Cryptic Prophages Contribution for Campylobacter jejuni and Campylobacter coli Introgression
Abstract
1. Introduction
2. Materials and Methods
2.1. Campylobacter Genomes
2.2. Prophage Screening Using Bioinformatic Tools
2.3. Prophage Induction
2.4. Phylogenetic Analysis
2.5. Testing Introgression Using ABBA-BABA Statistics
2.6. Prophage Nuclease Screening
3. Results
3.1. Identification of Prophages
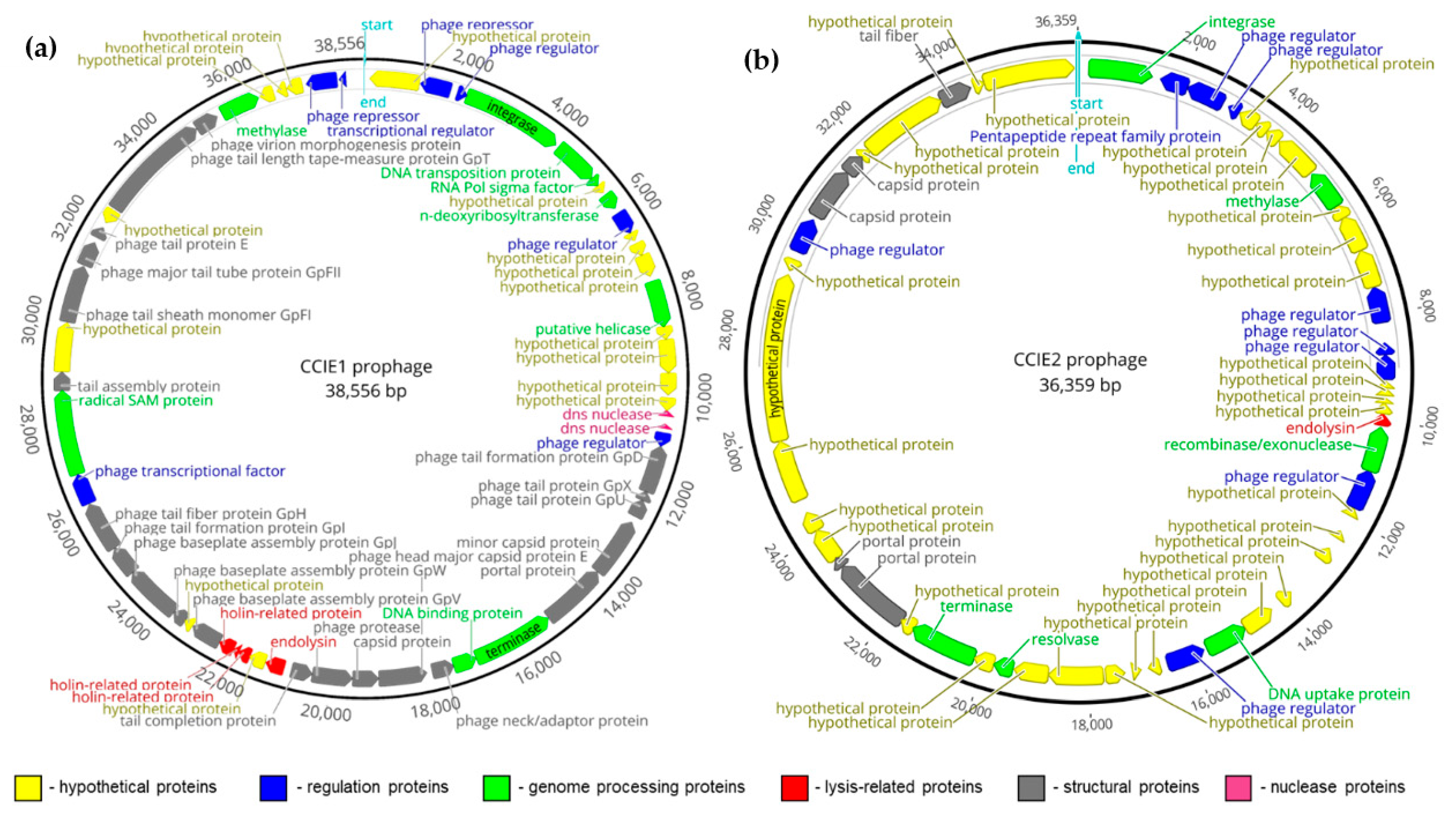
3.2. Characterization of the New CCIE1 and CCIE2 Prophages
| Characteristics | CCIE1 Prophage | CCIE2 Prophage | |
|---|---|---|---|
| Genome length | 38,556 bp | 36,356 bp | |
| GC content | 30.20% | 28.50% | |
| GenBank Accession No. | MZ667635 | MZ667634 | |
| Closest homologue | CJIE1-2 (HM192820.1) [51] | CJIE2 [12] | |
| Coverage with the closest homologue | 79% | 54% | |
| Identity with the closest homologue | 96.33% | 93.85% | |
| Host strain 1 | C. coli Cc63-H-18 | C. coli Cco1598-H-13 | |
| Insertion site 2 | 5′ | Bis-ABC ATPase YbiT (N149_0417) | tRNA-Leu-GAG (N149_0910) |
| 3′ | putative lipoprotein (N149_01930) | putative NTPase (N149_01865) | |
| Number of CDS 3 | 59 | 54 | |
| Main phage genes detected 3 | Integrase, dns nuclease, endolysin, holin, methylase, terminase, several phage structural and regulation proteins. | Integrase, endolysin, recombinase/exonuclease, methylase, resolvase, terminase, several phage structural and regulation proteins. | |
| VIRFAM analysis | Almost complete Mu-like Myoviridae phage (head closure protein missing) | Incomplete phage (several structural proteins missing) | |
