The Gut Microbiota May Affect Personality in Mongolian Gerbils
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Experimental Procedures
2.3. Ethical Statement
2.4. Personality Assay
2.5. Gut Microbiome Community Composition
2.6. Fecal Microbiota Transplant (FMT)
2.7. Statistical Analysis
3. Results
3.1. Individual Boldness Behavior
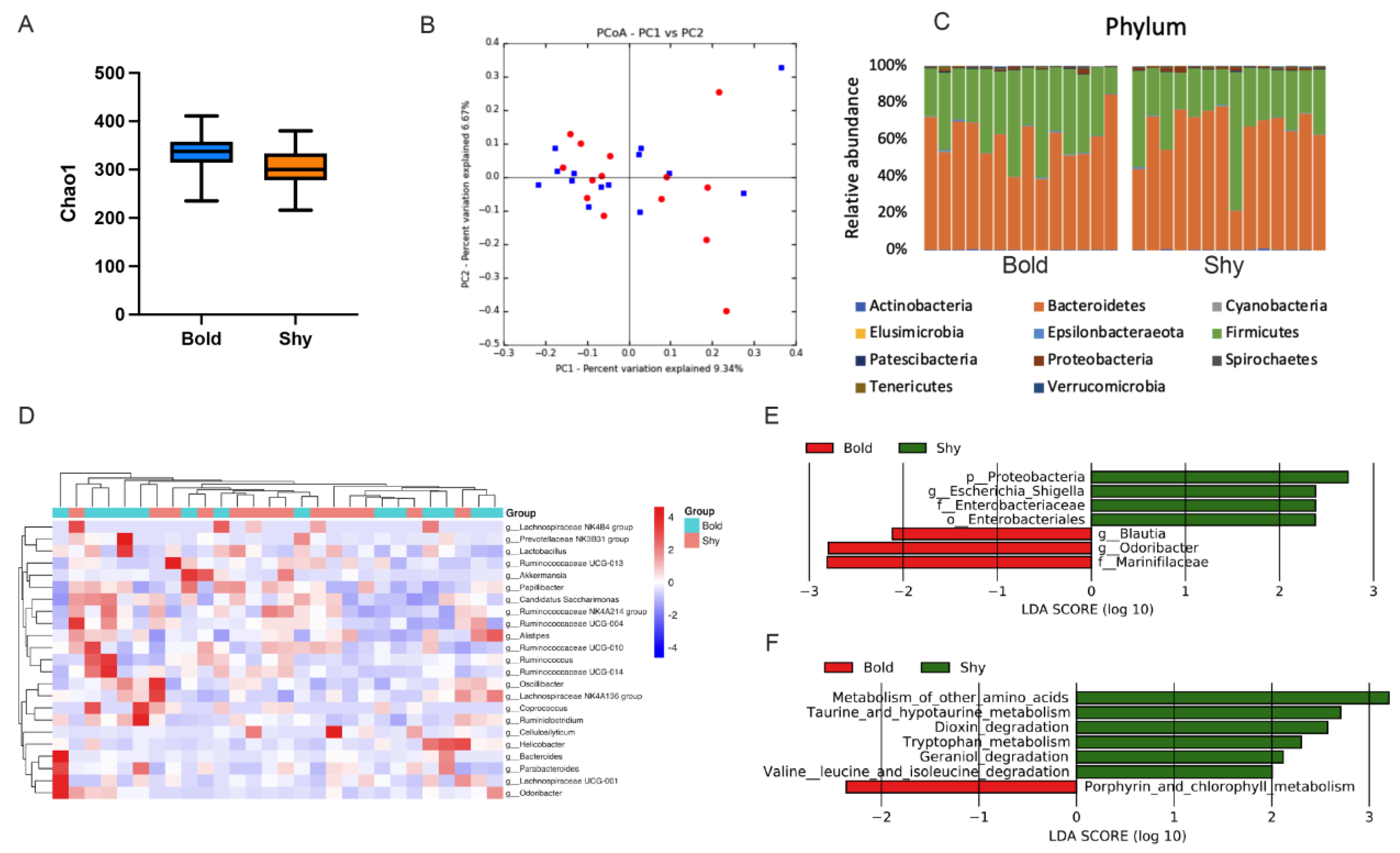
3.2. Gut Microbiota
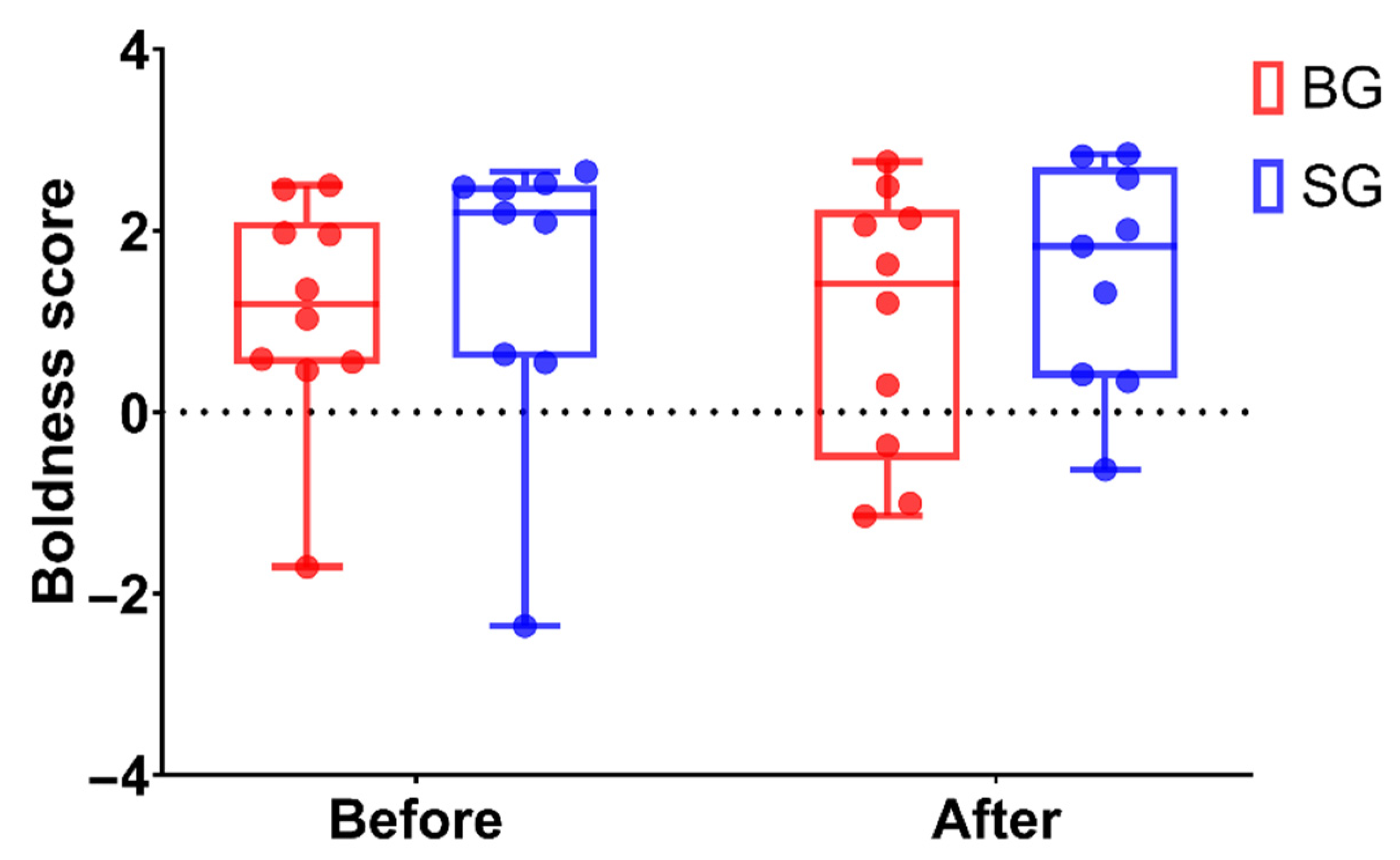
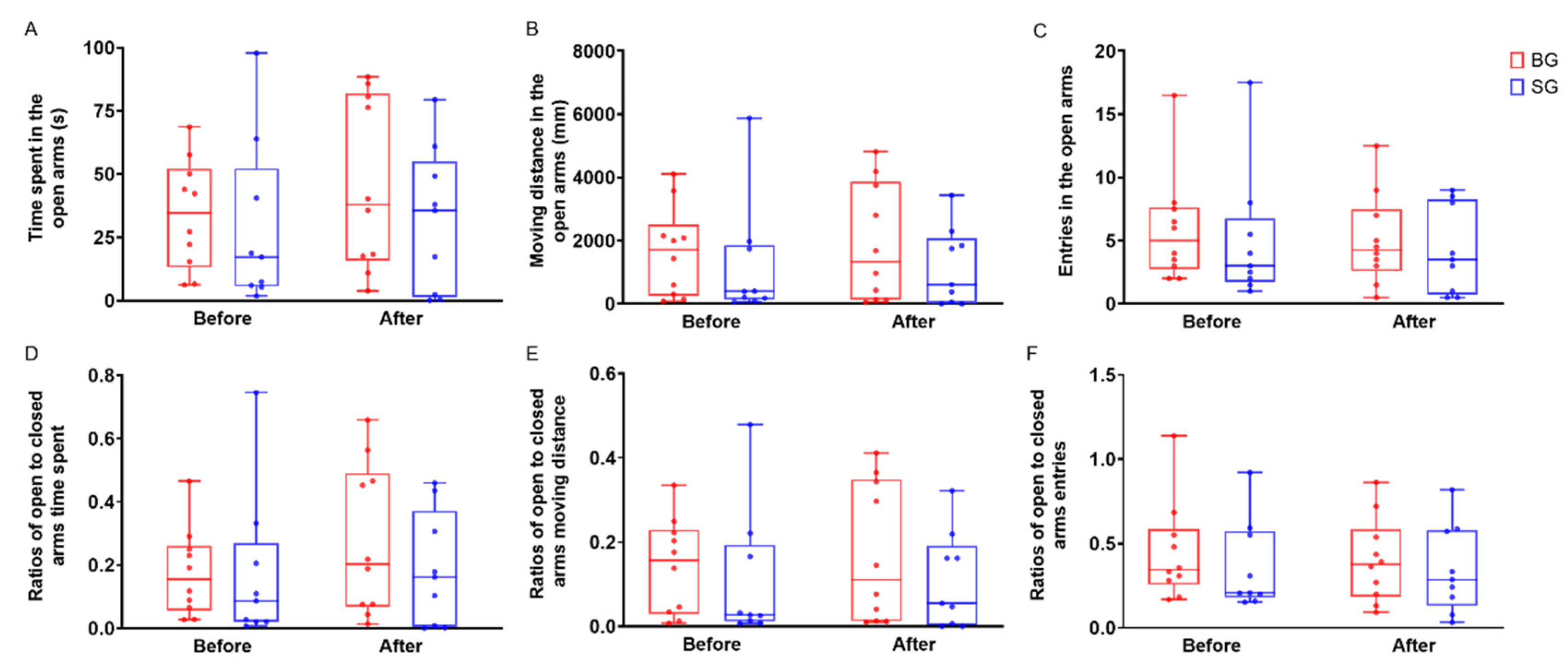
3.3. Boldness Behavior Changes after FMT
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Morais, L.H.; Schreiber, H.L.; Mazmanian, S.K. The Gut Microbiota–Brain Axis in Behaviour and Brain Disorders. Nat. Rev. Microbiol. 2021, 19, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.R.; Osadchiy, V.; Kalani, A.; Mayer, E.A. The Brain-Gut-Microbiome Axis. Cell Mol. Gastroenterol. Hepatol. 2018, 6, 133–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bo, T.-B.; Zhang, X.-Y.; Wen, J.; Deng, K.; Qin, X.-W.; Wang, D.-H. The Microbiota–Gut–Brain Interaction in Regulating Host Metabolic Adaptation to Cold in Male Brandt’s Voles (Lasiopodomys Brandtii). ISME J. 2019, 13, 3037–3053. [Google Scholar] [CrossRef] [PubMed]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between Microbiota and Immunity in Health and Disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Dabke, K.; Hendrick, G.; Devkota, S. The Gut Microbiome and Metabolic Syndrome. J. Clin. Investig. 2019, 129, 4050–4057. [Google Scholar] [CrossRef]
- Collins, J.; Borojevic, R.; Verdu, E.F.; Huizinga, J.D.; Ratcliffe, E.M. Intestinal Microbiota Influence the Early Postnatal Development of the Enteric Nervous System. Neurogastroenterol. Motil. 2014, 26, 98–107. [Google Scholar] [CrossRef]
- Wong, A.C.-N.; Wang, Q.-P.; Morimoto, J.; Senior, A.M.; Lihoreau, M.; Neely, G.G.; Simpson, S.J.; Ponton, F. Gut Microbiota Modifies Olfactory-Guided Microbial Preferences and Foraging Decisions in Drosophila. Curr. Biol. 2017, 27, 2397–2404.e4. [Google Scholar] [CrossRef]
