Bacteriocin Producing Streptococcus agalactiae Strains Isolated from Bovine Mastitis in Brazil
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains, Media, and Growth Conditions
2.2. Deferred Growth Inhibition Assays
2.3. DNA Isolation and Illumina Hiseq Whole-Genome Sequencing
2.4. Genome Assembly
2.5. Taxonomic Identification
2.6. Assessments for Virulence Factors and Antibiotic Resistance
2.7. Genome Mining for Antimicrobial Substances
2.8. Genome Comparison and Functional Annotation
3. Results
3.1. Antimicrobial Activity Spectrum
3.2. Genome Sequencing
3.3. Taxonomic Identification
3.4. Antibiotic Resistance Genes and Virulence Factors
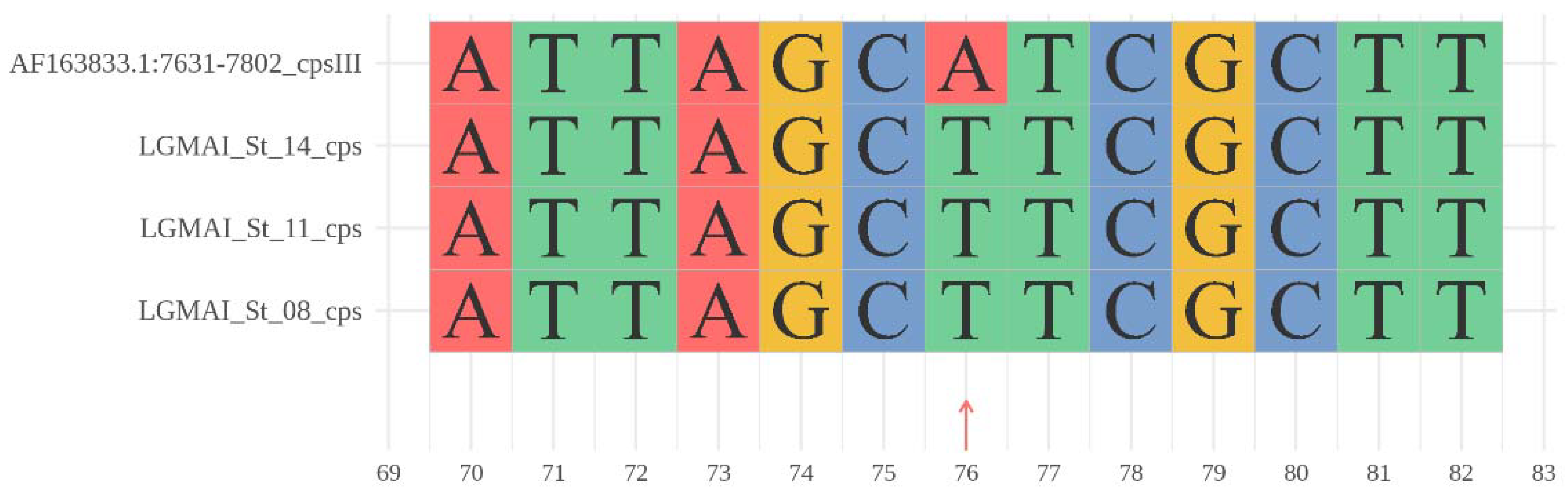
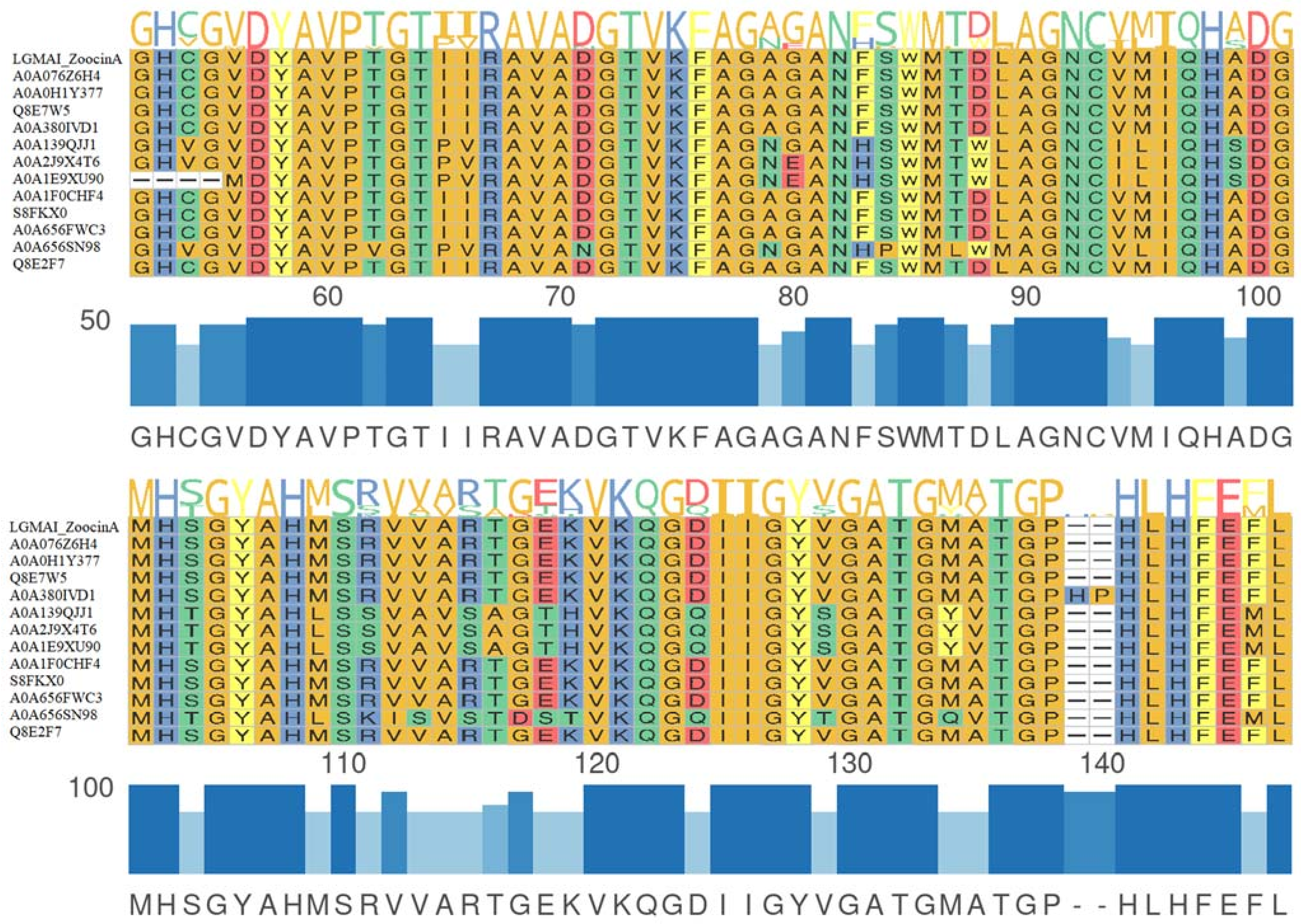
3.5. Gene Clusters of Antimicrobial Peptides
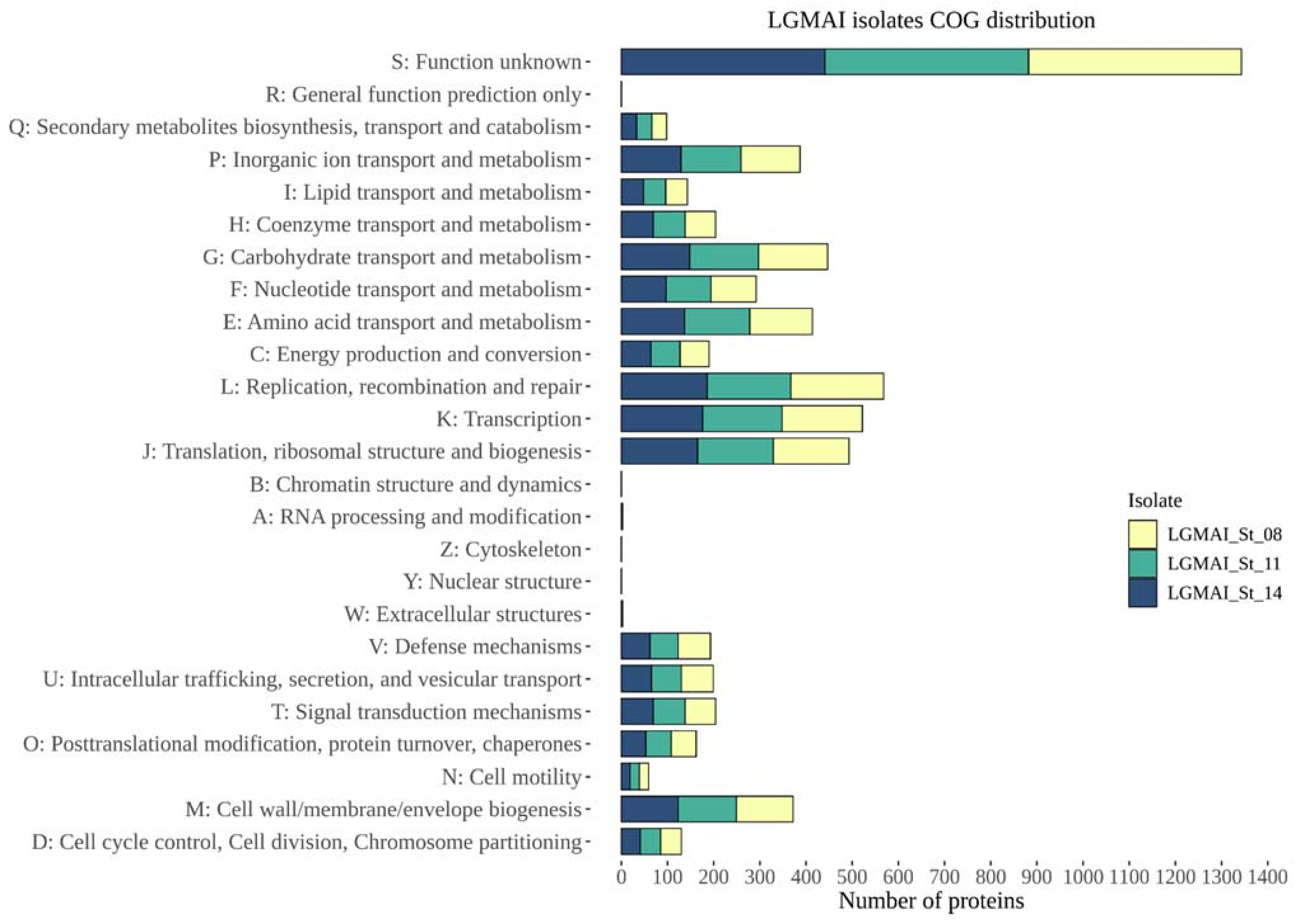
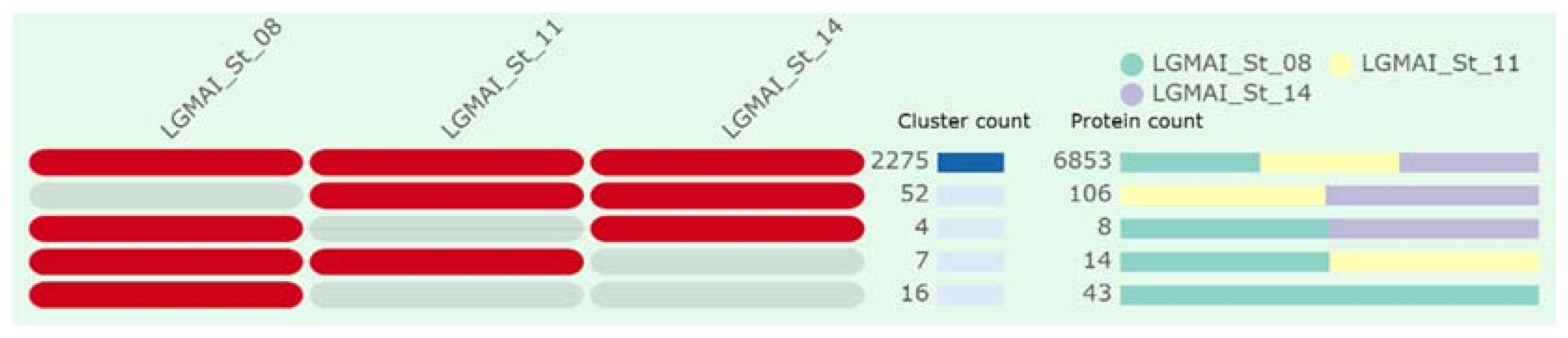
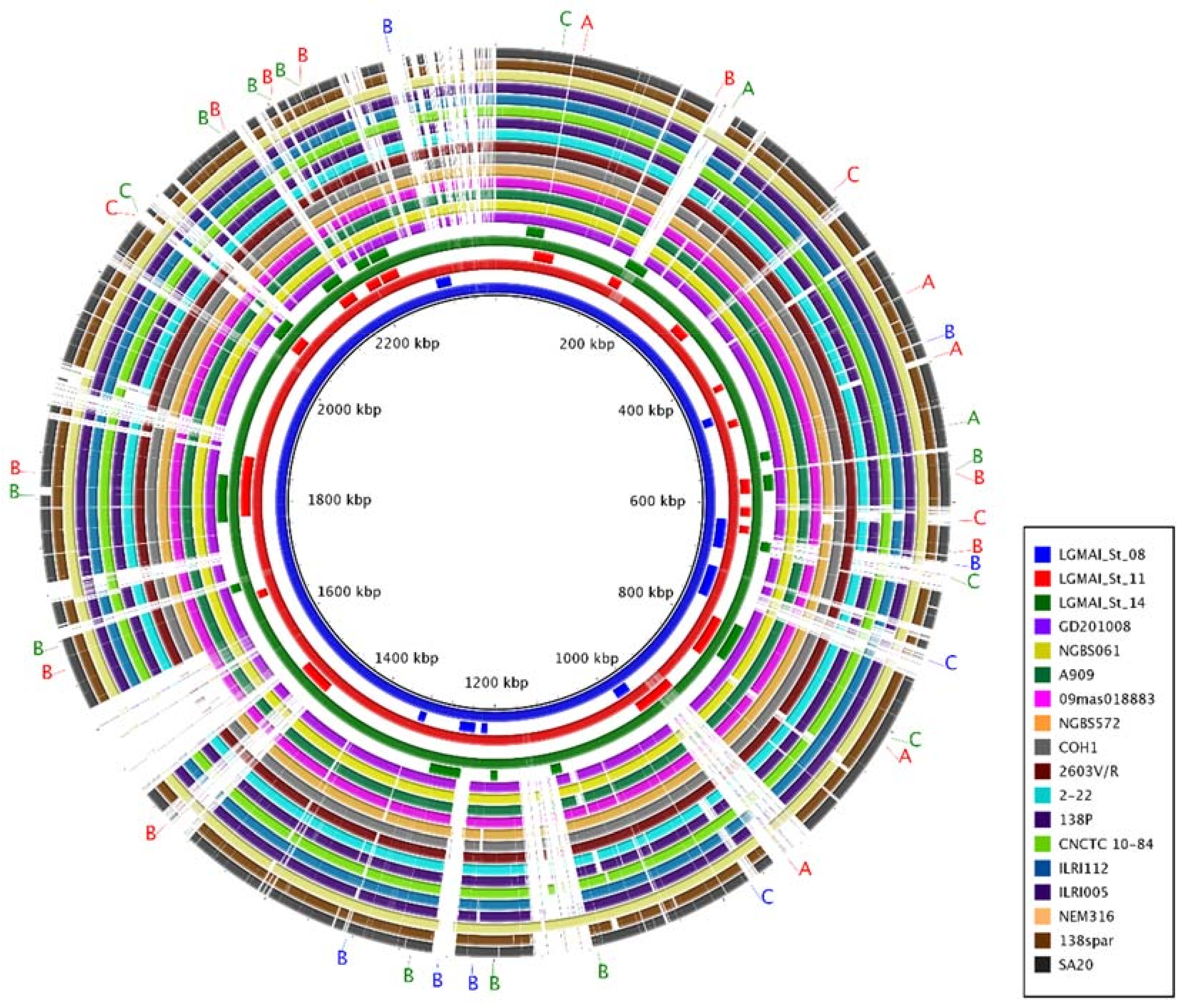
3.6. Genome Comparison and Functional Annotation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Streicher, L.M. Exploring the Future of Infectious Disease Treatment in a Post-Antibiotic Era: A Comparative Review of Alternative Therapeutics. J. Glob. Antimicrob. Resist. 2021, 24, 285–295. [Google Scholar] [CrossRef]
- Aminov, R. History of antimicrobial drug discovery: Major classes and health impact. Biochem. Pharmacol. 2016, 133, 4–19. [Google Scholar] [CrossRef]
- Roca, I.; Akova, M.; Baquero, F.; Carlet, J.; Cavaleri, M.; Coenen, S.; Cohen, J.; Findlay, D.; Gyssens, I.; Heure, O.E.; et al. The global threat of antimicrobial resistance: Science for intervention. New Microbes New Infect. 2015, 6, 22–29. [Google Scholar] [CrossRef]
- O’Neill, J. Review on Antimicrobial Resistance. 2014. Available online: https://amr-review.org (accessed on 23 March 2021).
- Hsu, J.-F.; Tsai, M.-H.; Lin, L.-C.; Chu, S.-M.; Lai, M.-Y.; Huang, H.-R.; Chiang, M.-C.; Yang, P.-H.; Lu, J.-J. Genomic Characterization of Serotype III/ST-17 Group B Streptococcus Strains with Antimicrobial Resistance Using Whole Genome Sequencing. Biomedicines 2021, 9, 1477. [Google Scholar] [CrossRef]
- Tulyaprawat, O.; Pharkjaksu, S.; Shrestha, R.K.; Ngamskulrungroj, P. Emergence of Multi-Drug Resistance and Its Association with Uncommon Serotypes of Streptococcus agalactiae Isolated from Non-neonatal Patients in Thailand. Front. Microbiol. 2021, 12. [Google Scholar] [CrossRef]
- Carvalho-Castro, G.A.; Silva, J.R.; Paiva, L.V.; Custódio, D.A.; Moreira, R.O.; Mian, G.F.; Prado, I.A.; Chalfun-Junior, A.; Costa, G.M. Molecular epidemiology of Streptococcus agalactiae isolated from mastitis in Brazilian dairy herds. Braz. J. Microbiol. 2017, 48, 551–559. [Google Scholar] [CrossRef]
- Lazzaro, B.P.; Zasloff, M.; Rolff, J. Antimicrobial peptides: Application informed by evolution. Science 2020, 368. [Google Scholar] [CrossRef]
- Koo, H.B.; Seo, J. Antimicrobial peptides under clinical investigation. Pept. Sci. 2019, 111, e24122. [Google Scholar] [CrossRef]
- Chernov, V.M.; Chernova, O.A.; Mouzykantov, A.A.; Lopukhov, L.L.; Aminov, R.I. Omics of antimicrobials and antimicrobial resistance. Expert Opin. Drug Hist. 2019, 14, 455–468. [Google Scholar] [CrossRef]
- Venegas-Ortega, M.G.; Flores-Gallegos, A.C.; Martínez-Hernández, J.L.; Aguilar, C.N.; Nevárez-Moorillón, G.V. Production of Bioactive Peptides from Lactic Acid Bacteria: A Sustainable Approach for Healthier Foods. Compr. Rev. Food Sci. Food Saf. 2019, 18, 1039–1051. [Google Scholar] [CrossRef]
- Alvarez-Sieiro, P.; Montalbán-López, M.; Mu, D.; Kuipers, O.P. Bacteriocins of Lactic Acid Bacteria: Extending the Family. Appl. Microbiol. Biotechnol. 2016, 100, 2939–2951. [Google Scholar] [CrossRef]
- Müller-Auffermann, K.; Grijalva, F.; Jacob, F.; Hutzler, M. Nisin and its usage in breweries: A review and discussion. J. Inst. Brew. 2015, 121, 309–319. [Google Scholar] [CrossRef]
- Xiao, H.; Chen, X.; Chen, M.; Tang, S.; Zhao, X.; Huan, L. Bovicin HJ50, a novel lantibiotic produced by Streptococcus bovis HJ50. Microbiology 2004, 150, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Mota-Meira, M.; Lacroix, C.; Lapointe, G.; Lavoie, M.C. Purification and structure of mutacin B-Ny266: A new lantibiotic produced byStreptococcus mutans. FEBS Lett. 1997, 410, 275–279. [Google Scholar] [CrossRef]