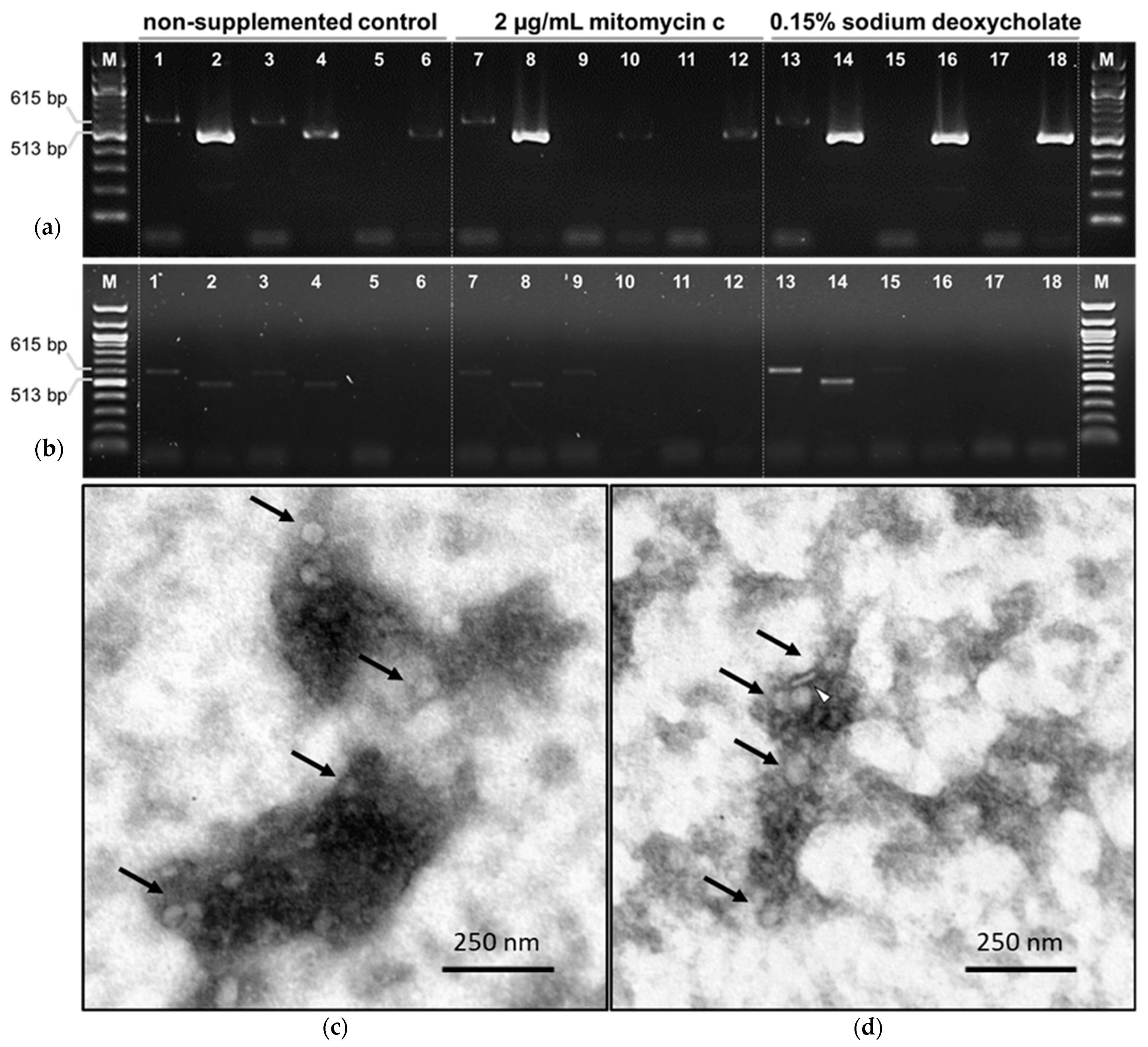
3.3. Prophage Induction Assays
3.4. Identification of CJIE1-, CJIE4-, and CCIE2-like Prophages within Campylobacter spp.
3.5. Insertion Sites of the Prophages Identified
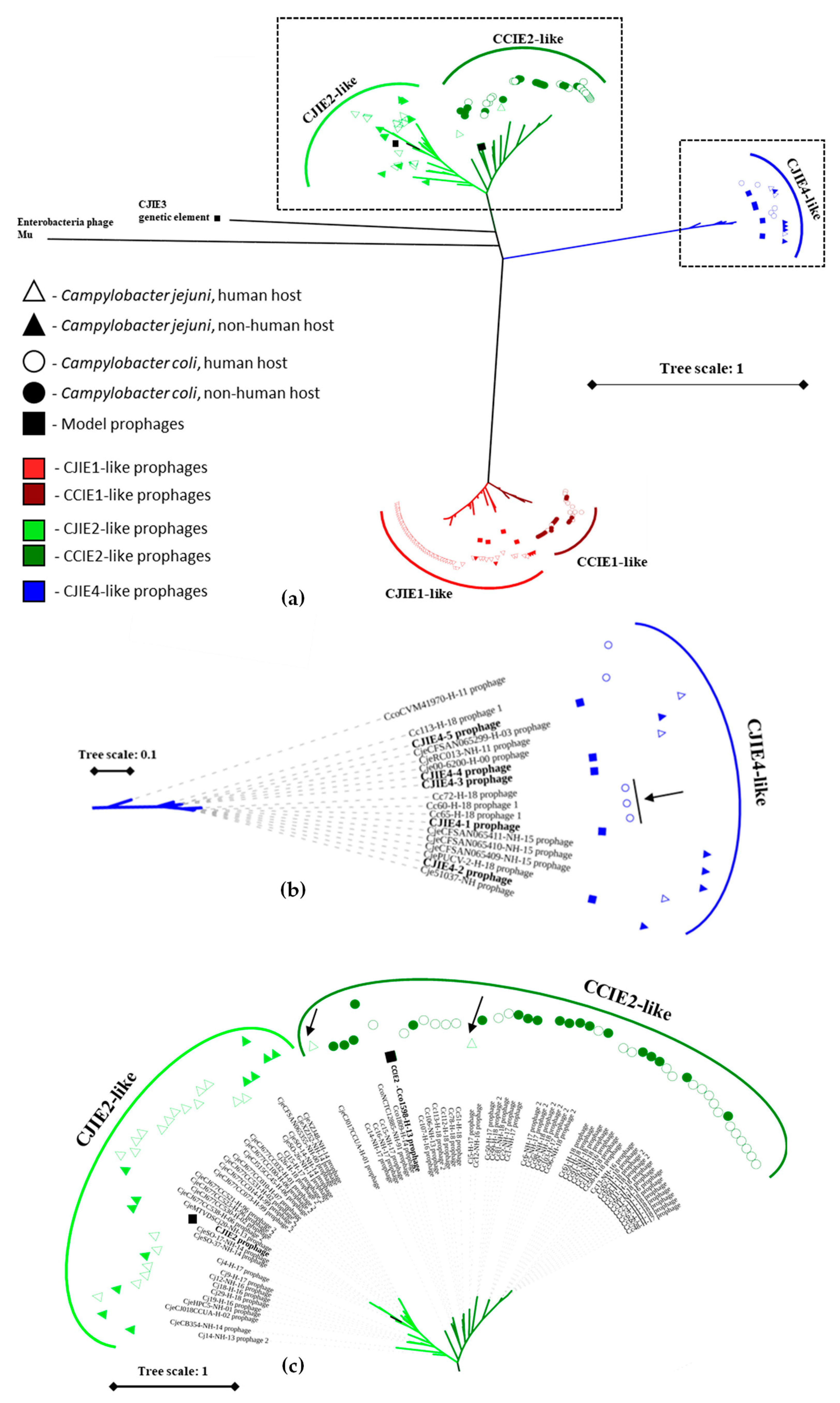
3.6. Phylogenetic Analysis of the Prophages Identified
3.7. Newly Identified MLST Allelic Variations and MLST Profiles
3.8. Nucleases Encoded by Campylobacter Prophages
4. Discussion
Supplementary Materials
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- European Food Safety Authority; European Centre for Disease Prevention and Control. The European Union One Health 2018 Zoonoses Report. EFSA J. 2019, 17, 5926. [Google Scholar] [CrossRef]
- Fitzgerald, C. Campylobacter. Clin. Lab. Med. 2015, 35, 289–298. [Google Scholar] [CrossRef]
- Kaakoush, N.O.; Castaño-Rodríguez, N.; Mitchell, H.M.; Ming-Man, S. Global Epidemiology of Campylobacter Infection. Clin. Microbiol. Rev. 2015, 28, 687–720. [Google Scholar] [CrossRef]
- Sheppard, S.K.; Maiden, M.C.J. The Evolution of Campylobacter jejuni and Campylobacter coli. Cold Spring Harb. Perspect. Biol. 2015, 7, a018119. [Google Scholar] [CrossRef]
- Wingstrand, A.; Neimann, J.; Engberg, J.; Nielsen, E.M.; Gerner-Smidt, P.; Wegener, H.C.; Mølbak, K. Fresh Chicken as Main Risk Factor for Campylobacteriosis, Denmark. Emerg. Infect. Dis. 2006, 12, 280–284. [Google Scholar] [CrossRef]
- Harris, N.V.; Weiss, N.S.; Nolan, C.M. The Role of Poultry and Meats in the Etiology of Campylobacter jejuni/coli Enteritis. Am. J. Public Health 1986, 76, 407–411. [Google Scholar] [CrossRef]
- Wilson, D.J.; Gabriel, E.; Leatherbarrow, A.J.H.; Cheesbrough, J.; Gee, S.; Bolton, E.; Fox, A.; Hart, C.A.; Diggle, P.J.; Fearnhead, P. Rapid evolution and the importance of recombination to the gastroenteric pathogen Campylobacter jejuni. Mol. Biol. Evol. 2009, 26, 385–397. [Google Scholar] [CrossRef]
- Beauchamp, J.M.; Leveque, R.M.; Dawid, S.; DiRita, V.J. Methylation-dependent DNA discrimination in natural transformation of Campylobacter jejuni. Proc. Natl. Acad. Sci. USA 2017, 114, E8053–E8061. [Google Scholar] [CrossRef]
- Sheppard, S.K.; Didelot, X.; Jolley, K.A.; Darling, A.E.; Pascoe, B.; Meric, G.; Kelly, D.J.; Cody, A.; Colles, F.M.; Strachan, N.J.C.; et al. Progressive genome-wide introgression in agricultural Campylobacter coli. Mol. Ecol. 2013, 22, 1051–1064. [Google Scholar] [CrossRef]
- Sheppard, S.K.; McCarthy, N.D.; Falush, D.; Maiden, M.C.J. Convergence of Campylobacter species: Implications for bacterial evolution. Science 2008, 320, 237–239. [Google Scholar] [CrossRef] [PubMed]
