrossRef] [PubMed]
- Abdelli, N.; Pérez, J.F.; Vilarrasa, E.; Luna, I.C.; Melo-Duran, D.; D’Angelo, M.; Solà-Oriol, D. Targeted-Release Organic Acids and Essential Oils Improve Performance and Digestive Function in Broilers under a Necrotic Enteritis Challenge. Animals 2020, 10, 259. [Google Scholar] [CrossRef] [PubMed]
- Pham, V.H.; Kan, L.; Huang, J.; Geng, Y.; Zhen, W.; Guo, Y.; Abbas, W.; Wang, Z. Dietary encapsulated essential oils and organic acids mixture improves gut health in broiler chickens challenged with necrotic enteritis. J. Anim. Sci. Biotechnol. 2020, 11, 18. [Google Scholar] [CrossRef] [PubMed]
- Thibodeau, A.; Fravalo, P.; Yergeau, E.; Arsenault, J.; Lahaye, L.; Letellier, A. Chicken Caecal Microbiome Modifications Induced by Campylobacter jejuni Colonization and by a Non-Antibiotic Feed Additive. PLoS ONE 2015, 10, e0131978. [Google Scholar] [CrossRef]
- Bauer, B.W.; Gangadoo, S.; Bajagai, Y.S.; Van, T.T.H.; Moore, R.J.; Stanley, D. Oregano powder reduces Streptococcus and increases SCFA concentration in a mixed bacterial culture assay. PLoS ONE 2019, 14, e0216853. [Google Scholar] [CrossRef]
- Choi, J.-H.; Lee, K.; Kim, D.-W.; Kil, D.Y.; Kim, G.-B.; Cha, C.-J. Influence of dietary avilamycin on ileal and cecal microbiota in broiler chickens. Poult. Sci. 2018, 97, 970–979. [Google Scholar] [CrossRef]
- Yang, C.; Kennes, Y.M.; Lepp, D.; Yin, X.; Wang, Q.; Yu, H.; Yang, C.; Gong, J.; Diarra, M.S. Effects of encapsulated cinnamaldehyde and citral on the performance and cecal microbiota of broilers vaccinated or not vaccinated against coccidiosis. Poult. Sci. 2020, 99, 936–948. [Google Scholar] [CrossRef]
- Stanley, D.; Hughes, R.J.; Moore, R.J. Microbiota of the chicken gastrointestinal tract: Influence on health, productivity and disease. Appl. Microbiol. Biotechnol. 2014, 98, 4301–4310. [Google Scholar] [CrossRef]
- Xu, S.; Lin, Y.; Zeng, D.; Zhou, M.; Zeng, Y.; Wang, H.; Zhou, Y.; Zhu, H.; Pan, K.; Jing, B.; et al. Bacillus licheniformis normalize the ileum microbiota of chickens infected with necrotic enteritis. Sci. Rep. 2018, 8, 1744. [Google Scholar] [CrossRef] [PubMed]
- Proszkowiec-Weglarz, M.; Miska, K.B.; Schreier, L.L.; Grim, C.J.; Jarvis, K.G.; Shao, J.; Vaessen, S.; Sygall, R.; Jenkins, M.C.; Kahl, S.; et al. Research Note: Effect of butyric acid glycerol esters on ileal and cecal mucosal and luminal microbiota in chickens challenged with Eimeria maxima. Poult. Sci. 2020, 99, 5143–5148. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-C.; Kil, D.Y.; Sul, W.J. Cecal microbiome divergence of broiler chickens by sex and body weight. J. Microbiol. 2017, 55, 939–945. [Google Scholar] [CrossRef] [PubMed]
- Stanley, D.; Hughes, R.J.; Geier, M.S.; Moore, R.J. Bacteria within the Gastrointestinal Tract Microbiota Correlated with Improved Growth and Feed Conversion: Challenges Presented for the Identification of Performance Enhancing Probiotic Bacteria. Front. Microbiol. 2016, 7, 187. [Google Scholar] [CrossRef] [PubMed]
- Meehan, C.; Beiko, R.G. A Phylogenomic View of Ecological Specialization in the Lachnospiraceae, a Family of Digestive Tract-Associated Bacteria. Genome Biol. Evol. 2014, 6, 703–713. [Google Scholar] [CrossRef]
- Yacoubi, N.; Saulnier, L.; Bonnin, E.; Devillard, E.; Eeckhaut, V.; Rhayat, L.; Ducatelle, R.; Van Immerseel, F. Short-chain arabinoxylans prepared from enzymatically treated wheat grain exert prebiotic effects during the broiler starter period. Poult. Sci. 2018, 97, 412–424. [Google Scholar] [CrossRef] [PubMed]