- Novák, J.; Caufield, P.W.; Miller, E.J. Isolation and Biochemical Characterization of a Novel Lantibiotic Mutacin from Strep-tococcus mutans. J. Bacteriol. 1994, 176, 4316–4320. [Google Scholar] [CrossRef] [PubMed]
- Qi, F.; Chen, P.; Caufield, P.W. Purification of Mutacin III from Group III Streptococcus mutans UA787 and Genetic Analyses of Mutacin III Biosynthesis Genes. Appl. Environ. Microbiol. 1999, 65, 3880–3887. [Google Scholar] [CrossRef] [PubMed]
- Vogel, V.; Spellerberg, B. Bacteriocin Production by Beta-Hemolytic Streptococci. Pathogens 2021, 10, 867. [Google Scholar] [CrossRef]
- Ross, K.F.; Ronson, C.W.; Tagg, J.R. Isolation and characterization of the lantibiotic salivaricin A and its structural gene salA from Streptococcus salivarius 20P3. Appl. Environ. Microbiol. 1993, 59, 2014–2021. [Google Scholar] [CrossRef]
- Wescombe, P.A.; Upton, M.; Dierksen, K.P.; Ragland, N.L.; Sivabalan, S.; Wirawan, R.E.; Inglis, M.A.; Moore, C.J.; Walker, G.V.; Chilcott, C.N.; et al. Production of the Lantibiotic Salivaricin A and Its Variants by Oral Streptococci and Use of a Specific Induction Assay to Detect Their Presence in Human Saliva. Appl. Environ. Microbiol. 2006, 72, 1459–1466. [Google Scholar] [CrossRef]
- Wescombe, P.A.; Upton, M.; Renault, P.; Wirawan, R.E.; Power, D.; Burton, J.P.; Chilcott, C.N.; Tagg, J.R. Salivaricin 9, a new lantibiotic produced by Streptococcus salivarius. Microbiology 2011, 157, 1290–1299. [Google Scholar] [CrossRef]
- Villani, F.; Pepe, O.; Mauriello, G.; Salzano, G.; Moschetti, G.; Coppola, S. Antilisterial Activity of Thermophilin 347, a Bacte-riocin Produced by Streptococcus thermophilus. Int. J. Food Microbiol. 1995, 25, 179–190. [Google Scholar] [CrossRef]
- Heng, N.C.K.; Burtenshaw, G.A.; Jack, R.W.; Tagg, J.R. Ubericin A, a Class IIa Bacteriocin Produced by Streptococcus uberis. Appl. Environ. Microbiol. 2007, 73, 7763–7766. [Google Scholar] [CrossRef] [PubMed]
- Wirawan, R.E.; Swanson, K.M.; Kleffmann, T.; Jack, R.W.; Tagg, J.R. Uberolysin: A novel cyclic bacteriocin produced by Streptococcus uberis. Microbiology 2007, 153, 1619–1630. [Google Scholar] [CrossRef]
- Razzak, L.A.; Sabri, M. Isolation and Characterization of Streptococcus agalactiae from Woman Patients with Vaginitis in Hilla Province. Med. J. Babylon 2005, 2, 3. [Google Scholar]
- Duarte, R.S.; Miranda, O.P.; Bellei, B.C.; Brito, M.A.V.P.; Teixeira, L.M. Phenotypic and Molecular Characteristics of Streptococcus agalactiae Isolates Recovered from Milk of Dairy Cows in Brazil. J. Clin. Microbiol. 2004, 42, 4214–4222. [Google Scholar] [CrossRef]
- Stoyancheva, G.; Marzotto, M.; Dellaglio, F.; Torriani, S. Bacteriocin production and gene sequencing analysis from vaginal Lactobacillus strains. Arch. Microbiol. 2014, 196, 645–653. [Google Scholar] [CrossRef]
- Chachaty, E.; Saulnier, P. Isolating Chromosomal DNA from Bacteria. In The Nucleic Acids Protocols Handbook, 1st ed.; Rapley, R., Ed.; Humana Press: Totowa, NJ, USA, 2000; pp. 29–32. [Google Scholar]
- Andrews, S.; Babraham, I. FastQC: A Quality Control Tool for High Throughput Sequence Data. 2013. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 21 September 2020).
- Schubert, M.; Lindgreen, S.; Orlando, L. AdapterRemoval v2: Rapid adapter trimming, identification, and read merging. BMC Res. Notes 2016, 9, 88. [Google Scholar] [CrossRef]
- Melsted, P.; Halldórsson, B. KmerStream: Streaming algorithms for k -mer abundance estimation. Bioinformatics 2014, 30, 3541–3547. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, D.; François, P.; Farinelli, L.; Østerås, M.; Schrenzel, J. De novo bacterial genome sequencing: Millions of very short reads assembled on a desktop computer. Genome Res. 2008, 18, 802–809. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Ge-nome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Metcalf, B.J.; Chochua, S.; Gertz, R.E., Jr.; Hawkins, P.A.; Ricaldi, J.; Li, Z.; Walker, H.; Tran, T.; Rivers, J.; Mathis, S.; et al. Short-Read Whole Genome Sequencing for Determination of Antimicrobial Resistance Mechanisms and Capsular Serotypes of Current Invasive Streptococcus Agalactiae Recovered in the USA. Clin. Microbiol. Infect. 2017, 23, 574.e7–574.e14. [Google Scholar] [CrossRef] [PubMed]