- Bobay, L.M. The prokaryotic species concept and challenges. Pangenome Divers. Dyn. Evol. Genomes 2020, 21–49. [Google Scholar] [CrossRef]
- Fouts, D.E.; Mongodin, E.F.; Mandrell, R.E.; Miller, W.G.; Rasko, D.A.; Ravel, J.; Brinkac, L.M.; Deboy, R.T.; Parker, C.T.; Daugherty, S.C.; et al. Major Structural Differences and Novel Potential Virulence Mechanisms from the Genomes of Multiple Campylobacter Species. PLoS Biol. 2005, 3, e15. [Google Scholar] [CrossRef]
- Skarp, C.P.A.; Akinrinade, O.; Nilsson, A.J.E.; Ellström, P.; Myllykangas, S.; Rautelin, H. Comparative genomics and genome biology of invasive Campylobacter jejuni. Sci. Rep. 2015, 5, 17300. [Google Scholar] [CrossRef]
- Clark, C.G.; Chong, P.M.; Mccorrister, S.J.; Mabon, P.; Walker, M.; Westmacott, G.R. DNA Sequence Heterogeneity of Campylobacter jejuni CJIE4 Prophages and Expression of Prophage Genes. PLoS ONE 2014, 9, e95349. [Google Scholar] [CrossRef] [PubMed]
- Negretti, N.M.; Gourley, C.R.; Clair, G.; Adkins, J.N.; Konkel, M.E. The food-borne pathogen Campylobacter jejuni responds to the bile salt deoxycholate with countermeasures to reactive oxygen species. Sci. Rep. 2017, 7, 15455. [Google Scholar] [CrossRef] [PubMed]
- Clark, C.G.; Chong, P.M.; McCorrister, S.J.; Simon, P.; Walker, M.; Lee, D.M.; Nguy, K.; Cheng, K.; Gilmour, M.W.; Westmacott, G.R. The CJIE1 prophage of Campylobacter jejuni affects protein expression in growth media with and without bile salts. BMC Microbiol. 2014, 14, 70. [Google Scholar] [CrossRef]
- Clark, C.G.; Grant, C.C.R.; Pollari, F.; Marshall, B.; Moses, J.; Tracz, D.M.; Gilmour, M.W. Effects of the Campylobacter jejuni CJIE1 prophage homologs on adherence and invasion in culture, patient symptoms, and source of infection. BMC Microbiol. 2012, 12, 269. [Google Scholar] [CrossRef]
- Gaasbeek, E.J.; Wagenaar, J.A.; Guilhabert, M.R.; Wo, M.M.S.M.; van Putten, J.P.M.; van der Graaf-van Bloois, L.; Parker, C.T.; van der Wal, F.J. A DNase Encoded by Integrated Element CJIE1 Inhibits Natural Transformation of Campylobacter jejuni. J. Bacteriol. 2009, 191, 2296–2306. [Google Scholar] [CrossRef]
- Gaasbeek, E.J.; Wagenaar, J.A.; Guilhabert, M.R.; van Putten, J.P.M.; Parker, C.T.; van der Wal, F.J. Nucleases Encoded by the Integrated Elements CJIE2 and CJIE4 Inhibit Natural Transformation of Campylobacter jejuni. J. Bacteriol. 2010, 192, 936–941. [Google Scholar] [CrossRef]
