COVID-19 Genetic Variants and Their Potential Impact in Vaccine Development
Abstract
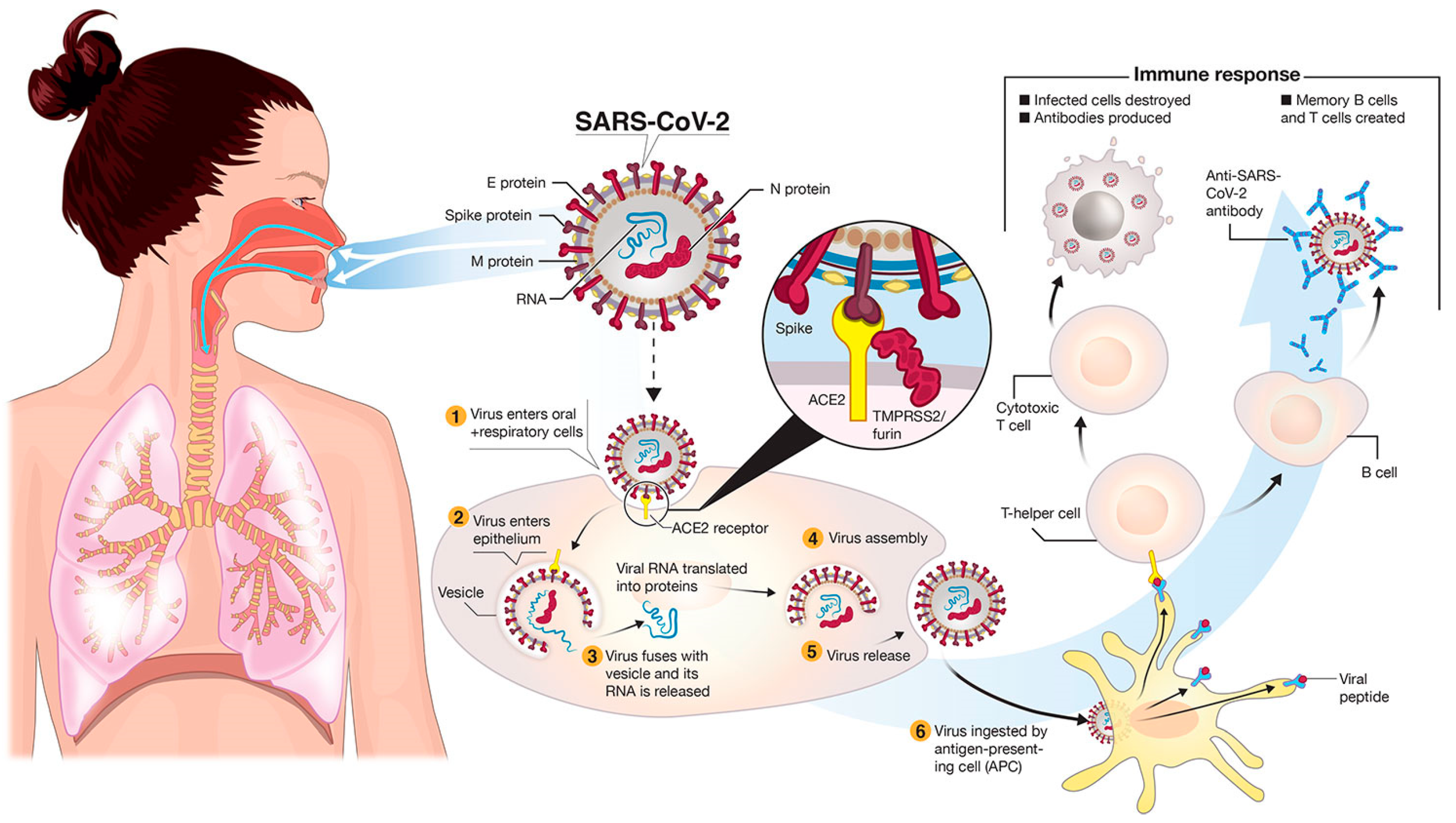
:1. Introduction
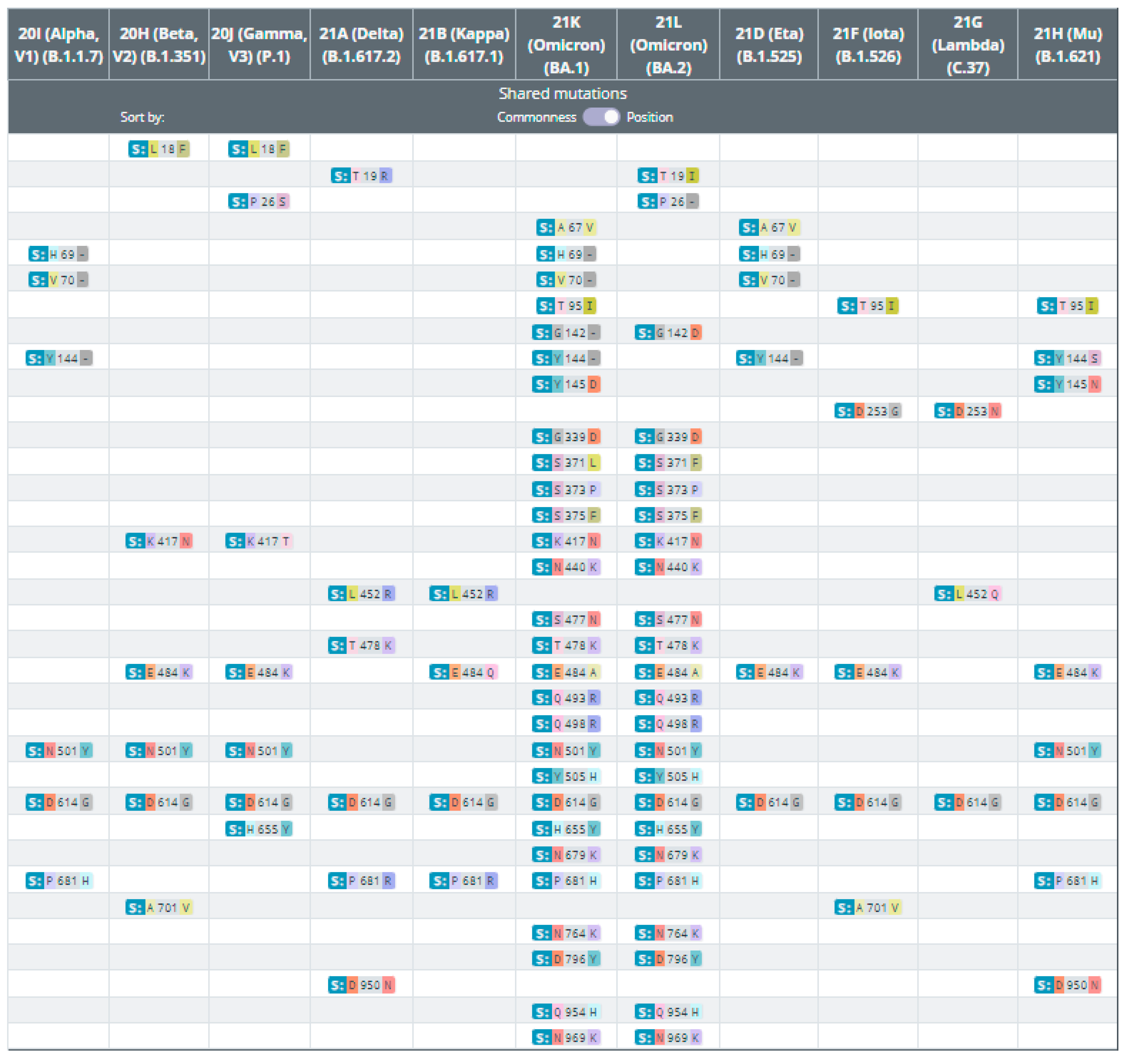
2. Genetic Variants of SARS-CoV-2
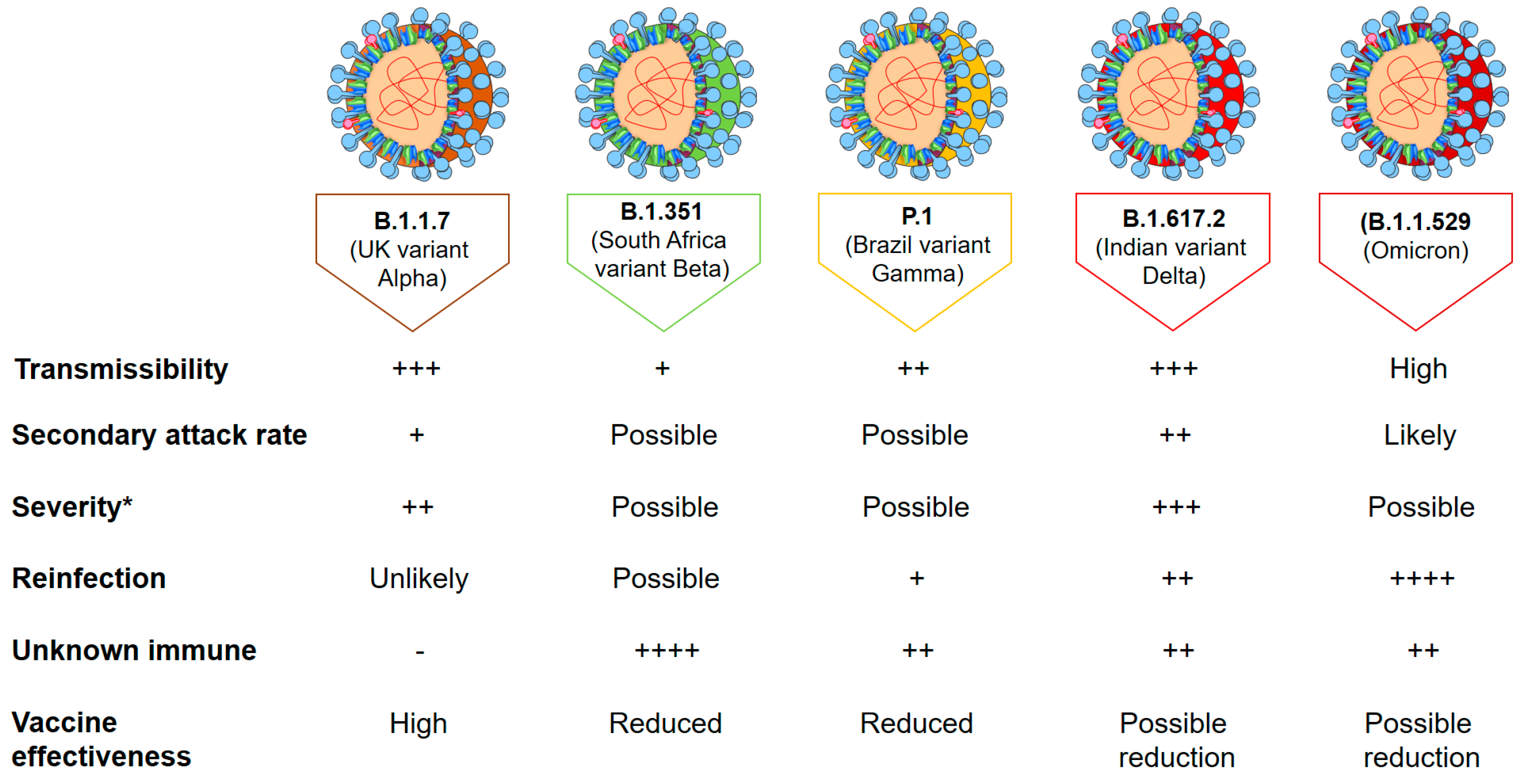
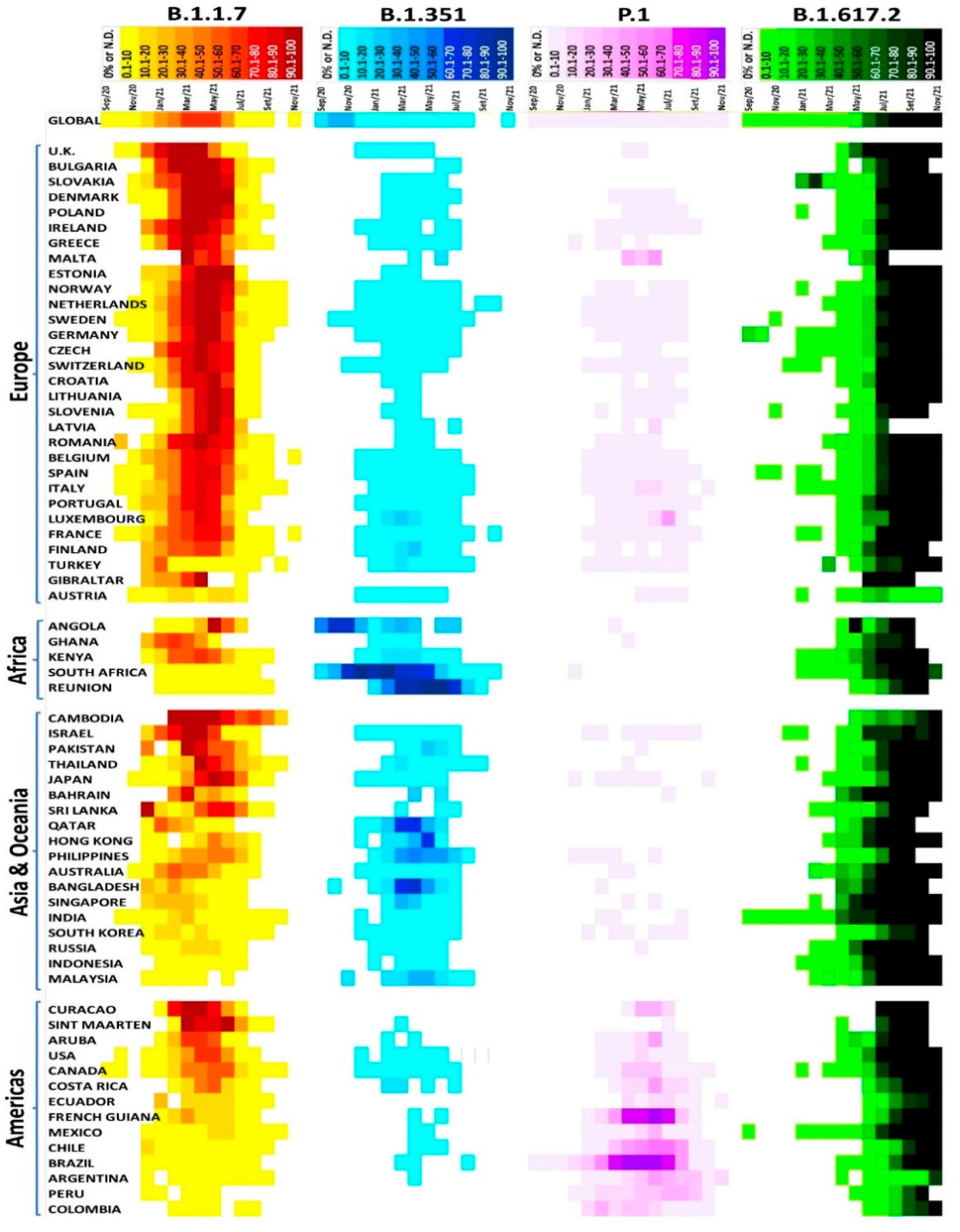
3. Virus Variants of Concern (VOCs) and Former VOCs
4. Variants of Interest (VOIs) and Former VOIs
5. Variants under Monitoring (VUMs)
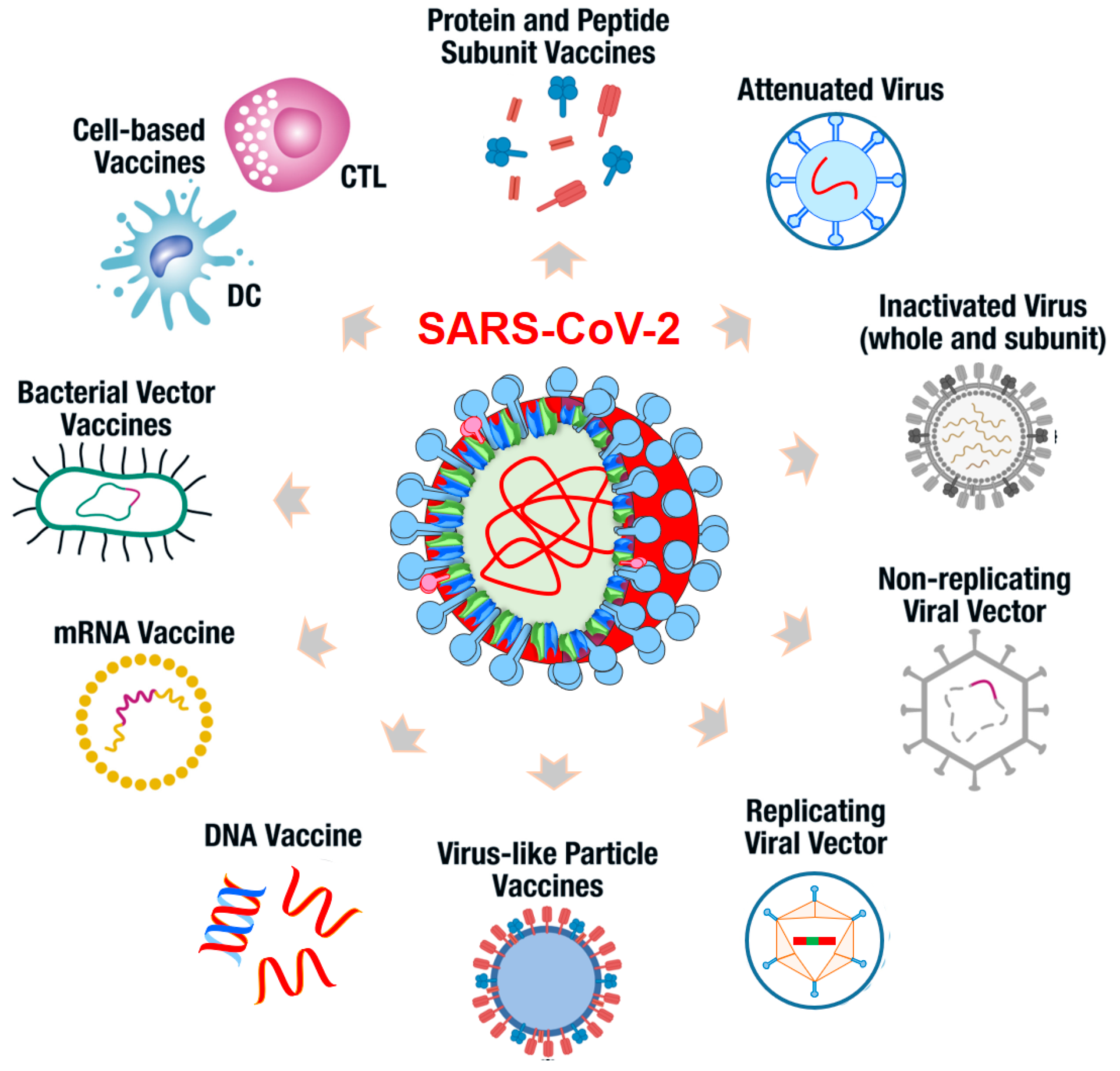
6. Current SARS-CoV-2 Vaccine Platforms
7. Potential Mixing, Matching SARS-CoV-2 Doses and Booster Vaccines
8. Potential SARS-CoV-2 Variant Evolution Trend
9. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef]
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y.; et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, M.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach, Biochimica et biophysica acta. Mol. Basis Dis. 2020, 1866, 165878. [Google Scholar] [CrossRef] [PubMed]
- Masters, P.S. The molecular biology of coronaviruses. Adv. Virus Res. 2006, 66, 193–292. [Google Scholar]
- Chen, Y.; Liu, Q.; Guo, D. Emerging coronaviruses: Genome structure, replication, and pathogenesis. J. Med. Virol. 2020, 92, 418–423. [Google Scholar] [CrossRef] [PubMed]
- Funk, C.D.; Laferrière, C.; Ardakani, A. A Snapshot of the Global Race for Vaccines Targeting SARS-CoV-2 and the COVID-19 Pandemic. Front. Pharmacol. 2020, 11, 937. [Google Scholar] [CrossRef] [PubMed]
- Kuhlmann, C.; Mayer, C.K.; Claassen, M.; Maponga, T.; Burgers, W.A.; Keeton, R.; Riou, C.; Sutherland, A.D.; Suliman, T.; Shaw, M.L.; et al. Breakthrough infections with SARS-CoV-2 omicron despite mRNA vaccine booster dose. Lancet 2022, 399, 625–626. [Google Scholar] [CrossRef]
- Lauring, A.S.; Hodcroft, E.B. Genetic Variants of SARS-CoV-2-What Do They Mean? JAMA 2021, 325, 529–531. [Google Scholar] [CrossRef] [PubMed]
- NGrubaugh, D.; Petrone, M.E.; Holmes, E.C. We shouldn’t worry when a virus mutates during disease outbreaks. Nat. Microbiol. 2020, 5, 529–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, Y.W.; Chao, T.L.; Li, C.L.; Wang, S.H.; Kao, H.C.; Tsai, Y.M.; Wang, H.Y.; Hsieh, C.L.; Lin, Y.Y.; Chen, P.J.; et al. D614G Substitution of SARS-CoV-2 Spike Protein Increases Syncytium Formation and Virus Titer via Enhanced Furin-Mediated Spike Cleavage. mBio 2021, 12, e0058721. [Google Scholar] [CrossRef]
- Li, J.; Lai, S.; Gao, G.F.; Shi, W. The emergence, genomic diversity and global spread of SARS-CoV-2. Nature 2021, 600, 408–418. [Google Scholar] [CrossRef] [PubMed]
- Korber, B.; Fischer, W.M.; Gnanakaran, S.; Yoon, H.; Theiler, J.; Abfalterer, W.; Hengartner, N.; Giorgi, E.E.; Bhattacharya, T.; Foley, B.; et al. Tracking Changes in SARS-CoV-2 Spike: Evidence that D614G Increases Infectivity of the COVID-19 Virus. Cell 2020, 182, 812–827.e19. [Google Scholar] [CrossRef]
- Plante, J.A.; Liu, Y.; Liu, J.; Xia, H.; Johnson, B.A.; Lokugamage, K.G.; Zhang, X.; Muruato, A.E.; Zou, J.; Fontes-Garfias, C.R.; et al. Spike mutation D614G alters SARS-CoV-2 fitness. Nature 2021, 592, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.Y.; Kung, Y.A.; Huang, P.N.; Chang, S.Y.; Gong, Y.N.; Han, Y.J.; Chiang, H.J.; Liu, K.T.; Lee, K.M.; Chang, C.Y.; et al. Stability of SARS-CoV-2 Spike G614 Variant Surpasses That of the D614 Variant after Cold Storage. mSphere 2021, 6, e00104-21. [Google Scholar] [CrossRef] [PubMed]
- Gamage, A.M.; Tan, K.S.; Chan, W.O.Y.; Liu, J.; Tan, C.W.; Ong, Y.K.; Thong, M.; Andiappan, A.K.; Anderson, D.E.; Wang, Y.; et al. Infection of human Nasal Epithelial Cells with SARS-CoV-2 and a 382-nt deletion isolate lacking ORF8 reveals similar viral kinetics and host transcriptional profiles. PLoS Pathog. 2020, 16, e1009130. [Google Scholar] [CrossRef]
- Davies, N.G.; Abbott, S.; Barnard, R.C.; Jarvis, C.I.; Kucharski, A.J.; Munday, J.D.; Pearson, C.A.B.; Russell, T.W.; Tully, D.C.; Washburne, A.D.; et al. Estimated transmissibility and impact of SARS-CoV-2 lineage B.1.1.7 in England. Science 2021, 372, eabg3055. [Google Scholar] [CrossRef]
- Rajah, M.M.; Hubert, M.; Bishop, E.; Saunders, N.; Robinot, R.; Grzelak, L.; Planas, D.; Dufloo, J.; Gellenoncourt, S.; Bongers, A.; et al. SARS-CoV-2 Alpha, Beta, and Delta variants display enhanced Spike-mediated syncytia formation. EMBO J. 2021, 40, e108944. [Google Scholar] [CrossRef] [PubMed]