- Śmiałek, J.; Nowakowski, M.; Bzowska, M.; Bocheńska, O.; Wlizło, A.; Kozik, A.; Dubin, G.; Mak, P. Structure, Biosynthesis, and Biological Activity of Succinylated Forms of Bacteriocin BacSp222. Int. J. Mol. Sci. 2021, 22, 6256. [Google Scholar] [CrossRef] [PubMed]
- Younus, Z.; Goyal, S.M.; Singh, V.; Ikram, A.; Imran, M. Genomic-based characterization of Enterococcus spp.: An emerging pathogen isolated from human gut. Mol. Biol. Rep. 2021, 48, 5371–5376. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Abricate, Github. Available online: https://github.com/tseemann/abricate (accessed on 15 November 2020).
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.-M. ARG-ANNOT, a New Bioinformatic Tool to Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob. Agents Chemother. 2013, 58, 212–220. [Google Scholar] [CrossRef]
- Jia, B.; Raphenya, A.R.; Alcock, B.; Waglechner, N.; Guo, P.; Tsang, K.K.; Lago, B.A.; Dave, B.M.; Pereira, S.; Sharma, A.N.; et al. CARD 2017: Expansion and Model-Centric Curation of the Comprehensive Antibiotic Resistance Database. Nucleic Acids Res. 2017, 45, D566–D573. [Google Scholar] [CrossRef]
- Feldgarden, M.; Brover, V.; Haft, D.H.; Prasad, A.B.; Slotta, D.J.; Tolstoy, I.; Tyson, G.H.; Zhao, S.; Hsu, C.-H.; McDermott, P.F.; et al. Validating the AMRFinder Tool and Resistance Gene Database by Using Antimicrobial Resistance Genotype-Phenotype Correlations in a Collection of Isolates. Antimicrob. Agents Chemother. 2019, 63, e00483-19. [Google Scholar] [CrossRef]
- Zankari, E.; Hasman, H.; Cosentino, S.; Vestergaard, M.; Rasmussen, S.; Lund, O.; Aarestrup, F.M.; Larsen, M.V. Identification of Acquired Antimicrobial Resistance Genes. J. Antimicrob. Chemother. 2012, 67, 2640–2644. [Google Scholar] [CrossRef]
- Chen, L.; Zheng, D.; Liu, B.; Yang, J.; Jin, Q. VFDB 2016: Hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res. 2015, 44, D694–D697. [Google Scholar] [CrossRef] [PubMed]
- Soares, S.C.; Geyik, H.; Ramos, R.T.; de Sá, P.H.; Barbosa, E.G.; Baumbach, J.; Figueiredo, H.C.; Miyoshi, A.; Tauch, A.; Silva, A.; et al. GIPSy: Genomic island prediction software. J. Biotechnol. 2015, 232, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Medema, M.H.; Blin, K.; Cimermancic, P.; De Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef] [PubMed]
- Van Heel, A.J.; De Jong, A.; Song, C.; Viel, J.; Kok, J.; Kuipers, O.P. BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Res. 2018, 46, W278–W281. [Google Scholar] [CrossRef]
- Hammami, R.; Zouhir, A.; Ben Hamida, J.; Fliss, I. BACTIBASE: A New Web-Accessible Database for Bacteriocin Character-ization. BMC Microbiol. 2007, 7, 89. [Google Scholar] [CrossRef]
- Huerta-Cepas, J.; Forslund, K.; Coelho, L.P.; Szklarczyk, D.; Jensen, L.J.; von Mering, C.; Bork, P. Fast Genome-Wide Functional Annotation through Orthology Assignment by eggNOG-Mapper. Mol. Biol. Evol. 2017, 34, 2115–2122. [Google Scholar] [CrossRef]
- Xu, L.; Dong, Z.; Fang, L.; Luo, Y.; Wei, Z.; Guo, H.; Zhang, G.; Gu, Y.Q.; Coleman-Derr, D.; Xia, Q.; et al. OrthoVenn2: A web server for whole-genome comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res. 2019, 47, W52–W58. [Google Scholar] [CrossRef]
- Alikhan, N.-F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple Prokaryote Genome Comparisons. BMC Genom. 2011, 12, 1–10. [Google Scholar] [CrossRef]
- Ogaki, M.B.; Furlaneto, M.C.; Maia, L.F. Revisão: Aspectos gerais das bacteriocinas. Braz. J. Food Technol. 2015, 18, 267–276. [Google Scholar] [CrossRef]
- Peters, J.; Price, J.; Llewelyn, M. Staphylococcal and streptococcal infections. Medicine 2017, 45, 727–734. [Google Scholar] [CrossRef]
- Duarte, R.S.; Barros, R.R.; Facklam, R.R.; Teixeira, L.M. Phenotypic and Genotypic Characteristics of Streptococcus porcinus Isolated from Human Sources. J. Clin. Microbiol. 2005, 43, 4592–4601. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Wang, Y.; Guo, H.; Bai, Y.; Li, T.; Xu, R.; Sun, T.; Lu, J.; Song, Q. Isolation and Characteristics of Multi-Drug Resistant Strep-tococcus porcinus from the Vaginal Secretions of Sow with Endometritis. BMC Vet. Res. 2020, 16, 1–8. [Google Scholar] [CrossRef]