- Neufeld, K.M.; Kang, N.; Bienenstock, J.; Foster, J.A. Reduced Anxiety-like Behavior and Central Neurochemical Change in Germ-Free Mice. Neurogastroenterol. Motil. 2011, 23, 255-e119. [Google Scholar] [CrossRef]
- Nishino, R.; Mikami, K.; Takahashi, H.; Tomonaga, S.; Furuse, M.; Hiramoto, T.; Aiba, Y.; Koga, Y.; Sudo, N. Commensal Microbiota Modulate Murine Behaviors in a Strictly Contamination-Free Environment Confirmed by Culture-Based Methods. Neurogastroenterol. Motil. 2013, 25, 521-e371. [Google Scholar] [CrossRef]
- Vuong, H.E.; Yano, J.M.; Fung, T.C.; Hsiao, E.Y. The Microbiome and Host Behavior. Annu. Rev. Neurosci. 2017, 40, 21–49. [Google Scholar] [CrossRef]
- Aatsinki, A.-K.; Lahti, L.; Uusitupa, H.-M.; Munukka, E.; Keskitalo, A.; Nolvi, S.; O’Mahony, S.; Pietilä, S.; Elo, L.L.; Eerola, E.; et al. Gut Microbiota Composition Is Associated with Temperament Traits in Infants. Brain Behav. Immunity 2019, 80, 849–858. [Google Scholar] [CrossRef] [PubMed]
- Gosling, S.D. Personality in Non-Human Animals: Animal Personality. Soc. Personal. Psychol. Compass 2008, 2, 985–1001. [Google Scholar] [CrossRef]
- Keiser, C.N.; Howell, K.A.; Pinter-Wollman, N.; Pruitt, J.N. Personality Composition Alters the Transmission of Cuticular Bacteria in Social Groups. Biol. Lett. 2016, 12, 20160297. [Google Scholar] [CrossRef] [PubMed]
- Kortet, R.; Hedrick, A.V.; Vainikka, A. Parasitism, Predation and the Evolution of Animal Personalities. Ecol. Lett. 2010, 13, 1449–1458. [Google Scholar] [CrossRef]
- Careau, V.; Montiglio, P.-O.; Garant, D.; Pelletier, F.; Speakman, J.R.; Humphries, M.M.; Réale, D. Energy Expenditure and Personality in Wild Chipmunks. Behav. Ecol. Sociobiol. 2015, 69, 653–661. [Google Scholar] [CrossRef]
- Myles-Gonzalez, E.; Burness, G.; Yavno, S.; Rooke, A.; Fox, M.G. To Boldly Go Where No Goby Has Gone before: Boldness, Dispersal Tendency, and Metabolism at the Invasion Front. Behav. Ecol. 2015, 26, 1083–1090. [Google Scholar] [CrossRef] [Green Version]