- Cetin, M.; Balci, P.O.; Sivgin, H.; Cetin, S.; Ulgen, A.; Demir, H.D.; Li, W. Alpha variant (B.1.1.7) of SARS-CoV-2 increases fatality-rate for patients under age of 70 years and hospitalization risk overall. Acta Microbiol. Immunol. Hung. 2021, 68, 153–161. [Google Scholar] [CrossRef]
- Meyer, M.; Holfter, A.; Ruebsteck, E.; Gruell, H.; Dewald, F.; Koerner, R.W.; Klein, F.; Lehmann, C.; Huenseler, C.; Weber, L.T. The Alpha Variant (B.1.1.7) of SARS-CoV-2 in Children: First Experience from 3544 Nucleic Acid Amplification Tests in a Cohort of Children in Germany. Viruses 2021, 13, 1600. [Google Scholar] [CrossRef] [PubMed]
- Tegally, H.; Wilkinson, E.; Giovanetti, M.; Iranzadeh, A.; Fonseca, V.; Giandhari, J.; Doolabh, D.; Pillay, S.; San, E.J.; Msomi, N.; et al. Detection of a SARS-CoV-2 variant of concern in South Africa. Nature 2021, 592, 438–443. [Google Scholar] [CrossRef]
- Istifli, E.S.; Netz, P.A.; Tepe, A.S.; Sarikurkcu, C.; Tepe, B. Understanding the molecular interaction of SARS-CoV-2 spike mutants with ACE2 (angiotensin converting enzyme 2). J. Biomol. Struct. Dyn. 2021, 1–12. [Google Scholar] [CrossRef]
- Greaney, A.J.; Loes, A.N.; Crawford, K.H.D.; Starr, T.N.; Malone, K.D.; Chu, H.Y.; Bloom, J.D. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host Microbe 2021, 29, 463–476.e6. [Google Scholar] [CrossRef] [PubMed]
- Collier, D.A.; de Marco, A.; Ferreira, I.; Meng, B.; Datir, R.P.; Walls, A.C.; Kemp, S.A.; Bassi, J.; Pinto, D.; Silacci-Fregni, C.; et al. Sensitivity of SARS-CoV-2 B.1.1.7 to mRNA vaccine-elicited antibodies. Nature 2021, 593, 136–141. [Google Scholar] [CrossRef]
- Chen, Q.; Huang, X.Y.; Tian, Y.; Fan, C.; Sun, M.; Zhou, C.; Li, R.; Zhang, R.R.; Wu, G.; Qin, C.F. The Infection and Pathogenicity of SARS-CoV-2 Variant B.1.351 in hACE2 Mice. Virol. Sin. 2021, 36, 1232–1235. [Google Scholar] [CrossRef]
- Buss, L.F.; Prete, C.A., Jr.; Abrahim, C.M.M.; Mendrone, A., Jr.; Salomon, T.; de Almeida-Neto, C.; França, R.F.O.; Belotti, M.C.; Carvalho, M.; Costa, A.G.; et al. Three-quarters attack rate of SARS-CoV-2 in the Brazilian Amazon during a largely unmitigated epidemic. Science 2021, 371, 288–292. [Google Scholar] [CrossRef]
- Faria, N.R.; Mellan, T.A.; Whittaker, C.; Claro, I.M.; Candido, D.D.S.; Mishra, S.; Crispim, M.A.E.; Sales, F.C.S.; Hawryluk, I.; McCrone, J.T.; et al. Genomics and epidemiology of the P.1 SARS-CoV-2 lineage in Manaus, Brazil. Science 2021, 372, 815–821. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, V.; Lucas, A.; Panja, S.; Miyauchi, R.; Mallela, K.M.G. Receptor binding, immune escape, and protein stability direct the natural selection of SARS-CoV-2 variants. J. Biol. Chem. 2021, 297, 101208. [Google Scholar] [CrossRef] [PubMed]
- He, X.; He, C.; Hong, W.; Zhang, K.; Wei, X. The challenges of COVID-19 Delta variant: Prevention and vaccine development. MedComm 2021, 2, 846–854. [Google Scholar] [CrossRef] [PubMed]
- Alhasan, K.; Aljamaan, F.; Temsah, M.H.; Alshahrani, F.; Bassrawi, R.; Alhaboob, A.; Assiri, R.; Alenezi, S.; Alaraj, A.; Alhomoudi, R.I.; et al. COVID-19 Delta Variant: Perceptions, Worries, and Vaccine-Booster Acceptability among Healthcare Workers. Healthcare 2021, 9, 1566. [Google Scholar] [CrossRef] [PubMed]
- Shiehzadegan, S.; Alaghemand, N.; Fox, M.; Venketaraman, V. Analysis of the Delta Variant B.1.617.2 COVID-19. Clin. Pract. 2021, 11, 778–784. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Xiao, T.; Cai, Y.; Lavine, C.L.; Peng, H.; Zhu, H.; Anand, K.; Tong, P.; Gautam, A.; Mayer, M.L.; et al. Membrane fusion and immune evasion by the spike protein of SARS-CoV-2 Delta variant. Science 2021, 374, 1353–1360. [Google Scholar] [CrossRef] [PubMed]
- Luo, C.H.; Morris, C.P.; Sachithanandham, J.; Amadi, A.; Gaston, D.; Li, M.; Swanson, N.J.; Schwartz, M.; Klein, E.Y.; Pekosz, A.; et al. Infection with the SARS-CoV-2 Delta Variant is Associated with Higher Infectious Virus Loads Compared to the Alpha Variant in both Unvaccinated and Vaccinated Individuals. medRxiv 2021. [Google Scholar] [CrossRef]
- Hendaus, M.A.; Jomha, F.A. Delta variant of COVID-19: A simple explanation. Qatar Med. J. 2021, 2021, 49. [Google Scholar] [CrossRef]
- Baraniuk, C. Covid-19: How effective are vaccines against the delta variant? BMJ 2021, 374, n1960. [Google Scholar] [CrossRef] [PubMed]