- Leelahapongsathon, K.; Schukken, Y.; Srithanasuwan, A.; Suriyasathaporn, W. Molecular epidemiology of Streptococcus uberis intramammary infections: Persistent and transient patterns of infection in a dairy herd. J. Dairy Sci. 2020, 103, 3565–3576. [Google Scholar] [CrossRef]
- Raabe, V.N.; Shane, A.L. Group B Streptococcus (Streptococcus agalactiae). Microbiol. Spectr. 2019, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Ruegg, P.L. A 100-Year Review: Mastitis detection, management, and prevention. J. Dairy Sci. 2017, 100, 10381–10397. [Google Scholar] [CrossRef] [PubMed]
- Angelopoulou, A.; Warda, A.K.; Hill, C.; Ross, R.P. Non-Antibiotic Microbial Solutions for Bovine Mastitis—Live Biotherapeutics, Bacteriophage, and Phage Lysins. Crit. Rev. Microbiol. 2019, 45, 564–580. [Google Scholar] [CrossRef] [PubMed]
- Ijaz, M.; Ameen, F.; Alfoteih, Y.A.; Shamim, S.; Alshehri, W.A.; Murtaza, G. Dissecting Streptococcus pyogenes interaction with human. Arch. Microbiol. 2020, 202, 2023–2032. [Google Scholar] [CrossRef] [PubMed]
- Thompson, C.C.; Emmel, V.E.; Fonseca, E.L.; Marin, M.A.; Vicente, A.C.P. Streptococcal Taxonomy Based on Genome Sequence Analyses. F1000Res. 2013, 2, 67. [Google Scholar] [CrossRef]
- Haslam, D.B.; Geme, J.W.S. Classification of Streptococci. In Principles and Practice of Pediatric Infectious Diseases, 5th ed.; Long, S.S., Prober, C.G., Fischer, M., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; pp. 712–714. [Google Scholar]
- Gao, X.-Y.; Zhi, X.-Y.; Li, H.-W.; Klenk, H.-P.; Li, W.-J. Comparative Genomics of the Bacterial Genus Streptococcus Illuminates Evolutionary Implications of Species Groups. PLoS ONE 2014, 9, e101229. [Google Scholar] [CrossRef]
- Clarebout, G.; Villers, C.; Leclercq, R. Macrolide Resistance Gene mreA of Streptococcus agalactiae Encodes a Flavokinase. Antimicrob. Agents Chemother. 2001, 45, 2280–2286. [Google Scholar] [CrossRef]
- Clancy, J.; Dib-Hajj, F.; Petitpas, J.W.; Yuan, W. Cloning and characterization of a novel macrolide efflux gene, mreA, from Streptococcus agalactiae. Antimicrob. Agents Chemother. 1997, 41, 2719–2723. [Google Scholar] [CrossRef]
- Oku, Y.; Kurokawa, K.; Ichihashi, N.; Sekimizu, K. Characterization of the Staphylococcus aureus mprF gene, involved in lysinylation of phosphatidylglycerol. Microbiology 2004, 150, 45–51. [Google Scholar] [CrossRef]
- Staubitz, P.; Neumann, H.; Schneider, T.; Wiedemann, I.; Peschel, A. MprF-mediated biosynthesis of lysylphosphatidylglycerol, an important determinant in staphylococcal defensin resistance. FEMS Microbiol. Lett. 2004, 231, 67–71. [Google Scholar] [CrossRef]
- Biggest Threats and Data: 2019 AR Threats Report. Available online: https://www.cdc.gov/drugresistance/biggest-threats.html (accessed on 8 April 2021).
- Kapatai, G.; Patel, D.; Efstratiou, A.; Chalker, V.J. Comparison of Molecular Serotyping Approaches of Streptococcus Agalactiae from Genomic Sequences. BMC Genomics 2017, 18, 429. [Google Scholar] [CrossRef]
- Armistead, B.; Oler, E.; Waldorf, K.A.; Rajagopal, L. The Double Life of Group B Streptococcus: Asymptomatic Colonizer and Potent Pathogen. J. Mol. Biol. 2019, 431, 2914–2931. [Google Scholar] [CrossRef]
- Vornhagen, J.; Waldorf, K.M.A.; Rajagopal, L. Perinatal Group B Streptococcal Infections: Virulence Factors, Immunity, and Prevention Strategies. Trends Microbiol. 2017, 25, 919–931. [Google Scholar] [CrossRef]
- Lang, S.; Palmer, M. Characterization of Streptococcus agalactiae CAMP Factor as a Pore-forming Toxin. J. Biol. Chem. 2003, 278, 38167–38173. [Google Scholar] [CrossRef]
- Jürgens, D.; Sterzik, B.; Fehrenbach, F.J. Unspecific Binding of Group B Streptococcal Cocytolysin (CAMP Factor) to Immunoglobulins and Its Possible Role in Pathogenicity. J. Exp. Med. 1987, 165, 720–732. [Google Scholar] [CrossRef]