- Dhellemmes, F.; Finger, J.-S.; Smukall, M.J.; Gruber, S.H.; Guttridge, T.L.; Laskowski, K.L.; Krause, J. Personality-Driven Life History Trade-Offs Differ in Two Subpopulations of Free-Ranging Predators. J. Anim. Ecol. 2021, 90, 260–272. [Google Scholar] [CrossRef]
- Kishida, O.; Trussell, G.C.; Ohno, A.; Kuwano, S.; Ikawa, T.; Nishimura, K. Predation Risk Suppresses the Positive Feedback between Size Structure and Cannibalism. J. Anim. Ecol. 2011, 80, 1278–1287. [Google Scholar] [CrossRef]
- Blaszczyk, M.B. Primates Got Personality, Too: Toward an Integrative Primatology of Consistent Individual Differences in Behavior. Evol. Anthropol. 2020, 29, 56–67. [Google Scholar] [CrossRef]
- Pederson, A.K.; King, J.E.; Landau, V.I. Chimpanzee (Pan Troglodytes) Personality Predicts Behavior. J. Res. Per. 2005, 39, 534–549. [Google Scholar] [CrossRef]
- Budaev, S.V. “Personality” in the Guppy (Poecilia Reticulata): A Correlational Study of Exploratory Behavior and Social Tendency. J. Comp. Psychol. 1997, 111, 399–411. [Google Scholar] [CrossRef]
- Sih, A.; Mathot, K.J.; Moirón, M.; Montiglio, P.-O.; Wolf, M.; Dingemanse, N.J. Animal Personality and State–Behaviour Feedbacks: A Review and Guide for Empiricists. Trends Ecol. Evol. 2015, 30, 50–60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalile, B.; Van Oudenhove, L.; Vervliet, B.; Verbeke, K. The Role of Short-Chain Fatty Acids in Microbiota–Gut–Brain Communication. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 461–478. [Google Scholar] [CrossRef] [PubMed]
- Strandwitz, P.; Kim, K.H.; Terekhova, D.; Liu, J.K.; Sharma, A.; Levering, J.; McDonald, D.; Dietrich, D.; Ramadhar, T.R.; Lekbua, A.; et al. GABA Modulating Bacteria of the Human Gut Microbiota. Nat. Microbiol. 2019, 4, 396–403. [Google Scholar] [CrossRef] [PubMed]
- Batsaikhan, N.; Tsytsulina, K. Iucn Red List of Threatened Species: Meriones Unguiculatus. IUCN Red List Threat. Species 2016, e.T13171A115110851. [Google Scholar] [CrossRef]