- Baj, A.; Novazzi, F.; Pasciuta, R.; Genoni, A.; Ferrante, F.D.; Valli, M.; Partenope, M.; Tripiciano, R.; Ciserchia, A.; Catanoso, G.; et al. Breakthrough Infections of E484K-Harboring SARS-CoV-2 Delta Variant, Lombardy, Italy. Emerg. Infect. Dis. 2021, 27, 3180–3182. [Google Scholar] [CrossRef]
- Rahman, F.I.; Ether, S.A.; Islam, M.R. The “Delta Plus” COVID-19 variant has evolved to become the next potential variant of concern: Mutation history and measures of prevention. J. Basic Clin. Physiol. Pharmacol. 2021, 33, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Ferré, V.M.; Peiffer-Smadja, N.; Visseaux, B.; Descamps, D.; Ghosn, J.; Charpentier, C. Omicron SARS-CoV-2 variant: What we know and what we don’t. Anaesth. Crit. Care Pain Med. 2021, 41, 100998. [Google Scholar] [CrossRef]
- Kannan, S.R.; Spratt, A.N.; Sharma, K.; Chand, H.S.; Byrareddy, S.N.; Singh, K. Omicron SARS-CoV-2 variant: Unique features and their impact on pre-existing antibodies. J. Autoimmun. 2022, 126, 102779. [Google Scholar] [CrossRef] [PubMed]
- Quarleri, J.; Galvan, V.; Delpino, M.V. Omicron variant of the SARS-CoV-2: A quest to define the consequences of its high mutational load. GeroScience 2022, 44, 53–56. [Google Scholar] [CrossRef]
- Kumar, S.; Thambiraja, T.S.; Karuppanan, K.; Subramaniam, G. Omicron and Delta variant of SARS-CoV-2: A comparative computational study of spike protein. J. Med. Virol. 2022, 94, 1641–1649. [Google Scholar] [CrossRef] [PubMed]
- Shanmugaraj, B.; Malla, A.; Khorattanakulchai, N.; Phoolcharoen, W. SARS-CoV-2 omicron variant: Could it be another threat? J. Med. Virol. 2022, 94, 1284–1288. [Google Scholar] [CrossRef] [PubMed]
- Ai, J.; Zhang, H.; Zhang, Y.; Lin, K.; Zhang, Y.; Wu, J.; Wan, Y.; Huang, Y.; Song, J.; Fu, Z.; et al. Omicron variant showed lower neutralizing sensitivity than other SARS-CoV-2 variants to immune sera elicited by vaccines after boost. Emerg. Microbes Infect. 2021, 11, 337–343. [Google Scholar] [CrossRef]
- Diamond, M.; Halfmann, P.; Maemura, T.; Iwatsuki-Horimoto, K.; Iida, S.; Kiso, M.; Scheaffer, S.; Darling, T.; Joshi, A.; Loeber, S.; et al. The SARS-CoV-2 B.1.1.529 Omicron virus causes attenuated infection and disease in mice and hamsters. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Nealon, J.; Cowling, B.J. Omicron severity: Milder but not mild. Lancet 2022, 399, 412–413. [Google Scholar] [CrossRef]
- Tchesnokova, V.; Kulasekara, H.; Larson, L.; Bowers, V.; Rechkina, E.; Kisiela, D.; Sledneva, Y.; Choudhury, D.; Maslova, I.; Deng, K.; et al. Acquisition of the L452R Mutation in the ACE2-Binding Interface of Spike Protein Triggers Recent Massive Expansion of SARS-CoV-2 Variants. J. Clin. Microbiol. 2021, 59, e0092121. [Google Scholar] [CrossRef]
- Motozono, C.; Toyoda, M.; Zahradnik, J.; Saito, A.; Nasser, H.; Tan, T.S.; Ngare, I.; Kimura, I.; Uriu, K.; Kosugi, Y.; et al. SARS-CoV-2 spike L452R variant evades cellular immunity and increases infectivity. Cell Host Microbe 2021, 29, 1124–1136.e11. [Google Scholar] [CrossRef]
- Yang, S.; Hemarajata, P.; Hilt, E.E.; Price, T.K.; Garner, O.B.; Green, N.M. Investigation of SARS-CoV-2 Epsilon Variant and Hospitalization Status by Genomic Surveillance in a Single Large Health System During the 2020–2021 Winter Surge in Southern California. Am. J. Clin. Pathol. 2021, aqab203. [Google Scholar] [CrossRef] [PubMed]
- Romero, P.E.; Dávila-Barclay, A.; Salvatierra, G.; González, L.; Cuicapuza, D.; Solís, L.; Marcos-Carbajal, P.; Huancachoque, J.; Maturrano, L.; Tsukayama, P. The Emergence of Sars-CoV-2 Variant Lambda (C.37) in South America. Microbiol. Spectr. 2021, 9, e0078921. [Google Scholar] [CrossRef]
- Chen, J.; Wang, R.; Wang, M.; Wei, G.W. Mutations Strengthened SARS-CoV-2 Infectivity. J. Mol. Biol. 2020, 432, 5212–5226. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; VanBlargan, L.A.; Bloyet, L.M.; Rothlauf, P.W.; Chen, R.E.; Stumpf, S.; Zhao, H.; Errico, J.M.; Theel, E.S.; Liebeskind, M.J.; et al. Identification of SARS-CoV-2 spike mutations that attenuate monoclonal and serum antibody neutralization. Cell Host Microbe 2021, 29, 477–488.e4. [Google Scholar] [CrossRef]
- Vargas-Herrera, N.; Araujo-Castillo, R.V.; Mestanza, O.; Galarza, M.; Rojas-Serrano, N.; Solari-Zerpa, L. SARS-CoV-2 Lambda and Gamma variants competition in Peru, a country with high seroprevalence, Lancet Regional Health. Americas 2022, 6, 100112. [Google Scholar]