- Lemos, M.L.; Nunes, A.; Ancora, M.; Cammà, C.; Martins da Costa, P.; Oleastro, M. Campylobacter jejuni in Different Canine Populations: Characteristics and Zoonotic Potential. Microorganisms 2021, 9, 2231. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Sun, H.; Zhang, C.; Cheng, L.; Peng, Y.; Deng, Z.; Wang, D.; Wang, Y.; Hu, M.; Liu, W.; et al. Prophage Hunter: An integrative hunting tool for active prophages. Nucleic Acids Res. 2019, 47, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Pearson, B.; Rokney, A.; Crossman, L.; Miller, W.; Wain, J.; Vliet, A. Complete Genome Sequence of The Campylobacter coli Clinical Isolate 15-537360. Genome Announc. 2013, 1, e01056-13. [Google Scholar] [CrossRef]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef]
- Boratyn, G.M.; Camacho, C.; Cooper, P.S.; Coulouris, G.; Fong, A.; Ma, N.; Madden, T.L.; Matten, W.T.; McGinnis, S.D.; Merezhuk, Y.; et al. BLAST: A more efficient report with usability improvements. Nucleic Acids Res. 2013, 41, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef]
- Vale, F.F.; Matos, A.P.A.; Carvalho, P.; Vtor, J.M.B. Helicobacter pylori phage screening. Microsc. Microanal. 2008, 14, 150–151. [Google Scholar] [CrossRef]
- Lehours, P.; Vale, F.F.; Bjursell, M.K.; Melefors, O.; Advani, R.; Glavas, S.; Guegueniat, J.; Gontier, E.; Lacomme, S.; Matos, A.A.; et al. Genome sequencing reveals a phage in Helicobacter pylori. mBio 2011, 2, e00239-11. [Google Scholar] [CrossRef] [PubMed]
- Vale, F.F.; Vadivelu, J.; Oleastro, M.; Breurec, S.; Engstrand, L.; Perets, T.T.; Mégraud, F.; Lehours, P. Dormant phages of Helicobacter pylori reveal distinct populations in Europe. Sci. Rep. 2015, 5, 14333. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Jukes, T.H.; Cantor, C. Evolution of Protein Molecules; Academic Press: New York, NY, USA, 1969; Chapter 24. [Google Scholar]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Morgan, G.J.; Hatfull, G.F.; Casjens, S.; Hendrix, R.W. Bacteriophage Mu genome sequence: Analysis and comparison with Mu-like prophages in Haemophilus, Neisseria and Deinococcus. J. Mol. Biol. 2002, 317, 337–359. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Green, R.E.; Krause, J.; Briggs, A.W.; Maricic, T.; Stenzel, U.; Kircher, M.; Patterson, N.; Li, H.; Zhai, W.; Fritz, M.H.-Y.; et al. A draft sequence of the Neandertal genome. Science 2010, 328, 710–722. [Google Scholar] [CrossRef] [PubMed]
- Durand, E.Y.; Patterson, N.; Reich, D.; Slatkin, M. Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 2011, 28, 2239–2252. [Google Scholar] [CrossRef]