- Lichvariková, A.; Soltys, K.; Szemes, T.; Slobodnikova, L.; Bukovska, G.; Turna, J.; Drahovska, H. Characterization of Clinical and Carrier Streptococcus agalactiae and Prophage Contribution to the Strain Variability. Viruses 2020, 12, 1323. [Google Scholar] [CrossRef]
- Yu, D.; Xu, F.; Zeng, J.; Zhan, J. Type III polyketide synthases in natural product biosynthesis. IUBMB Life 2012, 64, 285–295. [Google Scholar] [CrossRef]
- Schöner, T.A.; Gassel, S.; Osawa, A.; Tobias, N.J.; Okuno, Y.; Sakakibara, Y.; Shindo, K.; Sandmann, G.; Bode, H.B. Aryl Polyenes, a Highly Abundant Class of Bacterial Natural Products, Are Functionally Related to Antioxidative Carotenoids. ChemBioChem 2015, 17, 247–253. [Google Scholar] [CrossRef] [PubMed]
- Repka, L.M.; Chekan, J.R.; Nair, S.K.; van der Donk, W.A. Mechanistic Understanding of Lanthipeptide Biosynthetic Enzymes. Chem. Rev. 2017, 117, 5457–5520. [Google Scholar] [CrossRef] [PubMed]
- Heather, Z.; Holden, M.T.G.; Steward, K.F.; Parkhill, J.; Song, L.; Challis, G.L.; Robinson, C.; Davis-Poynter, N.; Waller, A.S. A novel streptococcal integrative conjugative element involved in iron acquisition. Mol. Microbiol. 2008, 70, 1274–1292. [Google Scholar] [CrossRef] [PubMed]
- Miethke, M.; Marahiel, M.A. Siderophore-Based Iron Acquisition and Pathogen Control. Microbiol. Mol. Biol. Rev. 2007, 71, 413–451. [Google Scholar] [CrossRef]
- Heath, L.S.; Heath, H.E.; LeBlanc, P.A.; Smithberg, S.R.; Dufour, M.; Simmonds, R.S.; Sloan, G.L. The Streptococcolytic Enzyme Zoocin A Is a Penicillin-Binding Protein. FEMS Microbiol. Lett. 2004, 236, 205–211. [Google Scholar] [CrossRef]
- Akesson, M.; Dufour, M.; Sloan, G.L.; Simmonds, R.S. Targeting of streptococci by zoocin A. FEMS Microbiol. Lett. 2007, 270, 155–161. [Google Scholar] [CrossRef]
- Chen, Y.; Simmonds, R.S.; Timkovich, R. Proposed docking interface between peptidoglycan and the target recognition domain of zoocin A. Biochem. Biophys. Res. Commun. 2013, 441, 297–300. [Google Scholar] [CrossRef]
- Gargis, S.R.; Gargis, A.S.; Heath, H.E.; Heath, L.S.; LeBlanc, P.A.; Senn, M.M.; Berger-Bächi, B.; Simmonds, R.S.; Sloan, G.L. Zif, the Zoocin A Immunity Factor, Is a FemABX-like Immunity Protein with a Novel Mode of Action. Appl. Environ. Microbiol. 2009, 75, 6205–6210. [Google Scholar] [CrossRef]


| Strains | Source |
|---|---|
| 3725, 3726, 3727, 3728, 3729, 3730, 3731, 3739, 3740, 3741, 3742, 3743, 3744, 3745, 3751, 3752 | Farm 1 *—Minas Gerais, Brazil |
| 3770, 3771, 3772, 3773, 3774, 3775, 3776 | Farm 2 *—São Paulo, Brazil |
| 3797, 3798, 3799, 3800, 3801, 3802, 3804 | Farm 3 *—Minas Gerais, Brazil |
| 3891, 3892, 3893, 3894, 3895, 3896, 3897, 3898, 3899 | Farm 4 *—Minas Gerais, Brazil |
| 129, 194, 835, 962, 1312, 1315, 3191, 3327, 3333, 3339, 3340, 3345, 3370, 3834, 3835 | NA |
| Genomes | Accession Numbers |
|---|---|
| Streptococcus acidominimus NCTC 12957 | GCA_900459045.1 |
| Streptococcus agalactiae NCTC 8181 | GCA_900458965.1 |
| Streptococcus anginosus subsp. anginosus NCTC 10713 | GCA_900636475.1 |
| Streptococcus anginosus subsp. whileyi CCUG 39159 | GCA_000257765.1 |
| Streptococcus constellatus subsp. constellatus NCTC 11325 | GCA_900459125.1 |
| Streptococcus constellatus subsp. pharyngis SK 1060 | GCA_000223295.2 |
| Streptococcus criceti HS 6 | GCA_000187975.3 |
| Streptococcus downei NCTC 11391 | GCA_900459175.1 |
| Streptococcus dysgalactiae subsp. dysgalactiae NCTC 13731 | GCA_900459225.1 |
| Streptococcus dysgalactiae subsp. equisimilis NCTC 13762 | GCA_900459095.1 |
| Streptococcus equi subsp. equi ATCC 33398 | GCA_900156215.1 |
| Streptococcus equi subsp. ruminatorum CECT 5772 | GCA_000706805.1 |
| Streptococcus equi subsp. zooepidemicus NCTC 4676 | GCA_900459475.1 |
| Streptococcus equinus NCTC 12969 | GCA_900459295.1 |
| Streptococcus gallolyticus subsp. gallolyticus DSM 16831 | GCA_002000985.1 |
| Streptococcus gallolyticus subsp. macedonicus NCTC 13767 | GCA_900459545.1 |
| Streptococcus gallolyticus subsp. pasteurianus NCTC 13784 | GCA_900478025.1 |
| Streptococcus gordonii NCTC 7865 | GCA_900475015.1 |
| Streptococcus intermedius NCTC 11324 | GCA_900475975.1 |
| Streptococcus mitis NCTC 12261 | GCA_000148585.3 |
| Streptococcus mutans NCTC 10449 | GCA_900475095.1 |