- McCallum, M.; Walls, A.C.; Sprouse, K.R.; Bowen, J.E.; Rosen, L.E.; Dang, H.V.; de Marco, A.; Franko, N.; Tilles, S.W.; Logue, J.; et al. Molecular basis of immune evasion by the Delta and Kappa SARS-CoV-2 variants. Science 2021, 374, 1621–1626. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Huynh, T.; Stauft, C.B.; Wang, T.T.; Luan, B. Structure-Function Analysis of Resistance to Bamlanivimab by SARS-CoV-2 Variants Kappa, Delta, and Lambda. J. Chem. Inf. Model. 2021, 61, 5133–5140. [Google Scholar] [CrossRef] [PubMed]
- Singh, J.; Malhotra, A.G.; Biswas, D.; Shankar, P.; Lokhande, L.; Yadav, A.K.; Raghuvanshi, A.; Kale, D.; Nema, S.; Saigal, S.; et al. Relative Consolidation of the Kappa Variant Pre-Dates the Massive Second Wave of COVID-19 in India. Genes 2021, 12, 1803. [Google Scholar] [CrossRef] [PubMed]
- Pereira, F.; Tosta, S.; Lima, M.M.; Reboredo de Oliveira da Silva, L.; Nardy, V.B.; Gómez, M.K.A.; Lima, J.G.; Fonseca, V.; de Oliveira, T.; Lourenço, J.; et al. Genomic surveillance activities unveil the introduction of the SARS-CoV-2 B.1.525 variant of interest in Brazil: Case report. J. Med. Virol. 2021, 93, 5523–5526. [Google Scholar] [CrossRef] [PubMed]
- Annavajhala, M.K.; Mohri, H.; Wang, P.; Nair, M.; Zucker, J.E.; Sheng, Z.; Gomez-Simmonds, A.; Kelley, A.L.; Tagliavia, M.; Huang, Y.; et al. Emergence and expansion of SARS-CoV-2 B.1.526 after identification in New York. Nature 2021, 597, 703–708. [Google Scholar] [CrossRef] [PubMed]
- Uriu, K.; Kimura, I.; Shirakawa, K.; Takaori-Kondo, A.; Nakada, T.A.; Kaneda, A.; Nakagawa, S.; Sato, K. Neutralization of the SARS-CoV-2 Mu Variant by Convalescent and Vaccine Serum. N. Engl. J. Med. 2021, 385, 2397–2399. [Google Scholar] [CrossRef]
- Laiton-Donato, K.; Franco-Muñoz, C.; Álvarez-Díaz, D.A.; Ruiz-Moreno, H.A.; Usme-Ciro, J.A.; Prada, D.A.; Reales-González, J.; Corchuelo, S.; Herrera-Sepúlveda, M.T.; Naizaque, J.; et al. Characterization of the emerging B.1.621 variant of interest of SARS-CoV-2. Infect. Genet. Evol. 2021, 95, 105038. [Google Scholar] [CrossRef] [PubMed]
- Lou, F.; Li, M.; Pang, Z.; Jiang, L.; Guan, L.; Tian, L.; Hu, J.; Fan, J.; Fan, H. Understanding the Secret of SARS-CoV-2 Variants of Concern/Interest and Immune Escape. Front. Immunol. 2021, 12, 744242. [Google Scholar] [CrossRef] [PubMed]
- Messali, S.; Bertelli, A.; Campisi, G.; Zani, A.; Ciccozzi, M.; Caruso, A.; Caccuri, F. A cluster of the new SARS-CoV-2 B.1.621 lineage in Italy and sensitivity of the viral isolate to the BNT162b2 vaccine. J. Med. Virol. 2021, 93, 6468–6470. [Google Scholar] [CrossRef]
- Tablizo, F.A.; Kim, K.M.; Lapid, C.M.; Castro, M.J.R.; Yangzon, M.S.L.; Maralit, B.A.; Ayes, M.E.C.; la Paz, E.M.C.; de Guzman, A.R.; Yap, J.M.C.; et al. Genome sequencing and analysis of an emergent SARS-CoV-2 variant characterized by multiple spike protein mutations detected from the Central Visayas Region of the Philippines. medRxiv 2021. [Google Scholar] [CrossRef]
- Rose, R.; Nolan, D.J.; LaFleur, T.M.; Lamers, S.L. Outbreak of P.3 (Theta) SARS-CoV-2 emerging variant of concern among service workers in Louisiana. J. Infect. Public Health 2022, 15, 7–9. [Google Scholar] [CrossRef]
- Voloch, C.M.; da Silva Francisco, R., Jr.; de Almeida, L.G.P.; Cardoso, C.C.; Brustolini, O.J.; Gerber, A.L.; Guimarães, A.P.C.; Mariani, D.; da Costa, R.M.; Ferreira, O.C.; et al. Genomic characterization of a novel SARS-CoV-2 lineage from Rio de Janeiro, Brazil. J. Virol. 2021, 95, e00119-21. [Google Scholar] [CrossRef] [PubMed]
- Kannan, S.R.; Spratt, A.N.; Cohen, A.R.; Naqvi, S.H.; Chand, H.S.; Quinn, T.P.; Lorson, C.L.; Byrareddy, S.N.; Singh, K. Evolutionary analysis of the Delta and Delta Plus variants of the SARS-CoV-2 viruses. J. Autoimmun. 2021, 124, 102715. [Google Scholar] [CrossRef] [PubMed]
- Angeletti, S.; Giovanetti, M.; Fogolari, M.; Cella, E.; de Florio, L.; Lintas, C.; Veralli, R.; Francesconi, M.; Caccuri, F.; de Cesaris, M.; et al. SARS-CoV-2 AY.4.2 variant circulating in Italy: Genomic preliminary insight. J. Med. Virol. 2021, 94, 1689–1692. [Google Scholar] [CrossRef]
- le Page, M. New variant gains ground. New Sci. 2021, 252, 8. [Google Scholar] [CrossRef]