- Martin, S.H.; Davey, J.W.; Jiggins, C.D. Evaluating the use of ABBA-BABA statistics to locate introgressed loci. Mol. Biol. Evol. 2015, 32, 244–257. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.H.; Davey, J.W.; Salazar, C.; Jiggins, C.D. Recombination rate variation shapes barriers to introgression across butterfly genomes. PLoS Biol. 2019, 17, e2006288. [Google Scholar] [CrossRef]
- Hibbins, M.S.; Hahn, M.W. The Timing and Direction of Introgression Under the Multispecies Network Coalescent. Genetics 2019, 211, 1059–1073. [Google Scholar] [CrossRef]
- Page, A.J.; Taylor, B.; Delaney, A.J.; Soares, J.; Seemann, T.; Keane, J.A.; Harris, S.R. SNP-sites: Rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2016, 2, e000056. [Google Scholar] [CrossRef] [PubMed]
- Pfeifer, B.; Wittelsbürger, U.; Ramos-Onsins, S.E.; Lercher, M.J. PopGenome: An efficient Swiss army knife for population genomic analyses in R. Mol. Biol. Evol. 2014, 31, 1929–1936. [Google Scholar] [CrossRef]
- Parker, C.T.; Quiñones, B.; Miller, W.G.; Horn, S.T.; Mandrell, R.E. Comparative genomic analysis of Campylobacter jejuni strains reveals diversity due to genomic elements similar to those present in C. jejuni strain RM1221. J. Clin. Microbiol. 2006, 44, 4125–4135. [Google Scholar] [CrossRef] [PubMed]
- Kovanen, S.; Rossi, M.; Pohja-Mykra, M.; Nieminen, T.; Raunio-Saarnisto, M.; Sauvala, M.; Fredriksson-Ahomaa, M.; Hanninen, M.; Kivisto, R. Population Genetics and Characterization of Campylobacter jejuni isolates drom Western Jackdaws and Game Birds in Finland. Appl. Environ. Microbiol. 2019, 85, e02365-18. [Google Scholar] [CrossRef] [PubMed]
- Revez, J.; Llarena, A.; Schott, T.; Kuusi, M.; Hakkinen, M.; Kivistö, R.; Hänninen, M.; Rossi, M. Genome analysis of Campylobacter jejuni strains isolated from a waterborne outbreak. BMC Genom. 2014, 15, 768. [Google Scholar] [CrossRef]
- Kivistö, R.; Kovanen, S.; Haan, A.S.; Schott, T.; Rahkio, M.; Rossi, M.; Hänninen, M. Evolution and Comparative Genomics of Campylobacter jejuni. Genome Biol. Evol. 2014, 6, 2424–2438. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.; Jerome, J.; Plovanich-Jones, A.; Smith, E.; Gettings, J.; Kim, H.; Landgraf, J.; Lefébure, T.; Kopper, J.; Rathinam, V.; et al. Outcome of infection of C57BL/6 IL-10−/− mice with Campylobacter jejuni strains is correlated with genome content of open reading frames up- and down-regulated in vivo. Microb. Pathog. 2014, 54, 1–19. [Google Scholar] [CrossRef]