| Streptococcus oralis subsp. dentisani CECT 7747 | GCA_000382825.1 |
| Streptococcus oralis subsp. oralis NCTC 11427 | GCA_900637025.1 |
| Streptococcus oralis subsp. tigurinus AZ 3a | GCA_000344275.1 |
| Streptococcus parasanguinis ATCC 15912 | GCA_000164675.2 |
| Streptococcus pneumoniae NCTC 7465 | GCA_001457635.1 |
| Streptococcus pseudopneumoniae CCUG 49455 | GCA_002087075.1 |
| Streptococcus pseudoporcinus NCTC 13786 | GCA_900637075.1 |
| Streptococcus pyogenes NCTC 8198 | GCA_002055535.1 |
| Streptococcus salivarius subsp. salivarius NCTC 8618 | GCA_900636435.1 |
| Streptococcus sanguinis NCTC 7863 | GCA_900475505.1 |
| Streptococcus sobrinus NCTC 12279 | GCA_900475395.1 |
| Streptococcus suis S735 | GCA_000294495.1 |
| Streptococcus thermophilus NCTC 12958 | GCA_900474985.1 |
| Streptococcus uberis NCTC 3858 | GCA_900475595.1 |
| Streptococcus vestibularis ATCC 49124 | GCA_000188295.1 |
| Lactococcus lactis subsp. cremoris ATCC 19257 (outgroup) | GCA_004354515.1 |
| Lactococcus lactis subsp. lactis ATCC 19435 (outgroup) | GCA_900099625.1 |
| UniProt Entry | Organism | Protein | Size |
|---|---|---|---|
| A0A076Z6H4 | Streptococcus agalactiae | Peptidase (M23/M37) | 299 |
| A0A0H1Y377 | Streptococcus agalactiae | Peptidase (M23/M37) | 299 |
| Q8E7W5 | Streptococcus agalactiae serotype III (NEM316) | Uncharacterized | 299 |
| A0A380IVD1 | Streptococcus agalactiae | Peptidase (M23/M37) | 301 |
| A0A139QJJ1 | Streptococcus constellatus | Peptidase (M23/M37) | 285 |
| A0A2J9X4T6 | Streptococcus sp. FDAARGOS_146 | Zoocin A | 255 |
| A0A1E9XU90 | Streptococcus sp. HMSC034B05 | Zoocin A | 238 |
| A0A1F0CHF4 | Streptococcus sp. HMSC069D09 | Zoocin A | 299 |
| S8FKX0 | Streptococcus agalactiae FSL S3-277 | Zoocin A | 299 |
| A0A656FWC3 | Streptococcus agalactiae (ATCC 13813/DSM 2134/JCM 5671/NCIMB 701348/NCTC 8181) | Peptidase (M23/M37) | 299 |
| A0A656SN98 | Streptococcus equi subsp. zooepidemicus SzAM60 | Peptidase (M24/M37) | 285 |
| Q8E2F7 | Streptococcus agalactiae serotype V (ATCC BAA-611/2603 V/R) | Peptidase (M23/M37) | 299 |
| Genome | NCBI Accession Number | Size (bp) |
|---|---|---|
| Streptococcus agalactiae 2-22 | GCA_000967445.1 | 1.838.867 |
| Streptococcus agalactiae 09mas018883 | GCF_000427035.1 | 2.138.694 |
| Streptococcus agalactiae 138P | GCA_000599965.1 | 1.838.701 |
| Streptococcus agalactiae 138spar | GCA_000636115.1 | 1.838.126 |
| Streptococcus agalactiae 2603V/R | GCF_000007265.1 | 2.160.267 |
| Streptococcus agalactiae A909 | GCF_000012705.1 | 2.127.839 |
| Streptococcus agalactiae CNCTC 10/84 | GCF_000782855.1 | 2.013.842 |
| Streptococcus agalactiae COH1 | GCF_000689235.1 | 2.065.074 |
| Streptococcus agalactiae GD201008-001 | GCF_000299135.1 | 2.063.112 |
| Streptococcus agalactiae ILRI005 | GCF_000427075.1 | 2.109.759 |
| Streptococcus agalactiae ILRI112 | GCA_000427055.1 | 2.029.198 |
| Streptococcus agalactiae NEM316 | GCF_000196055.1 | 2.211.485 |
| Streptococcus agalactiae NGBS061 | GCF_000730215.1 | 2.221.207 |
| Streptococcus agalactiae NGBS572 | GCF_000730255.1 | 2.061.426 |
| Streptococcus agalactiae SA20 | GCA_000302475.3 | 1.841.952 |
| Streptococcus agalactiae Strains | Indicator Strains | |||||
|---|---|---|---|---|---|---|
| Cellulomonas fimi NCTC 7547 | Listeria innocua | Staphylococcus aureus | Streptococcus agalactiae I14 | Streptococcus pyogenes | Lactococcus lactis | |
| 3770 | − | − | − | + | − | − |
| 3771 | − | − | − | + | + | − |
| 3772 | − | − | − | + | + | − |
| 3773 | − | − | − | + | + | − |
| 3774 | − | − | − | + | + | − |
| 3775 | − | − | − | + | + | − |
| 3776 | − | − | − | + | + | − |
| 3797 | − | − | − | + | + | − |
| 3798 | − | − | − | + | + | − |
| 3799 | − | − | − | + | + | − |
| 3800 | − | − | − | + | + | − |
| 3801 | − | − | − | + | + | − |
| 3802 | − | − | − | + | + | − |
| 3804 | − | − | − | + | + | − |
| Indicator Strains | Streptococcus agalactiae Strains | |||
|---|---|---|---|---|
| Species | Strains | LGMAI_St_08 | LGMAI_St_11 | LGMAI_St_14 |
| Streptococcus agalactiae | 3725 | + | + | + |
| 3726 | + | + | + | |
| 3727 | + | + | + | |
| 3728 | + | + | + | |
| 3729 | + | + | + | |
