Current Production Capability of Drug-Resistant Pathogen Enables Its Rapid Label-Free Detection Applicable to Wastewater-Based Epidemiology
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture Preparation
2.2. Whole-Cell Electrochemical Analysis
2.3. Scanning Electron Microscopy
2.4. Crystal Violet Staining
2.5. Antibiotic Solution
2.6. Live Dead Assay
3. Results and Discussion
3.1. K. pneumoniae Produces Electric Current via Glucose Oxidation
3.2. K. pneumoniae Grows on the ITO Electrode Surface
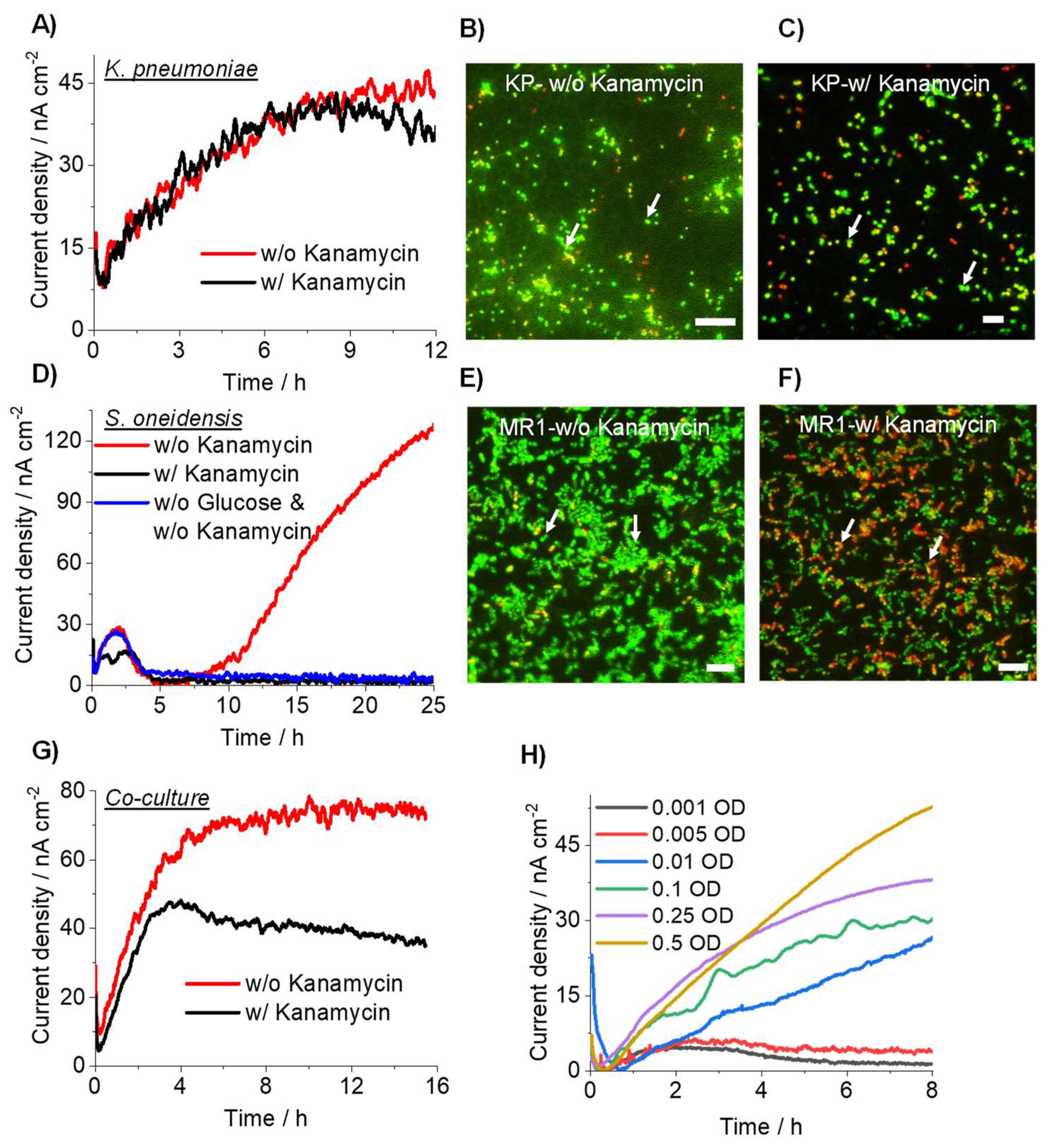
3.3. Detection of Drug-Resistant Pathogens Using Current Producing Capability
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Turolla, A.; Cattaneo, M.; Marazzi, F.; Mezzanotte, V.; Antonelli, M. Antibiotic resistant bacteria in urban sewage: Role of full-scale wastewater treatment plants on environmental spreading. Chemosphere 2018, 191, 761–769. [Google Scholar] [CrossRef] [PubMed]
- Popa, L.I.; Gheorghe, I.; Barbu, I.C.; Surleac, M.; Paraschiv, S.; Măruţescu, L.; Popa, M.; Pîrcălăbioru, G.G.; Talapan, D.; Niţă, M.; et al. Multidrug Resistant Klebsiella pneumoniae ST101 Clone Survival Chain from Inpatients to Hospital Effluent after Chlorine Treatment. Front. Microbiol. 2021, 11, 610296. [Google Scholar] [CrossRef]
- Zhang, Z.; Li, B.; Li, N.; Sardar, M.F.; Song, T.; Zhu, C.; Lv, X.; Li, H. Effects of UV disinfection on phenotypes and genotypes of antibiotic-resistant bacteria in secondary effluent from a municipal wastewater treatment plant. Water Res. 2019, 157, 546–554. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, P.; Adriana, A.; Jessica, S.; Carles, B.; Marinella, F.; Marta, L.; Luis, B.J.; Pierre, S. Antibiotic resistance in urban and hospital wastewaters and their impact on a receiving freshwater ecosystem. Chemosphere 2018, 206, 70–82. [Google Scholar] [CrossRef] [PubMed]