- Zhong, H.; Wang, X.-G.; Wang, J.; Chen, Y.-J.; Qin, H.-L.; Yang, R. Impact of probiotics supplement on the gut microbiota in neonates with antibiotic exposure: An open-label single-center randomized parallel controlled study. World J. Pediatr. 2021, 17, 385–393. [Google Scholar] [CrossRef]
- McKenna, A.; Ijaz, U.Z.; Kelly, C.; Linton, M.; Sloan, W.T.; Green, B.D.; Lavery, U.; Dorrell, N.; Wren, B.W.; Richmond, A.; et al. Impact of industrial production system parameters on chicken microbiomes: Mechanisms to improve performance and reduce Campylobacter. Microbiome 2020, 8, 128. [Google Scholar] [CrossRef]
- Wang, Y.; Xu, Y.; Xu, S.; Yang, J.; Wang, K.; Zhan, X. Bacillus subtilis DSM29784 Alleviates Negative Effects on Growth Performance in Broilers by Improving the Intestinal Health Under Necrotic Enteritis Challenge. Front. Microbiol. 2021, 12, 723187. [Google Scholar] [CrossRef]
- Yang, W.Y.; Lee, Y.; Lu, H.; Chou, C.H.; Wang, C. Analysis of contributory gut microbiota and lauric acid against necrotic enteritis in Clostridium perfringens and Eimeria side-by-side challenge model. bioRxiv 2018, 434449. [Google Scholar] [CrossRef]
- Eeckhaut, V.; Van Immerseel, F.; Croubels, S.; De Baere, S.; Haesebrouck, F.; Ducatelle, R.; Louis, P.; Vandamme, P. Butyrate production in phylogenetically diverse Firmicutes isolated from the chicken caecum. Microb. Biotechnol. 2011, 4, 503–512. [Google Scholar] [CrossRef] [PubMed]
- Polansky, O.; Sekelova, Z.; Faldynova, M.; Sebkova, A.; Sisak, F.; Rychlik, I. Important Metabolic Pathways and Biological Processes Expressed by Chicken Cecal Microbiota. Appl. Environ. Microbiol. 2016, 82, 1569–1576. [Google Scholar] [CrossRef] [PubMed]
- Jacquier, V.; Nelson, A.; Jlali, M.; Rhayat, L.; Brinch, K.; Devillard, E. Bacillus subtilis 29784 induces a shift in broiler gut microbiome toward butyrate-producing bacteria and improves intestinal histomorphology and animal performance. Poult. Sci. 2019, 98, 2548–2554. [Google Scholar] [CrossRef] [PubMed]
- Shang, Q.; Liu, S.; He, T.; Liu, H.; Mahfuz, S.; Ma, X.; Piao, X. Effects of wheat bran in comparison to antibiotics on growth performance, intestinal immunity, barrier function, and microbial composition in broiler chickens. Poult. Sci. 2020, 99, 4929–4938. [Google Scholar] [CrossRef] [PubMed]
- Xue, F.; Shi, L.; Li, Y.; Ni, A.; Ma, H.; Sun, Y.; Chen, J. Effects of replacing dietary Aureomycin with a combination of plant essential oils on production performance and gastrointestinal health of broilers. Poult. Sci. 2020, 99, 4521–4529. [Google Scholar] [CrossRef]
- Ma, X.; Wang, Q.; Li, H.; Xu, C.; Cui, N.; Zhao, X. 16S rRNA genes Illumina sequencing revealed differential cecal microbiome in specific pathogen free chickens infected with different subgroup of avian leukosis viruses. Veter. Microbiol. 2017, 207, 195–204. [Google Scholar] [CrossRef]
- Chen, H.-L.; Zhao, X.-Y.; Zhao, G.-X.; Huang, H.-B.; Li, H.-R.; Shi, C.-W.; Yang, W.-T.; Jiang, Y.-L.; Wang, J.-Z.; Ye, L.-P.; et al. Dissection of the cecal microbial community in chickens after Eimeria tenella infection. Parasites Vectors 2020, 13, 56. [Google Scholar] [CrossRef]
- Khan, S.; Chousalkar, K.K. Salmonella Typhimurium infection disrupts but continuous feeding of Bacillus based probiotic restores gut microbiota in infected hens. J. Anim. Sci. Biotechnol. 2020, 11, 29. [Google Scholar] [CrossRef]
- Hung, D.-Y.; Cheng, Y.-H.; Chen, W.-J.; Hua, K.-F.; Pietruszka, A.; Dybus, A.; Lin, C.-S.; Yu, Y.-H. Bacillus licheniformis-Fermented Products Reduce Diarrhea Incidence and Alter the Fecal Microbiota Community in Weaning Piglets. Animals 2019, 9, 1145. [Google Scholar] [CrossRef]