| 3730 | + | + | + | |
| 3731 | + | + | + | |
| 3739 | + | + | + | |
| 3740 | + | + | + | |
| 3741 | + | + | + | |
| 3742 | + | + | + | |
| 3743 | + | + | + | |
| 3744 | − | − | − | |
| 3745 | + | + | + | |
| 3751 | + | + | + | |
| 3752 | + | + | + | |
| Streptococcus porcinus | 628 | − | − | − |
| 662 | + | + | + | |
| 790 | − | − | − | |
| 857 | + | + | + | |
| 1058 | + | + | + | |
| 1124 | + | − | − | |
| 1217 | + | + | + | |
| 1451 | − | − | − | |
| 2378 | − | − | − | |
| 3123 | + | + | + | |
| 3176 | − | − | − | |
| 3658 | − | − | − | |
| Streptococcus pyogenes | 1465 | − | − | − |
| 1471 | − | − | − | |
| 1474 | − | − | − | |
| 1996 | − | − | − | |
| 2009 | − | − | − | |
| 2586 | + | + | + | |
| 2587 | + | + | + | |
| 2588 | + | + | + | |
| 2590 | − | + | + | |
| 2591 | + | − | + | |
| 2593 | − | − | − | |
| 2606 | − | − | − | |
| 2608 | − | − | − | |
| 2612 | − | − | − | |
| 2615 | − | − | − | |
| 2617 | − | − | − | |
| 2618 | + | + | + | |
| 2620 | − | − | − | |
| 2625 | + | − | − | |
| Streptococcus uberis | 602 | + | − | − |
| 752 | − | − | − | |
| 959 | − | − | − | |
| 2825 | + | − | − | |
| 3355 | + | + | − | |
| 3351 | + | + | + | |
| 3354 | + | + | + | |
| 3431 | − | − | − | |
| 3485 | + | − | + | |
| 3670 | + | + | + | |
| 3671 | + | + | + | |
| 3724 | − | − | − | |
| Cellulomonas fimi | + | + | + | |
| Micrococcus sp. | + | + | + | |
| Klebsiella pneumoniae | ATCC 13883 | − | − | − |
| KPC | − | − | − | |
| Staphylococcus aureus | ATCC 6538 | − | − | − |
| ATCC 29213 | − | − | − | |
| ATCC 25923 | − | − | − | |
| ATCC 33591 | − | − | − | |
| Parameter | Strains | ||
|---|---|---|---|
| LGMAI_St_08 | LGMAI_St_11 | LGMAI_St_14 | |
| GenBank accession number | JAIWPA000000000 | JAIWPB000000000 | JAIWPC000000000 |
| BioSample accession number | SAMN17072134 | SAMN17072135 | SAMN17072136 |
| BioProject accession number | PRJNA637496 | ||
| Total genome size | 2,397,674 | 2,292,224 | 2,289,478 |
| Number of contigs | 156 | 334 | 292 |
| N50 | 33,723 | 46,037 | 45,516 |
| L50 | 25 | 16 | 18 |
| GC content (%) | 35.7% | 35.6% | 35.6% |
| Number of genes | 2499 | 2502 | 2450 |
| Number of CDS | 2434 | 2452 | 2400 |
| Number of coding genes | 2334 | 2361 | 2310 |
| rRNA | 10 | 6 | 5 |
| tRNA | 52 | 41 | 42 |
| Pseudo Genes | 100 | 91 | 90 |
| Genes | Strains | |||
|---|---|---|---|---|
| LGMAI_St_08 | LGMAI_St_11 | LGMAI_St_14 | ||
| Locus cps | cpsA | 100% | 100% | 100% |
| cpsB | 100% | 100% | 100% | |
| cpsC | 100% | 100% | 100% | |
| cpsD | 100% | 100% | 100% | |
| cpsE | 99.35% | 99.35% | 99.35% | |
| cpsF | 100% | 100% | 100% | |
| cpsK | 100% | 100% | 100% | |
| cpsL | 100% | 100% | 100% | |
| neuA | 100% | 100% | 100% | |
| neuB | 100% | 100% | 100% | |
| neuC | 100% | 100% | 100% | |
| neuD | 100% | 100% | 100% | |
| Operon cyl | cylX | 100% | 100% | 100% |
| cylD | 100% | 100% | 100% | |
| cylG | 100% | 100% | 100% | |
| acpC | 100% | 100% | 100% | |
| cylZ | 100% | 100% | 100% | |
| cylA | 95.27% | 100% | 100% | |
| cylB | 100% | 100% | 100% | |
| cylF | 100% | 100% | 100% | |
| cylI | 100% | 100% | 100% | |
| cylJ | 100% | 100% | 100% | |
| cylK | 100% | 100% | 100% | |
| hylB | 100% | 100% | 100% | |
| cfa/cfb | 100% | NA | NA | |
| Strain | Genomic Island | GC Deviation | Codon Usage Deviation | Specific Proteins | Hypothetical Proteins | Position | Score |
|---|---|---|---|---|---|---|---|
| LGMAI_St_08 | Pat 1 | 50% | 25% | 41% | 66% | 456,093–470,295 | Normal |
| Pat 2 | 26% | 19% | 46% | 61% | 626,248–675,463 | Normal | |
| Pat 3 | 13% | 0% | 100% | 53% | 1,212,789–1,222,960 | Normal | |
| Pat 4 | 17% | 70% | 52% | 58% | 1,232,986–1,259,538 | Normal | |
| Pat 5 | 23% | 0% | 76% | 46% | 1,318,556–1,330,577 | Normal | |
| Pat 6 | 16% | 6% | 80% | 96% | 2,297,372–2,321,939 | Normal | |
| Misc 1 | 25% | 39% | 20% | 70% | 706,904–759,427 | Normal | |
| Misc 2 | 32% | 56% | 24% | 48% | 961,688–986,574 | Normal | |
| LGMAI_St_11 | Res 1 | 20% | 29% | 8% | 58% | 58,075–88,763 | Normal |
| Res 2 | 14% | 0% | 42% | 57% | 414,714–424,358 | Normal | |
| Res 3 | 0% | 21% | 21% | 71% | 471,775–483,184 | Normal | |
| Res 4 | 6% | 43% | 19% | 67% | 782,301–843,622 | Normal | |