- Arora, P.; Kempf, A.; Nehlmeier, I.; Graichen, L.; Winkler, M.S.; Lier, M.; Schulz, S.; Jäck, H.M.; Pöhlmann, S.; Hoffmann, M. No evidence for increased cell entry or antibody evasion by Delta sublineage AY.4.2. Cell. Mol. Immunol. 2022, 19, 449–452. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, F.; Abadi, A.T.B. Is Omicron the last SARS-CoV-2 Variant of Concern? Arch. Med. Res. 2022. [Google Scholar] [CrossRef]
- Yang, J.; Wang, W.; Chen, Z.; Lu, S.; Yang, F.; Bi, Z.; Bao, L.; Mo, F.; Li, X.; Huang, Y.; et al. A vaccine targeting the RBD of the S protein of SARS-CoV-2 induces protective immunity. Nature 2020, 586, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Loconsole, D.; Sallustio, A.; Accogli, M.; Centrone, F.; Capozzi, L.; del Sambro, L.; Parisi, A.; Chironna, M. Genome Sequence of a SARS-CoV-2 VUI 202012/01 Strain Identified from a Patient Returning from London, England, to the Apulia Region of Italy. Microbiol. Resour. Announc. 2021, 10, e01487-20. [Google Scholar] [CrossRef]
- Giovacchini, N.; Coppi, M.; Aiezza, N.; Baccani, I.; Malentacchi, F.; Pollini, S.; Antonelli, A.; Rossolini, G.M. Rapid screening for SARS-CoV-2 VOC-Alpha (202012/01, B.1.1.7) using the Allplex™ SARS-CoV-2/FluA/FluB/RSV Assay. Int. J. Infect. Dis. 2021, 113, 207–209. [Google Scholar] [CrossRef] [PubMed]
- Demoliner, M.; da Silva, M.S.; Gularte, J.S.; Hansen, A.W.; de Almeida, P.R.; Weber, M.N.; Heldt, F.H.; Silveira, F.; Filippi, M.; de Abreu Góes Pereira, V.M.; et al. Predominance of SARS-CoV-2 P.1 (Gamma) lineage inducing the recent COVID-19 wave in southern Brazil and the finding of an additional S: D614A mutation. Infect. Genet. Evol. 2021, 96, 105134. [Google Scholar] [CrossRef]
- España, G.; Cucunubá, Z.M.; Cuervo-Rojas, J.; Díaz, H.; González-Mayorga, M.; Ramírez, J.D. The impact of vaccination strategies for COVID-19 in the context of emerging variants and increasing social mixing in Bogotá, Colombia: A mathematical modelling study. medRxiv 2021. [Google Scholar] [CrossRef]
- Kaur, S.P.; Gupta, V. COVID-19 Vaccine: A comprehensive status report. Virus Res. 2020, 288, 198114. [Google Scholar] [CrossRef]
- Jeyanathan, M.; Afkhami, S.; Smaill, F.; Miller, M.S.; Lichty, B.D.; Xing, Z. Immunological considerations for COVID-19 vaccine strategies. Nat. Rev. Immunol. 2020, 20, 615–632. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Dai, T.; Wei, Y.; Zhang, L.; Zheng, M.; Zhou, F. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct. Target. Ther. 2020, 5, 237. [Google Scholar] [CrossRef]
- Lee, W.S.; Wheatley, A.K.; Kent, S.J.; DeKosky, B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020, 5, 1185–1191. [Google Scholar] [CrossRef] [PubMed]
- Samrat, S.K.; Tharappel, A.M.; Li, Z.; Li, H. Prospect of SARS-CoV-2 spike protein: Potential role in vaccine and therapeutic development. Virus Res. 2020, 288, 198141. [Google Scholar] [CrossRef]
- Tregoning, J.S.; Brown, E.S.; Cheeseman, H.M.; Flight, K.E.; Higham, S.L.; Lemm, N.M.; Pierce, B.F.; Stirling, D.C.; Wang, Z.; Pollock, K.M. Vaccines for COVID-19. Clin. Exp. Immunol. 2020, 202, 162–192. [Google Scholar] [CrossRef] [PubMed]
- Eyal, N.; Lipsitch, M.; Smith, P.G. Human Challenge Studies to Accelerate Coronavirus Vaccine Licensure. J. Infect. Dis. 2020, 221, 1752–1756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, N.; Shang, J.; Jiang, S.; Du, L. Subunit Vaccines against Emerging Pathogenic Human Coronaviruses. Front. Microbiol. 2020, 11, 298. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.A.; Islam, M.S. Early approval of COVID-19 vaccines: Pros and cons. Hum. Vaccines Immunother. 2021, 17, 3288–3296. [Google Scholar] [CrossRef]
- Dolgin, E. How protein-based COVID vaccines could change the pandemic. Nature 2021, 599, 359–360. [Google Scholar] [CrossRef]
- Cid, R.; Bolívar, J. Platforms for Production of Protein-Based Vaccines: From Classical to Next-Generation Strategies. Biomolecules 2021, 11, 1072. [Google Scholar] [CrossRef]
- Iversen, P.L.; Bavari, S. Inactivated COVID-19 vaccines to make a global impact. Lancet Infect. Dis. 2021, 21, 746–748. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, C.; Song, Y.; Coleman, J.R.; Stawowczyk, M.; Tafrova, J.; Tasker, S.; Boltz, D.; Baker, R.; Garcia, L.; et al. Scalable live-attenuated SARS-CoV-2 vaccine candidate demonstrates preclinical safety and efficacy. Proc. Natl. Acad. Sci. USA 2021, 118, e2102775118. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, C. First COVID-19 DNA vaccine approved, others in hot pursuit. Nat. Biotechnol. 2021, 39, 1479–1482. [Google Scholar] [CrossRef] [PubMed]