- Wilson, D.; Reeder, D.M. Mammal Species of the World. A Taxonomic and Geographic Reference, 3rd ed.; Academic Press: Johns Hopkins, MD, USA, 2005. [Google Scholar]
- Liu, W.; Wang, G.; Wang, Y.; Zhong, W.; Wan, X. Population Ecology of Wild Mongolian Gerbils Meriones Unguiculatus. J. Mammal. 2009, 90, 832–840. [Google Scholar] [CrossRef]
- Agren, G.; Zhou, Q.; Zhong, W. Ecology and Social Behaviour of Mongolian Gerbils, Meriones Unguiculatus, at Xilinhot, Inner Mongolia, China. Anim. Behav. 1989, 37, 11–27. [Google Scholar] [CrossRef]
- Agren, G.; Zhou, Q.; Zhong, W. Territoriality, Cooperation and Resource Priority: Hoarding in the Mongolian Gerbil, Meriones Unguiculatus. Anim. Behav. 1989, 37, 28–32. [Google Scholar] [CrossRef]
- Yang, H.-D.; Wang, Q.; Wang, D.-H. Food Hoarding, but Not Food Intake, Is Attenuated by Acute Diazepam Treatment in Female Mongolian Gerbils (Meriones Unguiculatus). Horm. Behav. 2014, 66, 186–195. [Google Scholar] [CrossRef]
- Khakisahneh, S.; Zhang, X.; Nouri, Z.; Wang, D.; Zhang, K. Gut Microbiota and Host Thermoregulation in Response to Ambient Temperature Fluctuations Downloaded From. mSystems 2020, 5, e00514-20. [Google Scholar] [CrossRef]
- Liu, Q.-S.; Wang, D.-H. Effects of Diet Quality on Phenotypic Flexibility of Organ Size and Digestive Function in Mongolian Gerbils (Meriones Unguiculatus). J. Comp. Physiol. B 2007, 177, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Qi, Y.; Yang, X.; Zhao, L.; Wen, S.; Liu, Y.; Tang, L. Association between Polycystic Ovary Syndrome and Gut Microbiota. PLoS ONE 2016, 11, e0153196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deloris Alexander, A.; Orcutt, R.P.; Henry, J.C.; Baker, J.; Bissahoyo, A.C.; Threadgill, D.W. Quantitative PCR Assays for Mouse Enteric Flora Reveal Strain-Dependent Differences in Composition That Are Influenced by the Microenvironment. Mamm. Genome 2006, 17, 1093–1104. [Google Scholar] [CrossRef] [PubMed]