- Kulshreshtha, G.; Rathgeber, B.; MacIsaac, J.; Boulianne, M.; Brigitte, L.; Stratton, G.; Thomas, N.A.; Critchley, A.T.; Hafting, J.; Prithiviraj, B. Feed Supplementation with Red Seaweeds, Chondrus crispus and Sarcodiotheca gaudichaudii, Reduce Salmonella Enteritidis in Laying Hens. Front. Microbiol. 2017, 8, 567. [Google Scholar] [CrossRef]
- Aydın, Ö.D.; Yıldız, G. Using of Essential Oil Mixture in Quail Breeders (Coturnix Coturnix Japonica) for Improving Cecal Short-Chain Fatty Acid Concentrations. Turk. J. Agric.-Food Sci. Technol. 2020, 8, 2021–2024. [Google Scholar] [CrossRef]
- Tiihonen, K.; Kettunen, H.; Bento, M.; Saarinen, M.; Lahtinen, S.; Ouwehand, A.; Schulze, H.; Rautonen, N. The effect of feeding essential oils on broiler performance and gut microbiota. Br. Poult. Sci. 2010, 51, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Vahjen, W.; Dadi, T.; Saliu, E.-M.; Boroojeni, F.G.; Zentek, J. Synergistic Effects of Probiotics and Phytobiotics on the Intestinal Microbiota in Young Broiler Chicken. Microorganisms 2019, 7, 684. [Google Scholar] [CrossRef] [PubMed]
- Mašek, T.; Starčević, K. Mikulec Einfluss des zusatzes von Thymol, Gerbsäure oder Gallussäure zum Futter auf Leis-tung, Malondialdehyd-Gehalt im serum und die fermentationsleistung im Blinddarm von Broilern. Eur. Poult. Sci. 2014, 78, 1–8. [Google Scholar] [CrossRef]
- Adewole, D.I.; Oladokun, S.; Santin, E. Effect of organic acids–essential oils blend and oat fiber combination on broiler chicken growth performance, blood parameters, and intestinal health. Anim. Nutr. 2021, 7, 1039–1051. [Google Scholar] [CrossRef]
- Sun, B.; Hou, L.; Yang, Y. Effects of Altered Dietary Fiber on the Gut Microbiota, Short-Chain Fatty Acids and Cecum of Chickens during Different Growth Periods. Anim. Sci. Zool.-Prepr. 2020. [Google Scholar] [CrossRef]
- Bajagai, Y.; Radovanovic, A.; Steel, J.; Stanley, D. The Effects of Continual Consumption of Origanum Vulgare on Liver Transcriptomics. Animals 2021, 11, 398. [Google Scholar] [CrossRef]
- You, X.; Xu, M.; Li, Q.; Zhang, K.; Hao, G.; Xu, H. Discovery of potential transcriptional biomarkers in broiler chicken for detection of amantadine abuse based on RNA sequencing technology. Food Addit. Contam. Part A Chem. Anal. Control. Expo. Risk Assess. 2019, 36, 254–269. [Google Scholar] [CrossRef]
- Hong, Y.; Cheng, Y.; Guan, L.; Zhou, Z.; Li, X.; Shi, D.; Xiao, Y. Bacillus amyloliquefaciens TL Downregulates the Ileal Expression of Genes Involved in Immune Responses in Broiler Chickens to Improve Growth Performance. Microorganisms 2021, 9, 382. [Google Scholar] [CrossRef]
- González-Mariscal, L.; Betanzos, A.; Nava, P.; Jaramillo, B. Tight junction proteins. Prog. Biophys. Mol. Biol. 2003, 81, 1–44. [Google Scholar] [CrossRef]
- Wada, A.M.; Reese, D.E.; Bader, D.M. Bves: Prototype of a new class of cell adhesion molecules expressed during coronary artery development. Development 2001, 128, 2085–2093. [Google Scholar] [CrossRef] [PubMed]
- Osler, M.E.; Chang, M.S.; Bader, D.M. Bves modulates epithelial integrity through an interaction at the tight junction. J. Cell Sci. 2005, 118, 4667–4678. [Google Scholar] [CrossRef] [PubMed]
- Russ, P.K.; Kupperman, A.I.; Presley, S.-H.; Haselton, F.R.; Chang, M.S. Inhibition of RhoA Signaling with Increased Bves in Trabecular Meshwork Cells. Investig. Opthalmol. Vis. Sci. 2010, 51, 223–230. [Google Scholar] [CrossRef] [PubMed]