- Hepworth, P.J.; Ashelford, K.E.; Hinds, J.; Gould, K.A.; Witney, A.A.; Williams, N.J.; Leatherbarrow, H.; French, N.P.; Birtles, R.J.; Mendonca, C.; et al. Genomic variations define divergence of water/wildlife-associated Campylobacter jejuni niche specialists from common clonal complexes. Environ. Microbiol. 2011, 13, 1549–1560. [Google Scholar] [CrossRef]
- Touchon, M.; Moura de Sousa, J.A.; Rocha, E.P. Embracing the enemy: The diversification of microbial gene repertoires by phage-mediated horizontal gene transfer. Curr. Opin. Microbiol. 2017, 38, 66–73. [Google Scholar] [CrossRef]
- Clark, C.G. Sequencing of CJIE1 prophages from Campylobacter jejuni isolates reveals the presence of inserted and (or) deleted genes. Can. J. Microbiol. 2011, 57, 795–809. [Google Scholar] [CrossRef]
- Olszak, T.; Latka, A.; Roszniowski, B.; Valvano, M.A.; Drulis-Kawa, Z. Phage Life Cycles Behind Bacterial Biodiversity. Curr. Med. Chem. 2017, 24, 3987–4001. [Google Scholar] [CrossRef]
- Clark, C.G.; Ng, L.K. Sequence variability of Campylobacter temperate bacteriophages. BMC Microbiol. 2008, 8, 49. [Google Scholar] [CrossRef] [PubMed]
- Feiner, R.; Argov, T.; Rabinovich, L.; Sigal, N.; Borovok, I.; Herskovits, A. A new perspective on lysogeny: Prophages as active regulatory switches of bacteria. Nat. Rev. Microbiol. 2015, 13, 641–650. [Google Scholar] [CrossRef] [PubMed]
- Harrison, E.; Brockhurst, M.A. Ecological and Evolutionary Benefits of Temperate Phage: What Does or Doesn’t Kill You Makes You Stronger. BioEssays 2017, 39, 1700112. [Google Scholar] [CrossRef] [PubMed]
- French, N.P.; Zhang, J.; Carter, G.P.; Midwinter, A.C.; Biggs, P.J.; Dyet, K.; Gilpin, B.J.; Ingle, D.J.; Mulqueen, K.; Rogers, L.E.; et al. Genomic analysis of fluoroquinolone- and tetracycline-resistant Campylobacter jejuni sequence type 6964 in humans and poultry, New Zealand, 2014–2016. Emerg. Infect. Dis. 2019, 25, 2226–2234. [Google Scholar] [CrossRef] [PubMed]
- Clark, C.G.; Berry, C.; Walker, M.; Petkau, A.; Barker, D.O.R.; Guan, C.; Reimer, A.; Taboada, E.N. Genomic insights from whole genome sequencing of four clonal outbreak Campylobacter jejuni assessed within the global C. jejuni population. BMC Genom. 2016, 17, 990. [Google Scholar] [CrossRef] [PubMed]
- Jolley, K.A.; Bray, J.E.; Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications [version 1; referees: 2 approved]. Wellcome Open Res. 2018, 3, 124. [Google Scholar] [CrossRef]
- Pratama, A.A.; Chaib De Mares, M.; van Elsas, J.D. Evolutionary History of Bacteriophages in the Genus Paraburkholderia. Front. Microbiol. 2018, 9, 835. [Google Scholar] [CrossRef]