- Réale, D.; Reader, S.M.; Sol, D.; McDougall, P.T.; Dingemanse, N.J. Integrating Animal Temperament within Ecology and Evolution. Biol. Rev. 2007, 82, 291–318. [Google Scholar] [CrossRef] [Green Version]
- Vobrubová, B.; Landová, E.; Frynta, D. Methods for Measuring Mammalian Personalities: In Which Animals and How Accurately Can We Quantify It? Metody Měření Personality Savců: U Kterých Zvířat a Jak Přesně Ji Dokážeme Měřit? Lynx New Ser. 2017, 48, 183–198. [Google Scholar] [CrossRef] [Green Version]
- Carobrez, A.P.; Bertoglio, L.J. Ethological and Temporal Analyses of Anxiety-like Behavior: The Elevated plus-Maze Model 20 Years On. Neurosci. Biobehav. Rev. 2005, 29, 1193–1205. [Google Scholar] [CrossRef]
- Carter, A.J.; Feeney, W.E.; Marshall, H.H.; Cowlishaw, G.; Heinsohn, R. Animal Personality: What Are Behavioural Ecologists Measuring? Biol. Rev. 2013, 88, 465–475. [Google Scholar] [CrossRef] [Green Version]
- Dochtermann, N.A. Behavioral Syndromes: Carryover Effects, False Discovery Rates, and a Priori Hypotheses. Behav. Ecol. 2010, 21, 437–439. [Google Scholar] [CrossRef] [Green Version]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [Green Version]
- Anderson, M.J. PERMANOVA: A FORTRAN Computer Program for Permutational Multivariate Analysis of Variance; University of Auckland: Auckland, New Zealand, 2005; pp. 1–23. [Google Scholar]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic Biomarker Discovery and Explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douglas, G.M.; Maffei, V.J.; Zaneveld, J.R.; Yurgel, S.N.; Brown, J.R.; Taylor, C.M.; Huttenhower, C.; Langille, M.G.I. PICRUSt2 for Prediction of Metagenome Functions. Nat. Biotechnol. 2020, 38, 685–688. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Peres-Neto, P.R.; Jackson, D.A.; Somers, K.M. How Many Principal Components? Stopping Rules for Determining the Number of Non-Trivial Axes Revisited. Comput. Stat. Data Anal. 2005, 49, 974–997. [Google Scholar] [CrossRef]