- A Hager, H.; Bader, D.M. Bves: Ten years after. Histol. Histopathol. 2009, 24, 777–787. [Google Scholar] [CrossRef]
- Vasavada, T.K.; DiAngelo, J.R.; Duncan, M.K. Developmental Expression of Pop1/Bves. J. Histochem. Cytochem. 2004, 52, 371–377. [Google Scholar] [CrossRef]
- Andrée, B.; Fleige, A.; Arnold, H.-H.; Brand, T. Mouse Pop1 Is Required for Muscle Regeneration in Adult Skeletal Muscle. Mol. Cell. Biol. 2002, 22, 1504–1512. [Google Scholar] [CrossRef]
- DiAngelo, J.R.; Vasavada, T.K.; Cain, W.; Duncan, M.K. Production of Monoclonal Antibodies Against Chicken Pop1 (BVES). Hybrid. Hybridomics 2001, 20, 377–381. [Google Scholar] [CrossRef]
- Gu, J.; Liang, Q.; Liu, C.; Li, S. Genomic Analyses Reveal Adaptation to Hot Arid and Harsh Environments in Native Chickens of China. Front. Genet. 2020, 11, 11. [Google Scholar] [CrossRef]
- Froese, A.; Breher, S.S.; Waldeyer, C.; Schindler, R.F.; Nikolaev, V.O.; Rinné, S.; Wischmeyer, E.; Schlueter, J.; Becher, J.; Simrick, S.; et al. Popeye domain containing proteins are essential for stress-mediated modulation of cardiac pacemaking in mice. J. Clin. Investig. 2012, 122, 1119–1130. [Google Scholar] [CrossRef]
- Torlopp, A.; Breher, S.S.; Schlüter, J.; Brand, T. Comparative analysis of mRNA and protein expression of Popdc1 (Bves) during early development in the chick embryo. Dev. Dyn. 2006, 235, 691–700. [Google Scholar] [CrossRef]
- Aengwanich, W.; Simaraks, S. Pathology of heart, lung, liver and kidney in broilers under chronic heat stress. Songklanakarin J. Sci. Technol. 2004, 26, 417–424. [Google Scholar]
- Glahn, R.P.; Mitsos, W.J.; Wideman, R.F. Evaluation of Sex Differences in Embryonic Heart Rates. Poult. Sci. 1987, 66, 1398–1401. [Google Scholar] [CrossRef] [PubMed]
- Ringer, R.K.; Weiss, H.S.; Sturkie, P.D. Heart Rate of Chickens as Influenced by Age and Gonadal Hormones. Am. J. Physiol. Content 1957, 191, 145–147. [Google Scholar] [CrossRef] [PubMed]
- Baillat, D.; Hakimi, M.-A.; Näär, A.M.; Shilatifard, A.; Cooch, N.; Shiekhattar, R. Integrator, a Multiprotein Mediator of Small Nuclear RNA Processing, Associates with the C-Terminal Repeat of RNA Polymerase II. Cell 2005, 123, 265–276. [Google Scholar] [CrossRef]
- Will, C.L.; Lührmann, R. Spliceosome structure and function. Cold Spring Harb. Perspect. Biol. 2011, 3, a003707. [Google Scholar] [CrossRef]
- Tsalikis, J.; Tattoli, I.; Ling, A.; Sorbara, M.T.; Croitoru, D.O.; Philpott, D.J.; Girardin, S.E. Intracellular Bacterial Pathogens Trigger the Formation of U Small Nuclear RNA Bodies (U Bodies) through Metabolic Stress Induction. J. Biol. Chem. 2015, 290, 20904–20918. [Google Scholar] [CrossRef]
- Chittka, D.; Banas, B.; Lennartz, L.; Putz, F.J.; Eidenschink, K.; Beck, S.; Stempfl, T.; Moehle, C.; Reichelt-Wurm, S.; Banas, M.C. Long-term expression of glomerular genes in diabetic nephropathy. Nephrol. Dial. Transplant. 2018, 33, 1533–1544. [Google Scholar] [CrossRef]
- Wright, E.M. Glucose transport families SLC5 and SLC50. Mol. Asp. Med. 2013, 34, 183–196. [Google Scholar] [CrossRef]
- Leturque, A.; Brot-Laroche, E.; Le Gall, M.; Stolarczyk, E.; Tobin, V. The role of GLUT2 in dietary sugar handling. J. Physiol. Biochem. 2005, 61, 529–537. [Google Scholar] [CrossRef]
- Fukuzawa, T.; Fukazawa, M.; Ueda, O.; Shimada, H.; Kito, A.; Kakefuda, M.; Kawase, Y.; Wada, N.A.; Goto, C.; Fukushima, N.; et al. SGLT5 Reabsorbs Fructose in the Kidney but Its Deficiency Paradoxically Exacerbates Hepatic Steatosis Induced by Fructose. PLoS ONE 2013, 8, e56681. [Google Scholar] [CrossRef]