| Res 5 | 20% | 57% | 11% | 70% | 905,416–968,419 | Normal | |
| Pat 1 | 6% | 6% | 43% | 68% | 181,057–198,487 | Normal | |
| Pat 2 | 4% | 0% | 100% | 26% | 564,594–586,997 | Normal | |
| Pat 3 | 0% | 90% | 45% | 72% | 634,882–646,520 | Strong | |
| Pat 4 | 18% | 23% | 46% | 55% | 1,477,345–1,527,804 | Normal | |
| Pat 5 | 15% | 0% | 76% | 46% | 1,650,399–1,662,421 | Normal | |
| Pat 6 | 1% | 33% | 43% | 78% | 1,776,966–1,868,151 | Normal | |
| Pat 7 | 23% | 53% | 42% | 69% | 2,144,963–2,170,746 | Normal | |
| Pat 8 | 14% | 35% | 67% | 85% | 2,192,667–2,214,594 | Normal | |
| Pat 9 | 7% | 0% | 40% | 88% | 2,217,596–2,244,976 | Normal | |
| Misc 1 | 6% | 100% | 20% | 53% | 302,365–327,227 | Strong | |
| Misc 2 | 26% | 86% | 26% | 53% | 607,341–620,962 | Strong | |
| Misc 3 | 20% | 45% | 12% | 58% | 2,042,812–2,067,825 | Normal | |
| LGMAI_St_14 | Res 1 | 23% | 29% | 8% | 58% | 190,826–221,514 | Normal |
| Res 2 | 0% | 33% | 26% | 66% | 528,604–541,172 | Normal | |
| Pat 1 | 4% | 0% | 100% | 26% | 561,516–583,919 | Normal | |
| Pat 2 | 21% | 0% | 78% | 42% | 1,105,918–1,120,919 | Normal | |
| Pat 3 | 41% | 50% | 41% | 66% | 1,195,840–1,205,898 | Normal | |
| Pat 4 | 25% | 29% | 51% | 65% | 1,247,996–1,291,657 | Normal | |
| Pat 5 | 15% | 0% | 76% | 46% | 1,670,544–1,682,566 | Normal | |
| Pat 6 | 0% | 11% | 52% | 71% | 1,770,456–1,836,354 | Normal | |
| Pat 7 | 32% | 21% | 46% | 67% | 2,139,499–2,166,899 | Normal | |
| Pat 8 | 15% | 33% | 66% | 84% | 2,193,909–2,213,031 | Normal | |
| Pat 9 | 11% | 0% | 46% | 92% | 2,216,033–2,242,629 | Normal | |
| Misc 1 | 6% | 100% | 20% | 53% | 43,216–68,078 | Strong | |
| Misc 2 | 20% | 93% | 20% | 60% | 655,073–668,583 | Strong | |
| Misc 3 | 15% | 51% | 23% | 71% | 778,157–830,840 | Normal | |
| Misc 4 | 25% | 42% | 10% | 53% | 2,044,364–2,073,403 | Normal |
| Region | Database | Accession Number | Position | e-Value |
|---|---|---|---|---|
| FemABX peptidyl transferase | Pfam | PF02388 | 6–406 | |
| Acyl-CoA N-acyltransferase | Superfamily | SSF55729 | 1–159 | |
| Acyl-CoA N-acyltransferase | 163–403 | |||
| Class I and II aminoacyl-tRNA synthetase | SSF46589 | 239–303 | ||
| (tRNA-binding arm) | Coils | Coil | 243–263 | NA |
| Category | Strains | ||
|---|---|---|---|
| LGMAI_St_08 | LGMAI_St_11 | LGMAI_St_14 | |
| Cellular processes and signaling | 448 | 441 | 433 |
| Information storage and processing | 540 | 518 | 528 |
| Metabolism | 720 | 730 | 725 |
| Poorly characterized | 461 | 441 | 441 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vidal Amaral, J.R.; Jucá Ramos, R.T.; Almeida Araújo, F.; Bentes Kato, R.; Figueira Aburjaile, F.; de Castro Soares, S.; Góes-Neto, A.; Matiuzzi da Costa, M.; Azevedo, V.; Brenig, B.; et al. Bacteriocin Producing Streptococcus agalactiae Strains Isolated from Bovine Mastitis in Brazil. Microorganisms 2022, 10, 588. https://doi.org/10.3390/microorganisms10030588
Vidal Amaral JR, Jucá Ramos RT, Almeida Araújo F, Bentes Kato R, Figueira Aburjaile F, de Castro Soares S, Góes-Neto A, Matiuzzi da Costa M, Azevedo V, Brenig B, et al. Bacteriocin Producing Streptococcus agalactiae Strains Isolated from Bovine Mastitis in Brazil. Microorganisms. 2022; 10(3):588. https://doi.org/10.3390/microorganisms10030588
Chicago/Turabian StyleVidal Amaral, João Ricardo, Rommel Thiago Jucá Ramos, Fabrício Almeida Araújo, Rodrigo Bentes Kato, Flávia Figueira Aburjaile, Siomar de Castro Soares, Aristóteles Góes-Neto, Mateus Matiuzzi da Costa, Vasco Azevedo, Bertram Brenig, and et al. 2022. "Bacteriocin Producing Streptococcus agalactiae Strains Isolated from Bovine Mastitis in Brazil" Microorganisms 10, no. 3: 588. https://doi.org/10.3390/microorganisms10030588
APA StyleVidal Amaral, J. R., Jucá Ramos, R. T., Almeida Araújo, F., Bentes Kato, R., Figueira Aburjaile, F., de Castro Soares, S., Góes-Neto, A., Matiuzzi da Costa, M., Azevedo, V., Brenig, B., Soares de Oliveira, S., & Soares Rosado, A. (2022). Bacteriocin Producing Streptococcus agalactiae Strains Isolated from Bovine Mastitis in Brazil. Microorganisms, 10(3), 588. https://doi.org/10.3390/microorganisms10030588