- Hadfield, J.D. MCMC Methods for Multi-Response Generalized Linear Mixed Models: The MCMCglmm R Package. J. Stat. Soft. 2010, 33, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Cryan, J.F.; O’Riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The Microbiota-Gut-Brain Axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef]
- Colston, T.J.; Jackson, C.R. Microbiome Evolution along Divergent Branches of the Vertebrate Tree of Life: What Is Known and Unknown. Mol. Ecol. 2016, 25, 3776–3800. [Google Scholar] [CrossRef] [Green Version]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut Microbiota Functions: Metabolism of Nutrients and Other Food Components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Feehley, T.; Plunkett, C.H.; Bao, R.; Hong, S.M.C.; Culleen, E.; Belda-Ferre, P.; Campbell, E.; Aitoro, R.; Nocerino, R.; Paparo, L.; et al. Healthy Infants Harbor Intestinal Bacteria That Protect against Food Allergy. Nat. Med. 2019, 25, 448–453. [Google Scholar] [CrossRef]
- Xing, C.; Wang, M.; Ajibade, A.A.; Tan, P.; Fu, C.; Chen, L.; Zhu, M.; Hao, Z.-Z.; Chu, J.; Yu, X.; et al. Microbiota Regulate Innate Immune Signaling and Protective Immunity against Cancer. Cell Host Microbe 2021, 29, 959–974.e7. [Google Scholar] [CrossRef]
- Vital, M.; Howe, A.C.; Tiedje, J.M. Revealing the Bacterial Butyrate Synthesis Pathways by Analyzing (Meta)Genomic Data. mBio 2014, 5, e00889-14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, X.; Mao, B.; Gu, J.; Wu, J.; Cui, S.; Wang, G.; Zhao, J.; Zhang, H.; Chen, W. Blautia—A New Functional Genus with Potential Probiotic Properties? Gut Microbes 2021, 13, 1875796. [Google Scholar] [CrossRef] [PubMed]
- Lai, Z.-L.; Tseng, C.-H.; Ho, H.J.; Cheung, C.K.Y.; Lin, J.-Y.; Chen, Y.-J.; Cheng, F.-C.; Hsu, Y.-C.; Lin, J.-T.; El-Omar, E.M.; et al. Fecal Microbiota Transplantation Confers Beneficial Metabolic Effects of Diet and Exercise on Diet-Induced Obese Mice. Sci. Rep. 2018, 8, 15625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.