- Gaby, A.R. Adverse effects of dietary fructose. Altern. Med. Rev. J. Clin. Ther. 2005, 10, 294–306. [Google Scholar]
- Boles, M.K.; Wilkinson, B.M.; Wilming, L.G.; Liu, B.; Probst, F.J.; Harrow, J.; Grafham, D.; Hentges, K.E.; Woodward, L.P.; Maxwell, A.; et al. Discovery of Candidate Disease Genes in ENU-Induced Mouse Mutants by Large-Scale Sequencing, Including a Splice-Site Mutation in Nucleoredoxin. PLoS Genet. 2009, 5, e1000759. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Yuan, C.-X.; Okano, H.J.; Darnell, R.; Roeder, R.G. Involvement of the TRAP220 Component of the TRAP/SMCC Coactivator Complex in Embryonic Development and Thyroid Hormone Action. Mol. Cell 2000, 5, 683–693. [Google Scholar] [CrossRef]
- Ito, M.; Okano, H.J.; Darnell, R.B.; Roeder, R.G. The TRAP100 component of the TRAP/Mediator complex is essential in broad transcriptional events and development. EMBO J. 2002, 21, 3464–3475. [Google Scholar] [CrossRef] [PubMed]
- Wolton, K.; Barnes, E.; Wright, J.; Hentges, K. Two alleles of Med31 provide a model to study delayed fetal growth and endochondral ossification. Toxicol. Lett. 2014, 229, S51. [Google Scholar] [CrossRef]
- Kim, S.K.; Brotslaw, E.; Thome, V.; Mitchell, J.; Ventrella, R.; Collins, C.; Mitchell, B. A role for Cep70 in centriole amplification in multiciliated cells. Dev. Biol. 2020, 471, 10–17. [Google Scholar] [CrossRef]
- Yang, Y.; Ran, J.; Liu, M.; Li, D.; Li, Y.; Shi, X.; Meng, D.; Pan, J.; Ou, G.; Aneja, R.; et al. CYLD mediates ciliogenesis in multiple organs by deubiquitinating Cep70 and inactivating HDAC6. Cell Res. 2014, 24, 1342–1353. [Google Scholar] [CrossRef]
- Shi, X.; Yao, Y.; Wang, Y.; Zhang, Y.; Huang, Q.; Zhou, J.; Liu, M.; Li, D. Cep70 regulates microtubule stability by interacting with HDAC6. FEBS Lett. 2015, 589, 1771–1777. [Google Scholar] [CrossRef]
- Shi, X.; Wang, J.; Yang, Y.; Ren, Y.; Zhou, J.; Li, D. Cep70 promotes microtubule assembly in vitro by increasing microtubule elongation. Acta Biochim. Biophys. Sin. 2012, 44, 450–454. [Google Scholar] [CrossRef]
- Nigg, E.A.; Raff, J. Centrioles, Centrosomes, and Cilia in Health and Disease. Cell 2009, 139, 663–678. [Google Scholar] [CrossRef]
- Libe, R.; Horvath, A.; Vezzosi, D.; Fratticci, A.; Coste, J.; Perlemoine, K.; Ragazzon, B.; Guillaud-Bataille, M.; Groussin, L.; Clauser, E.; et al. Frequent Phosphodiesterase 11A Gene (PDE11A) Defects in Patients with Carney Complex (CNC) Caused byPRKAR1AMutations:PDE11AMay Contribute to Adrenal and Testicular Tumors in CNC as a Modifier of the Phenotype. J. Clin. Endocrinol. Metab. 2011, 96, E208–E214. [Google Scholar] [CrossRef] [PubMed]
- Faucz, F.R.; Horvath, A.; Rothenbuhler, A.; Almeida, M.Q.; Libe, R.; Raffin-Sanson, M.-L.; Bertherat, J.; Carraro, D.; Soares, F.A.; Molina, G.D.C.; et al. Phosphodiesterase 11A (PDE11A) Genetic Variants May Increase Susceptibility to Prostatic Cancer. J. Clin. Endocrinol. Metab. 2011, 96, E135–E140. [Google Scholar] [CrossRef] [PubMed]
- Dietert, K.; Reppe, K.; Mundhenk, L.; Witzenrath, M.; Gruber, A.D. mCLCA3 Modulates IL-17 and CXCL-1 Induction and Leukocyte Recruitment in Murine Staphylococcus aureus Pneumonia. PLoS ONE 2014, 9, e102606. [Google Scholar] [CrossRef]
- Mamber, S.W.; Gurel, V.; Lins, J.; Ferri, F.; Beseme, S.; McMichael, J. Effects of cannabis oil extract on immune response gene expression in human small airway epithelial cells (HSAEpC): Implications for chronic obstructive pulmonary disease (COPD). J. Cannabis Res. 2020, 2, 5. [Google Scholar] [CrossRef] [PubMed]