- Silveira, M.M.; Moreira, G.M.S.G.; Mendonça, M. DNA vaccines against COVID-19: Perspectives and challenges. Life Sci. 2021, 267, 118919. [Google Scholar] [CrossRef] [PubMed]
- Xia, X. Detailed Dissection and Critical Evaluation of the Pfizer/BioNTech and Moderna mRNA Vaccines. Vaccines 2021, 9, 734. [Google Scholar] [CrossRef]
- Borobia, A.M.; Carcas, A.J.; Pérez-Olmeda, M.; Castaño, L.; Bertran, M.J.; García-Pérez, J.; Campins, M.; Portolés, A.; González-Pérez, M.; Morales, M.T.G.; et al. Immunogenicity and reactogenicity of BNT162b2 booster in ChAdOx1-S-primed participants (CombiVacS): A multicentre, open-label, randomised, controlled, phase 2 trial. Lancet 2021, 398, 121–130. [Google Scholar] [CrossRef]
- Shaw, R.H.; Stuart, A.; Greenland, M.; Liu, X.; Van-Tam, J.S.N.; Snape, M.D. Heterologous prime-boost COVID-19 vaccination: Initial reactogenicity data. Lancet 2021, 397, 2043–2046. [Google Scholar] [CrossRef]
- Nguyen, T.T.; Quach, T.H.T.; Tran, T.M.; Phuoc, H.N.; Nguyen, H.T.; Vo, T.K.; Vo, G.V. Reactogenicity and immunogenicity of heterologous prime-boost immunization with COVID-19 vaccine. Biomed. Pharmacother. 2022, 147, 112650. [Google Scholar] [CrossRef] [PubMed]
- Duong, D. Feds sign off on mixing and matching COVID-19 vaccines, but evidence gaps remain. Can. Med. Assoc. J. 2021, 193, e967–e968. [Google Scholar] [CrossRef]
- Schmidt, T.; Klemis, V.; Schub, D.; Mihm, J.; Hielscher, F.; Marx, S.; Abu-Omar, A.; Ziegler, L.; Guckelmus, C.; Urschel, R.; et al. Immunogenicity and reactogenicity of heterologous ChAdOx1 nCoV-19/mRNA vaccination. Nat. Med. 2021, 27, 1530–1535. [Google Scholar] [CrossRef] [PubMed]
- Pozzetto, B.; Legros, V.; Djebali, S.; Barateau, V.; Guibert, N.; Villard, M.; Peyrot, L.; Allatif, O.; Fassier, J.B.; Massardier-Pilonchéry, A.; et al. Immunogenicity and efficacy of heterologous ChAdOx1-BNT162b2 vaccination. Nature 2021, 600, 701–706. [Google Scholar] [CrossRef]
- Nordström, P.; Ballin, M.; Nordström, A. Effectiveness of heterologous ChAdOx1 nCoV-19 and mRNA prime-boost vaccination against symptomatic Covid-19 infection in Sweden: A nationwide cohort study, The Lancet regional health. Europe 2021, 11, 100249. [Google Scholar] [CrossRef] [PubMed]
- Andrews, N.; Stowe, J.; Kirsebom, F.; Gower, C.; Ramsay, M.; Bernal, J.L. Effectiveness of BNT162b2 (Comirnaty, Pfizer-BioNTech) COVID-19 booster vaccine against covid-19 related symptoms in England: Test negative case-control study. medRxiv 2021. [Google Scholar] [CrossRef]
- Patalon, T.; Gazit, S.; Pitzer, V.E.; Prunas, O.; Warren, J.L.; Weinberger, D.M. Odds of Testing Positive for SARS-CoV-2 Following Receipt of 3 vs 2 Doses of the BNT162b2 mRNA Vaccine. JAMA Intern. Med. 2021, 182, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Bar-On, Y.M.; Goldberg, Y.; Mandel, M.; Bodenheimer, O.; Freedman, L.; Kalkstein, N.; Mizrahi, B.; Alroy-Preis, S.; Ash, N.; Milo, R.; et al. Protection of BNT162b2 Vaccine Booster against Covid-19 in Israel. N. Engl. J. Med. 2021, 385, 1393–1400. [Google Scholar] [CrossRef]
- Li, X. Omicron: Call for updated vaccines. J. Med. Virol. 2022, 94, 1261–1263. [Google Scholar] [CrossRef] [PubMed]
- Khan, N.A.; Al-Thani, H.; El-Menyar, A. The emergence of new SARS-CoV-2 variant (Omicron) and increasing calls for COVID-19 vaccine boosters-The debate continues. Travel Med. Infect. Dis. 2022, 45, 102246. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Beltran, W.F.; Denis, K.J.S.; Hoelzemer, A.; Lam, E.C.; Nitido, A.D.; Sheehan, M.L.; Berrios, C.; Ofoman, O.; Chang, C.C.; Hauser, B.M.; et al. mRNA-based COVID-19 vaccine boosters induce neutralizing immunity against SARS-CoV-2 Omicron variant. Cell 2022, 185, 457–466.e4. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ginn, H.M.; Dejnirattisai, W.; Supasa, P.; Wang, B.; Tuekprakhon, A.; Nutalai, R.; Zhou, D.; Mentzer, A.J.; Zhao, Y.; et al. Reduced neutralization of SARS-CoV-2 B.1.617 by vaccine and convalescent serum. Cell 2021, 184, 4220–4236.e13. [Google Scholar] [CrossRef] [PubMed]