- Secka, O.; Vale, F.F.; Buissonnière, A.; Thomas, J.E.; Mégraud, F.; Lehours, P. Phylogeographic agreement between prophage and bacterial housekeeping genes in Helicobacter pylori strains from The Gambia. Helicobacter 2017, 22, e12394. [Google Scholar] [CrossRef]
- Vale, F.F.; Lehours, P. Relating phage genomes to Helicobacter pylori population structure: General steps using whole-genome sequencing data. Int. J. Mol. Sci. 2018, 19, 1831. [Google Scholar] [CrossRef]
- Vale, F.F.; Nunes, A.; Oleastro, M.; Gomes, J.P.; Sampaio, D.A.; Rocha, R.; Vítor, J.M.B.; Engstrand, L.; Pascoe, B.; Berthenet, E.; et al. Genomic structure and insertion sites of Helicobacter pylori prophages from various geographical origins. Sci. Rep. 2017, 7, 42471. [Google Scholar] [CrossRef]
- Bobay, L.M.; Touchon, M.; Rocha, E.P.C. Pervasive domestication of defective prophages by bacteria. Proc. Natl. Acad. Sci. USA 2014, 111, 12127–12132. [Google Scholar] [CrossRef]
- Rezaei Javan, R.; Ramos-Sevillano, E.; Akter, A.; Brown, J.; Brueggemann, A.B. Prophages and satellite prophages are widespread in Streptococcus and may play a role in pneumococcal pathogenesis. Nat. Commun. 2019, 10, 4852. [Google Scholar] [CrossRef] [PubMed]
- Kasman, M. Bacteriophage. In Brenner’s Encyclopedia of Genetics, 2nd ed.; Academic Press: Treasure Island, FL, USA, 2013; pp. 280–283. ISBN 9780080961569. [Google Scholar]
- Cavassim, M.I.A.; Moeskjær, S.; Moslemi, C.; Fields, B.; Bachmann, A.; Vilhjálmsson, B.J.; Schierup, M.H.; Young, J.P.W.; Andersen, S.U. Symbiosis genes show a unique pattern of introgression and selection within a Rhizobium leguminosarum species complex. Microb. Genom. 2020, 6, 4. [Google Scholar] [CrossRef]
- Harrison, R.G.; Larson, E.L. Hybridization, Introgression, and the Nature of Species Boundaries. J. Hered. 2014, 105, 795–809. [Google Scholar] [CrossRef]
- Wang, Y.; Taylor, D.E. Natural transformation in Campylobacter species. J. Bacteriol. 1990, 172, 949–955. [Google Scholar] [CrossRef] [PubMed]
- Fortier, L.; Sekulovic, O. Importance of prophages to evolution and virulence of bacterial pathogens. Virulence 2013, 4, 354–365. [Google Scholar] [CrossRef] [PubMed]
- Christie, G.E.; Dokland, T. Pirates of the Caudovirales. Virology 2012, 434, 210–221. [Google Scholar] [CrossRef]
- Dokland, T. Molecular Piracy: Redirection of Bacteriophage Capsid Assembly by Mobile Genetic Elements. Viruses 2019, 11, 1003. [Google Scholar] [CrossRef] [PubMed]
- Lefkowitz, E.J.; Dempsey, D.M.; Hendrickson, R.C.; Orton, R.J.; Siddell, S.G.; Smith, D.B. Virus taxonomy: The database of the International Committee on Taxonomy of Viruses (ICTV). Nucleic Acids Res. 2018, 46, D708–D717. [Google Scholar] [CrossRef]