-J.; Bakker, B.M. The Role of Short-Chain Fatty Acids in the Interplay between Diet, Gut Microbiota, and Host Energy Metabolism. J. Lipid. Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robles-Vera, I.; Callejo, M.; Ramos, R.; Duarte, J.; Perez-Vizcaino, F. Impact of Vitamin D Deficit on the Rat Gut Microbiome. Nutrients 2019, 11, 2564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, L.; Xiao, X.; Li, M.; Zhang, Q.; Yu, M.; Zheng, J.; Deng, M. Maternal Exercise Improves High-Fat Diet-Induced Metabolic Abnormalities and Gut Microbiota Profiles in Mouse Dams and Offspring. Front. Cell Infect. Microbiol. 2020, 10, 292. [Google Scholar] [CrossRef]
- Quiroga, R.; Nistal, E.; Estébanez, B.; Porras, D.; Juárez-Fernández, M.; Martínez-Flórez, S.; García-Mediavilla, M.V.; de Paz, J.A.; González-Gallego, J.; Sánchez-Campos, S.; et al. Exercise Training Modulates the Gut Microbiota Profile and Impairs Inflammatory Signaling Pathways in Obese Children. Exp. Mol. Med. 2020, 52, 1048–1061. [Google Scholar] [CrossRef]
- Behrens, J.W.; von Friesen, L.W.; Brodin, T.; Ericsson, P.; Hirsch, P.E.; Persson, A.; Sundelin, A.; van Deurs, M.; Nilsson, P.A. Personality- and Size-Related Metabolic Performance in Invasive Round Goby (Neogobius Melanostomus). Physiol. Behav. 2020, 215, 112777. [Google Scholar] [CrossRef]
- Réale, D.; Garant, D.; Humphries, M.M.; Bergeron, P.; Careau, V.; Montiglio, P.-O. Personality and the Emergence of the Pace-of-Life Syndrome Concept at the Population Level. Phil. Trans. R. Soc. B 2010, 365, 4051–4063. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Xu, J.; Xu, Y.; Xiao, J.; Bi, N.; Gu, X.; Wang, H.-L. Gut Microbiota Shapes Social Dominance through Modulating HDAC2 in the Medial Prefrontal Cortex. Cell Rep. 2022, 38. [Google Scholar] [CrossRef]