- Hirabara, S.M.; Serdan, T.D.A.; Gorjao, R.; Masi, L.N.; Pithon-Curi, T.C.; Covas, D.T.; Curi, R.; Durigon, E.L. SARS-COV-2 Variants: Differences and Potential of Immune Evasion. Front. Cell. Infect. Microbiol. 2022, 11, 1401. [Google Scholar] [CrossRef]
- Otto, S.P.; Day, T.; Arino, J.; Colijn, C.; Dushoff, J.; Li, M.; Mechai, S.; van Domselaar, G.; Wu, J.; Earn, D.J.D.; et al. The origins and potential future of SARS-CoV-2 variants of concern in the evolving COVID-19 pandemic. Curr. Biol. 2021, 31, r918–r929. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Hulme, J.; Tran, H.D.; Vo, T.K.; Vo, G.V. The potential impact of COVID-19 on male reproductive health. J. Endocrinol. Investig. 2022, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.T.; Nguyen, T.T.D.; Tran, N.M.; Nguyen, H.T.; Vo, G.V. Microneedles enable the development of skin-targeted vaccines against coronaviruses and influenza viruses. Pharm. Dev. Technol. 2022, 27, 83–94. [Google Scholar] [CrossRef]
- Van Vo, G.; Bagyinszky, E.; Park, Y.S.; Hulme, J.; An, S.S.A. SARS-CoV-2 (COVID-19): Beginning to Understand a New Virus. Adv. Exp. Med. Biol. 2021, 1321, 3–19. [Google Scholar]
- Vo, V.G.; Bagyinszky, E.; Shim, K.; Park, Y.S.; An, S.S.A. Additional diagnostic testing of the 2019 novel coronavirus (SARS-CoV-2). Mol. Cell Toxicol. 2020, 16, 355–357. [Google Scholar] [CrossRef] [PubMed]
| Virus Variant | Origin | Spike Mutations | Other Mutations | VOC/VOI | Properties of Significant Mutation | Refs |
|---|---|---|---|---|---|---|
| SARS-Wuhan | China | D614G | ORF1a: P323L, UTR: g.241C > T, g.23403A > G | VOC/ former VOC | D614G: enables rapid viral spreading | [11] |
| VOC 202012/01 or B.1.1.7 or Alpha variant | UK | A570D, D614G, D1118H, H69del, N501Y, P681H, S982A, T716I, del_V69-70, Y145del | N: D3L, S235F ORF8: Q27X, R52I, Y73C ORF1a: G465S, A890D, I1412T, T183I, del_S106, del_G107, del_F108, P323L, G204R, R203K | VOC/ former VOC | N501Y: increases of viral biding to ACE2 receptor P681H: enhances viral entering to host del_V69-70: putative immune escape variant | [70,71] |
| GH501Y.V2 or B.1.351 or Beta variant | South Africa | D614G, D80A, D215G, E484K, N501Y, A701V, L18F, R246I, K417N, del_aa242-244 | E: P71L N: T205I ORF1ab: K1655N, del_aa3675-3677, T265I, H2799Y, P4715L | VOC | E484K and N500Y: enhance the ACE2 binding to virus K417N: enhances further viral binding to ACE2 All of them: immune escape | [20] |
| P1, B.1.1.28; GR/501Y.V3 or Gamma variant | Brazil | L18F, T20N, P26S, D138Y, R190S, K417T, E484K, N501Y H655Y, T1027I, D614G | ORF1ab: N: P80R, R203K ORF3a: S235P ORF8: E92K ORF1ab: P323L, K977Q, del_aa106-107 UTR: g. 733T > C, g. ins28263AACA, g. 28877A > T, c.28878G > C, c.12778C > T, del11288-11296 | VOC | N501Y and E484K: Virus-ACE2 interaction enhancement K417T: immune escape | [72] |
| B.1.617 or Delta variant | India | T19R, del_aa157-158, L452R, T478K, D614G, P681R D950N | ORF1a: A1306S, P2046L, P2287S, V2930L, T3255I, T3646A ORF1b: P314L, G662S, P1000L, A1918V ORF3a: S26L M: I82T ORF7a: V82A, T120I ORF7b: T40I ORF8→del119/120 N: D63G, R203M, D377Y | VOC | L452R and T478K: immune escape P681R: enhances viral ability to enter to host, increases viral load | [32,33] |
| B.1.1.529 or Omicron variant | South Africa | A67V, del_V69-70, T95I, G142D/del_aa143-145, Δ211/L212I, ins_214EPE, G339D, S371L, S373P, S375F, K417N, N440K, G446S, S477N, T478K, E484A, Q493R, G496S, Q498R, N501Y, Y505H, T547K, D614G, H655Y, N679K, P681H, N764K, D796Y, N856K, Q954H, N969K, and L981F | ORF1a: del_L3674-S3675 and G3676 N: R203K G204R | VOC | N501Y, H655Y, P681H, N679K, and D614G: high transmissibility Q498R and N501Y: higher ACE2 binding K417N and T478K: ability for immune escape | [39] |
| B.1.427/B.1.429, or Epsilon variant | USA (California) | B.1.427: L452R, D614G; B.1.429: S13I, W152C, L452R, D614G | ORF1ab: T85I, I64V, P323L, D260Y ORF3a: Q57H N: T205I | Former VOI | L452R: enhance viral infectivity, fusion and viral binding to ACE2 | [45] |
| C.37 or Lambda | Peru | G75V, T76I, D253N, L452Q, F490S, D614G, T859N | ORF1ab: del3675 to 3677, T428I, P323L N: P13L, P10S, R203K, R204G, R214C, 246_253delinsN | VOI | L452Q, F490S: may enhance viral affinity to host | [48] |