- Farmahin, R.; Gannon, A.M.; Gagné, R.; Rowan-Carroll, A.; Kuo, B.; Williams, A.; Curran, I.; Yauk, C.L. Hepatic transcriptional dose-response analysis of male and female Fischer rats exposed to hexabromocyclododecane. Food Chem. Toxicol. 2019, 133, 110262. [Google Scholar] [CrossRef]
- Kwon, T.-J.; Kim, D.-B.; Bae, J.W.; Sagong, B.; Choi, S.-Y.; Cho, H.-J.; Kim, U.-K.; Lee, K.-Y. Molecular cloning, characterization, and expression of pannexin genes in chicken. Poult. Sci. 2014, 93, 2253–2261. [Google Scholar] [CrossRef][Green Version]
- Tang, W.; Ahmad, S.; Shestopalov, V.I.; Lin, X. Pannexins are new molecular candidates for assembling gap junctions in the cochlea. NeuroReport 2008, 19, 1253–1257. [Google Scholar] [CrossRef]
- Jiang, J.X.; Penuela, S. Connexin and pannexin channels in cancer. BMC Cell Biol. 2016, 17, 105–120. [Google Scholar] [CrossRef]
- Xie, C.-R.; Sun, H.; Wang, F.-Q.; Li, Z.; Yin, Y.-R.; Fang, Q.-L.; Sun, Y.; Zhao, W.-X.; Zhang, S.; Wang, X.-M.; et al. Integrated analysis of gene expression and DNA methylation changes induced by hepatocyte growth factor in human hepatocytes. Mol. Med. Rep. 2015, 12, 4250–4258. [Google Scholar] [CrossRef]
- Anand, P.; Kumar, S.V.; Ravi, K.; Simmi, T. Differential gene expression in duodenum of colored broiler chicken divergently selected for residual feed intake. Trop. Anim. Health Prod. 2021, 53, 59. [Google Scholar] [CrossRef]
- Ren, J.; Li, Y.; Xu, N.; Li, H.; Li, C.; Han, R.; Wang, Y.; Li, Z.; Kang, X.; Liu, X.; et al. Association of estradiol on expression of melanocortin receptors and their accessory proteins in the liver of chicken (Gallus gallus). Gen. Comp. Endocrinol. 2017, 240, 182–190. [Google Scholar] [CrossRef] [PubMed]
- Blankenship, K.; Gilley, A.; Piekarski, A.; Orlowski, S.; Greene, E.; Bottje, W.; Anthony, N.; Dridi, S. Differential expression of feeding-related hypothalamic neuropeptides in the first generation of quails divergently selected for low or high feed efficiency. Neuropeptides 2016, 58, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.; Lin, J.E.; Li, P.; Snook, A.; Gong, J.; Sato, T.; Liu, C.; Girondo, M.A.; Rui, H.; Hyslop, T.; et al. The Paracrine Hormone for the GUCY2C Tumor Suppressor, Guanylin, Is Universally Lost in Colorectal Cancer. Cancer Epidemiol. Biomark. Prev. 2014, 23, 2328–2337. [Google Scholar] [CrossRef]
- Steinbrecher, K.A.; Harmel-Laws, E.; Garin-Laflam, M.P.; Mann, E.A.; Bezerra, L.D.; Hogan, S.P.; Cohen, M.B. Murine Guanylate Cyclase C Regulates Colonic Injury and Inflammation. J. Immunol. 2011, 186, 7205–7214. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.; Giráldez, F.J.; Trevisi, E.; Lucini, L.; Frutos, J.; Andrés, S. Liver transcriptomic and plasma metabolomic profiles of fattening lambs are modified by feed restriction during the suckling period1. J. Anim. Sci. 2018, 96, 1495–1507. [Google Scholar] [CrossRef]
- Erwin, E.A.; Jaramillo, L.M.; Smith, B.; Kruszewski, P.G.; Kahwash, B.; Grayson, M.H.; Mejias, A.; Ramilo, O. Sex Differences in Blood Transcriptional Profiles and Clinical Phenotypes in Pediatric Patients with Eosinophilic Esophagitis. J. Allergy Clin. Immunol. Pract. 2021, 9, 3350–3358. [Google Scholar] [CrossRef]
- Shaik, A.P.; AlSaeed, A.H.; Kiranmayee, S.; Bammidi, V.; Sultana, A. Phylogenetic analysis of cubilin (CUBN) gene. Bioinformation 2013, 9, 29–36. [Google Scholar] [CrossRef]
- Christensen, E.I.; Birn, H. Megalin and cubilin: Multifunctional endocytic receptors. Nat. Rev. Mol. Cell Biol. 2002, 3, 258–267. [Google Scholar] [CrossRef]