- Chun, J.L.; Ji, S.Y.; Lee, S.D.; Lee, Y.K.; Kim, B.; Kim, K.H. Difference of Gut Microbiota Composition Based on the Body Condition Scores in Dogs. J. Anim. Sci. Technol. 2020, 62, 239–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, J.; Wang, T.; Xu, Y.; Gu, X.; Li, D.; Niu, K.; Wang, T.; Zhao, J.; Zhou, R.; Wang, H.-L. Long-Term Probiotic Intervention Mitigates Memory Dysfunction through a Novel H3K27me3-Based Mechanism in Lead-Exposed Rats. Transl. Psychiatry 2020, 10, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, K.; Mu, C.; Farzi, A.; Zhu, W. Tryptophan Metabolism: A Link Between the Gut Microbiota and Brain. Adv. Nutr. 2020, 11, 709–723. [Google Scholar] [CrossRef] [PubMed]
- Arnold, K.E.; Ramsay, S.L.; Donaldson, C.; Adam, A. Parental Prey Selection Affects Risk-Taking Behaviour and Spatial Learning in Avian Offspring. Proc. Biol. Sci. 2007, 274, 2563–2569. [Google Scholar] [CrossRef] [Green Version]
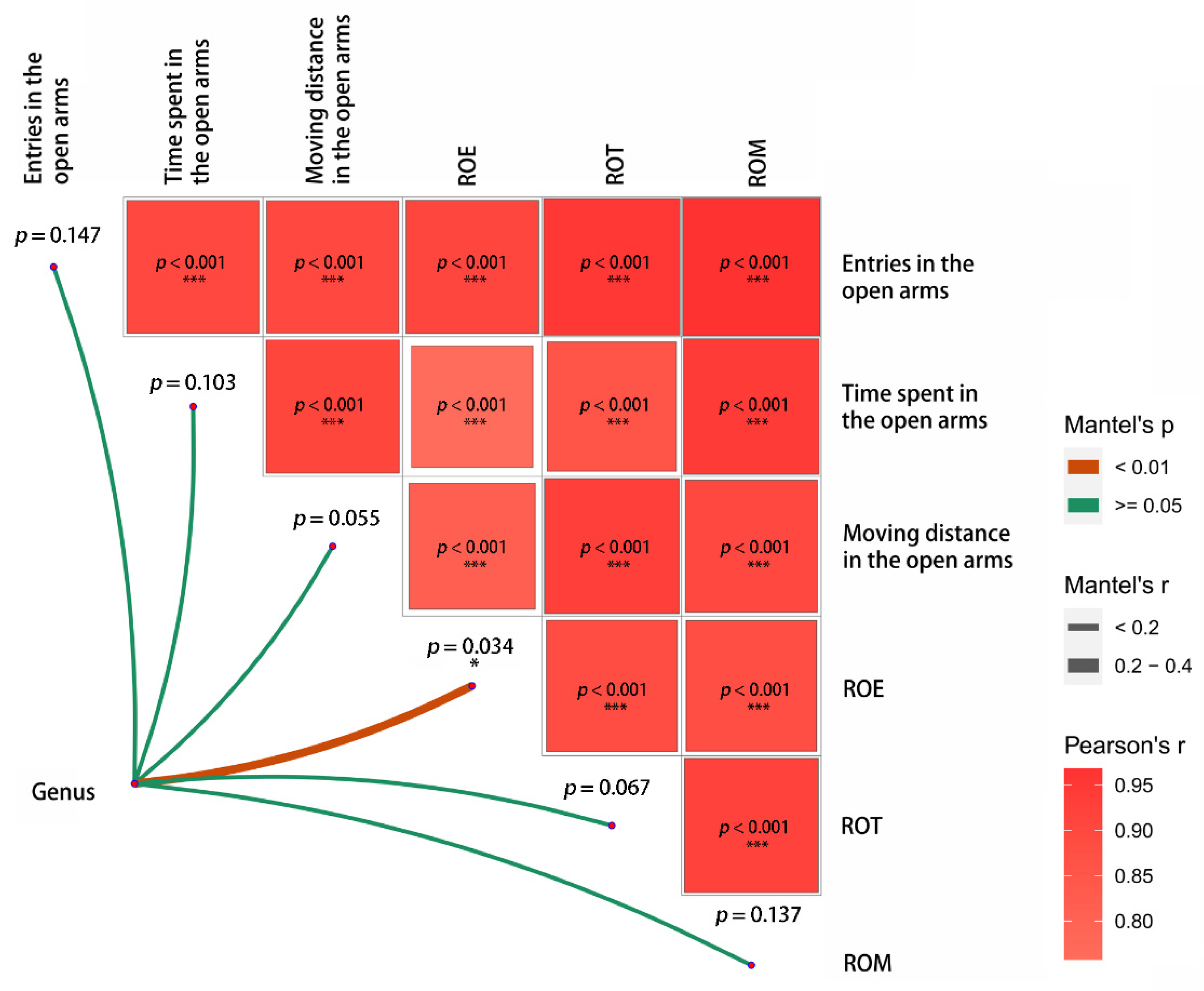
| Parameters | Component 1 (PC1) | Component 2 (PC2) |
|---|---|---|
| Entries in the open arms | −0.4207 | 0.2240 |
| Time spent in the open arms | −0.4160 | −0.1926 |
| Moving distance in the open arms | −0.3769 | −0.7419 |
| ROE | −0.4028 | 0.4844 |
| ROT | −0.4203 | 0.3209 |
| ROM | −0.4112 | −0.1569 |
| Eigenvalue | 5.04 | 0.47 |
| Total variance (%) | 84.04% | 7.83% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gan, L.; Bo, T.; Liu, W.; Wang, D. The Gut Microbiota May Affect Personality in Mongolian Gerbils. Microorganisms 2022, 10, 1054. https://doi.org/10.3390/microorganisms10051054
Gan L, Bo T, Liu W, Wang D. The Gut Microbiota May Affect Personality in Mongolian Gerbils. Microorganisms. 2022; 10(5):1054. https://doi.org/10.3390/microorganisms10051054
Chicago/Turabian StyleGan, Lin, Tingbei Bo, Wei Liu, and Dehua Wang. 2022. "The Gut Microbiota May Affect Personality in Mongolian Gerbils" Microorganisms 10, no. 5: 1054. https://doi.org/10.3390/microorganisms10051054
APA StyleGan, L., Bo, T., Liu, W., & Wang, D. (2022). The Gut Microbiota May Affect Personality in Mongolian Gerbils. Microorganisms, 10(5), 1054. https://doi.org/10.3390/microorganisms10051054