| B.1.617.1 or Kappa | India | T95I, G142D, E154K, L452R, E484Q, D614G, P681R, Q1071H | ORF1ab: T749I, T77A, P323L, M429I, K259R ORF3: S26L Orf7a: V82A N: R203M, N377I | Former VOI | L452R and E484Q: may enhance viral affinity to host P681R: Influence viral entry into host | [52] |
| B.1.526 or Iota | USA (New York) | L5F, T95I, D253G, S477N, L452R, E484K, D614G, A701V | Orf1ab: T85I, L438P, del_aa106-108, Q57H Orf3: P199L N: M231I | Former VOI | L452R, S477N and E484K: may enhance viral affinity to host | [56] |
| B.1.525 or Eta | USA (New York | A67V, del_aa69-70, del_aa144, E484K, D614G, Q677H, F888L | ORF1ab: T1189I, P323F E: L21F M: I82T N: del_aa3, A12G, T205I | Former VOI | Q677H: enhances viral transmission del_aa144: immune escape F888L: putative virulence functions | [55] |
| B.1.621 or Mu | Columbia | del_aa69-70, Y144T, Y145S, ins_146N, R346K, E484K, N501Y, D614G, P681H, and D950N | ORF1ab: T237A, T720I, T492I, Q160R ORF3a: Q57H, del_aa256-257 ORF8: T11K, P38S N: T205I | VOI | del_aa69-70, E484K and N501Y: impact the antibody neutralization, immune escape ins146N: ACE2 binding | [73] |
| P.3 or Theta | Philippines | del_aa141-143, E484K, N501Y, P681H, D614G, H1101Y, E1092K, V1176F, G593G and S875S | ORF1ab: D736G, L438P, L71F, A368V ORF8: K2Q N: R203K, G204R | Former VOI | E484K, N501Y, P681H: enhance viral infectivity, Viral ACE2 binding del_aa141-143: immune escape | [62] |
| P.2 or Zeta | Brazil | E484K, F565L, D614G, V1167F | ORF1ab: L205V, L71F, P323L N: A119S | Former VOI | E484K and D614G: rapid spread, immune escape | [63] |
| AY.4.2, B.1.617.2 or Delta plus | India, UK | Delta variant mutations + T95I, A222V, Y145H G142D, R158G, and K417N | Delta variant mutations + ORF1A: A1146T, P1604L, A3209V, V3718S, and T3750I). | VOI | Y145H and A222V: enables viral penetration to blood cells K417T: immune escape | [64] |
| Vaccine Platform | Description | Benefits | Disadvantages | Examples of SARS-CoV-2 Vaccines | Refs |
|---|---|---|---|---|---|
| Protein subunit | Use specific viral part (spike protein) for appropriate immune response | Safe, stable, lower risk for autoimmunity or other side effects | Production may be costly, difficult to purify, and growing pathogens may be difficult; long-term protection may be doubtful | Novavax | [83,84] |
| Inactivated virus | Vaccine contains killed virus, which induces immune system | Successful platform against different viruses, may speed up research | Production may be costly, difficult to purify, and growing pathogens may be difficult | Janssen/Johnson & Johnson, AstraZeneca, SinoVac. BBV152 | [85] |
| Live attenuated virus | Uses alive, but weakened viruses | May induce a broad degree of immune responses; human systems may adapt effectively | Vaccine is not approved yet; under development | COVI-VAC | [86] |
| DNA | Two or three dose plasmid DNA, which encodes the viral S-protein (or N-protein), with potential signal peptide | Easy to produce, Can be produced in large amount No pathogens are used More stable than RNA vaccines | Development is in early stage; not tested in humans DNA may insert into human DNA Risk of autoimmunity | ZyCoV-D, AG0302-COVID19 | [87,88] |
| RNA | Two doses of vaccine, using the synthetic viral RNA, inducing the cells producing S-protein, resulting in antibody production against SARS-CoV-2 | Easy to produce, Able to produce in large quantity No pathogen, generally safe No risk for inserting into human genome | Not too stable, need to be kept at low temperature (−60 °C) Risk for autoimmunity | Pfizer/BioNTech Moderna | [88,89] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vo, G.V.; Bagyinszky, E.; An, S.S.A. COVID-19 Genetic Variants and Their Potential Impact in Vaccine Development. Microorganisms 2022, 10, 598. https://doi.org/10.3390/microorganisms10030598
Vo GV, Bagyinszky E, An SSA. COVID-19 Genetic Variants and Their Potential Impact in Vaccine Development. Microorganisms. 2022; 10(3):598. https://doi.org/10.3390/microorganisms10030598
Chicago/Turabian StyleVo, Giau Van, Eva Bagyinszky, and Seong Soo A. An. 2022. "COVID-19 Genetic Variants and Their Potential Impact in Vaccine Development" Microorganisms 10, no. 3: 598. https://doi.org/10.3390/microorganisms10030598
APA StyleVo, G. V., Bagyinszky, E., & An, S. S. A. (2022). COVID-19 Genetic Variants and Their Potential Impact in Vaccine Development. Microorganisms, 10(3), 598. https://doi.org/10.3390/microorganisms10030598