- Lee, J.; Karnuah, A.B.; Rekaya, R.; Anthony, N.B.; Aggrey, S.E. Transcriptomic analysis to elucidate the molecular mechanisms that underlie feed efficiency in meat-type chickens. Mol. Gen. Genet. MGG 2015, 290, 1673–1682. [Google Scholar] [CrossRef]
- Sun, H.; Jiang, R.; Xu, S.; Zhang, Z.; Xu, G.; Zheng, J.; Qu, L. Transcriptome responses to heat stress in hypothalamus of a meat-type chicken. J. Anim. Sci. Biotechnol. 2015, 6, 6. [Google Scholar] [CrossRef]
- Balla, T. Phosphoinositides: Tiny Lipids with Giant Impact on Cell Regulation. Physiol. Rev. 2013, 93, 1019–1137. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Li, Z.; Lin, L.; Liu, G.; Ko, K.; Coetzee, W.A.; Skolnik, E.Y. The Phosphatidylinositol 3-Phosphate Phosphatase Myotubularin- Related Protein 6 (MTMR6) Is a Negative Regulator of the Ca2+ -Activated K+ Channel K Ca 3.1. Mol. Cell. Biol. 2005, 25, 3630–3638. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Gu, X.; Bednarczyk, P.; Wiedemann, F.; Haddad, G.; Siemen, D. Hypoxia Increases Activity of the BK-Channel in the Inner Mitochondrial Membrane and Reduces Activity of the Permeability Transition Pore. Cell. Physiol. Biochem. 2008, 22, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Lu, D.-Z.; Li, Y.-M.; Zhang, X.-Q.; Zhou, X.-X.; Jin, X. Proteomic analysis of liver mitochondria from rats with nonalcoholic steatohepatitis. World J. Gastroenterol. 2014, 20, 4778–4786. [Google Scholar] [CrossRef]
- Huang, Z.; Jin, S.; Lv, Z. Dietary genistein supplementation alters mRNA expression profile and alternative splicing signature in the thymus of chicks with lipopolysaccharide challenge. Poult. Sci. 2021, 101, 101561. [Google Scholar] [CrossRef]
- Smith, I.A.; Knezevic, B.R.; Ammann, J.U.; Rhodes, D.A.; Aw, D.; Palmer, D.B.; Mather, I.H.; Trowsdale, J. BTN1A1, the Mammary Gland Butyrophilin, and BTN2A2 Are Both Inhibitors of T Cell Activation. J. Immunol. 2010, 184, 3514–3525. [Google Scholar] [CrossRef]
- Yamazaki, T.; Goya, I.; Graf, D.; Craig, S.; Martin-Orozco, N.; Dong, C. A Butyrophilin Family Member Critically Inhibits T Cell Activation. J. Immunol. 2010, 185, 5907–5914. [Google Scholar] [CrossRef]
- Wu, C.; Liu, L.; Jinglong, H.; Lianjun, L.; Yongwang, M. Isolation, bioinformatic and tissue expression analysis of a novel water buffalo gene-BTN1A1. Buffalo Bull. 2014, 33, 449–461. [Google Scholar]
- Xu, Z.; Le, K.; Moghadasian, M.H. Long-term phytosterol treatment alters gene expression in the liver of apo E-deficient mice. J. Nutr. Biochem. 2008, 19, 545–554. [Google Scholar] [CrossRef]
- Moghadasian, M.H. Pharmacological properties of plant sterols: In vivo and in vitro observations. Life Sci. 2000, 67, 605–615. [Google Scholar] [CrossRef]
- Wolfs, M.; De Jong, N.; Ocké, M.C.; Verhagen, H.; Verschuren, W.M. Effectiveness of customary use of phytosterol/-stanol enriched margarines on blood cholesterol lowering. Food Chem. Toxicol. 2006, 44, 1682–1688. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yang, Y.; Zhang, Y.; Liu, Z. lncRNA ST8SIA6-AS1 facilitates proliferation and invasion in liver cancer by regulating miR-142-3p. Exp. Ther. Med. 2021, 22, 1348. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, L.; Karkeni, E.; Couturier, C.; Astier, J.; Dalifard, J.; Defoort, C.; Svilar, L.; Martin, J.-C.; Tourniaire, F.; Landrier, J.-F. Gene Expression Pattern in Response to Cholecalciferol Supplementation Highlights Cubilin as a Major Protein of 25(OH)D Uptake in Adipocytes and Male Mice White Adipose Tissue. Endocrinology 2018, 159, 957–966. [Google Scholar] [CrossRef] [PubMed]