| Detected Prophage Sequences * | C. jejuni (n = 22) | C. coli (n = 82) | Total (n = 104) |
|---|---|---|---|
| Genomes without prophage sequences | 1 | 5 | 6 |
| PHASTER (total) | 39 | 84 | 123 |
| PHASTER (intact) | 2 | 7 | 9 |
| Prophage Hunter Tool (total) | 84 | 195 | 279 |
| Prophage Hunter Tool (active) | 4 | 25 | 29 |
| Campylobacter spp. Genomes | CJIE1 | CCIE2 | CJIE4 |
|---|---|---|---|
| C. coli genomes from present study | 35.37% (29/82) | 52.44% (43/82) | 4.88% (4/82) |
| C. coli genomes from public databases | 0.00% (0/44) | 2.27% (1/44) | 2.27% (1/44) |
| C. jejuni genomes from present study | 45.45% (10/22) | 45.45% (10/22) | 0.00% (0/22) |
| C. jejuni genomes from public databases | 6.94% (48/692) | 3.32% (23/692) | 1.16% (8/692) |
| Total per model prophage (n = 177) | 87 | 77 | 13 |
| Prophage | Harboring-Species | Nuclease Gene * | |
|---|---|---|---|
| Complete | Partial | ||
| CJIE1-CCIE1-like prophages | C. coli (n = 29) | 2 | 27 |
| C. jejuni (n = 58) | 58 | n.d. | |
| CJIE2-CCIE2-like prophages | C. coli (n = 44) | 6 | n.d. |
| C. jejuni (n = 33) | 18 | 3 | |
| CJIE4-like prophages | C. coli (n = 5) | 1 | n.d. |
| C. jejuni (n = 8) | 3 | n.d. | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanoeiro, L.; Oleastro, M.; Nunes, A.; Marques, A.T.; Duarte, S.V.; Gomes, J.P.; Matos, A.P.A.; Vítor, J.M.B.; Vale, F.F. Cryptic Prophages Contribution for Campylobacter jejuni and Campylobacter coli Introgression. Microorganisms 2022, 10, 516. https://doi.org/10.3390/microorganisms10030516
Tanoeiro L, Oleastro M, Nunes A, Marques AT, Duarte SV, Gomes JP, Matos APA, Vítor JMB, Vale FF. Cryptic Prophages Contribution for Campylobacter jejuni and Campylobacter coli Introgression. Microorganisms. 2022; 10(3):516. https://doi.org/10.3390/microorganisms10030516
Chicago/Turabian StyleTanoeiro, Luís, Mónica Oleastro, Alexandra Nunes, Andreia T. Marques, Sílvia Vaz Duarte, João Paulo Gomes, António Pedro Alves Matos, Jorge M. B. Vítor, and Filipa F. Vale. 2022. "Cryptic Prophages Contribution for Campylobacter jejuni and Campylobacter coli Introgression" Microorganisms 10, no. 3: 516. https://doi.org/10.3390/microorganisms10030516
APA StyleTanoeiro, L., Oleastro, M., Nunes, A., Marques, A. T., Duarte, S. V., Gomes, J. P., Matos, A. P. A., Vítor, J. M. B., & Vale, F. F. (2022). Cryptic Prophages Contribution for Campylobacter jejuni and Campylobacter coli Introgression. Microorganisms, 10(3), 516. https://doi.org/10.3390/microorganisms10030516