- Nykjaer, A.; Fyfe, J.C.; Kozyraki, R.; Leheste, J.-R.; Jacobsen, C.; Nielsen, M.S.; Verroust, P.J.; Aminoff, M.; de la Chapelle, A.; Moestrup, S.K.; et al. Cubilin dysfunction causes abnormal metabolism of the steroid hormone 25(OH) vitamin D3. Proc. Natl. Acad. Sci. USA 2001, 98, 13895–13900. [Google Scholar] [CrossRef] [PubMed]
- Collison, K.S.; Maqbool, Z.; Saleh, S.M.; Inglis, A.; Makhoul, N.J.; Bakheet, R.; Al-Johi, M.; Al-Rabiah, R.; Zaidi, M.Z.; Al-Mohanna, F.A. Effect of dietary monosodium glutamate on trans fat-induced nonalcoholic fatty liver disease. J. Lipid Res. 2009, 50, 1521–1537. [Google Scholar] [CrossRef]



| Short-Chain Fatty Acid Concentration (mmol/kg) | Treatments 1 | SEM 2 p Value | ||||||
|---|---|---|---|---|---|---|---|---|
| Negative Control | In-Feed Antibiotics | In-Water Essential Oil | In Ovo Saline | In Ovo Essential Oil | In Ovo Essential Oil + In-Water Essential Oil | |||
| Acetic acid | 51.9 | 50.5 | 57.2 | 55.4 | 50.9 | 55.8 | 1.73 | 0.82 |
| Propionic acid | 4.24 | 3.79 | 4.31 | 4.43 | 3.81 | 4.36 | 0.18 | 0.86 |
| Butyric acid | 13.6 | 15.7 | 19.3 | 18.8 | 13.4 | 17.9 | 0.77 | 0.09 |
| Valeric acid | 1.05 | 0.79 | 0.87 | 1.17 | 1.08 | 1.07 | 0.07 | 0.67 |
| Lactic acid | 0.60 | 0.73 | 1.40 | 1.26 | 1.09 | 0.83 | 0.86 | 0.49 |
| Total SCFA | 74.1 | 74.2 | 89.7 | 84.1 | 74.4 | 82.4 | 2.65 | 0.41 |
| Branched-chain fatty acids | 2.25 | 1.63 | 1.94 | 2.28 | 1.77 | 1.91 | 0.11 | 0.46 |
| Volatile fatty acids | 73.1 | 72.4 | 83.6 | 82.1 | 70.9 | 81.0 | 2.40 | 0.49 |
| Total eubacteria (copies/gram of sample) | 2.3 × 1012 | 1.9 × 1012 | 3.0 × 1012 | 2.6 × 1012 | 2.5 × 1012 | 2.2 × 1012 | 2.06 × 1012 | 0.72 |
| Treatments | Gene Symbol | Gene Description | Expression Level | log2FoldChange | p Value |
|---|---|---|---|---|---|
| B vs. A | ALDH1L2 | aldehyde dehydrogenase 1 family member L2 | Down | −0.6 | <0.01 |
| C vs. A | BTN1A1 | butyrophilin subfamily 1 member A1-like | Up | 0.4 | 0.02 |
| D vs. A | AVD | Avidin | Up | 0.5 | <0.01 |
| E vs. A | ST8SIA6 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 6 | Up | 0.6 | <0.01 |
| CUBN | Cubilin | Down | −0.7 | 0.01 | |
| F vs. A | ALDH1L2 | aldehyde dehydrogenase 1 family member L2 | Down | −0.6 | 0.01 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oladokun, S.; Clark, K.F.; Adewole, D.I. Microbiota and Transcriptomic Effects of an Essential Oil Blend and Its Delivery Route Compared to an Antibiotic Growth Promoter in Broiler Chickens. Microorganisms 2022, 10, 861. https://doi.org/10.3390/microorganisms10050861
Oladokun S, Clark KF, Adewole DI. Microbiota and Transcriptomic Effects of an Essential Oil Blend and Its Delivery Route Compared to an Antibiotic Growth Promoter in Broiler Chickens. Microorganisms. 2022; 10(5):861. https://doi.org/10.3390/microorganisms10050861
Chicago/Turabian StyleOladokun, Samson, K. Fraser Clark, and Deborah I. Adewole. 2022. "Microbiota and Transcriptomic Effects of an Essential Oil Blend and Its Delivery Route Compared to an Antibiotic Growth Promoter in Broiler Chickens" Microorganisms 10, no. 5: 861. https://doi.org/10.3390/microorganisms10050861
APA StyleOladokun, S., Clark, K. F., & Adewole, D. I. (2022). Microbiota and Transcriptomic Effects of an Essential Oil Blend and Its Delivery Route Compared to an Antibiotic Growth Promoter in Broiler Chickens. Microorganisms, 10(5), 861. https://doi.org/10.3390/microorganisms10050861