- Madhusoodanan, J. Innovative tools take aim at antibiotic-resistant microbes. Nature 2021, 596, 611–613. [Google Scholar] [CrossRef]
- Archer, E.; Castrignanò, E.; Kasprzyk-Hordern, B.; Wolfaardt, G.M. Wastewater-based epidemiology and enantiomeric profiling for drugs of abuse in South African wastewaters. Sci. Total Environ. 2018, 625, 792–800. [Google Scholar] [CrossRef]
- Mao, K.; Zhang, H.; Pan, Y.; Yang, Z. Biosensors for wastewater-based epidemiology for monitoring public health. Water Res. 2021, 191, 116787. [Google Scholar] [CrossRef]
- El-Liethy, M.; El-Taweel, G.; El-Sonosy, W.; Samhan, F.; Moussa, T. Determination of pathogenic bacteria in wastewater using conventional and PCR techniques. Environ. Biotechnol. 2009, 5, 73–80. [Google Scholar]
- Naradasu, D.; Guionet, A.; Miran, W.; Okamoto, A. Microbial current production from Streptococcus mutans correlates with biofilm metabolic activity. Biosens. Bioelectron. 2020, 162, 112236. [Google Scholar] [CrossRef]
- Naradasu, D.; Miran, W.; Okamoto, A. Metabolic Current Production by an Oral Biofilm Pathogen Corynebacterium matruchotii. Molecules 2020, 25, 3141. [Google Scholar] [CrossRef]
- Miran, W.; Naradasu, D.; Okamoto, A. Pathogens electrogenicity as a tool for in-situ metabolic activity monitoring and drug assessment in biofilms. iScience 2021, 24, 102068. [Google Scholar] [CrossRef] [PubMed]
- Cesewski, E.; Johnson, B.N. Electrochemical biosensors for pathogen detection. Biosens. Bioelectron. 2020, 159, 112214. [Google Scholar] [CrossRef]
- Bassetti, M.; Giacobbe, D.R.; Giamarellou, H.; Viscoli, C.; Daikos, G.L.; Dimopoulos, G.; De Rosa, F.G.; Giamarellos-Bourboulis, E.J.; Rossolini, G.M.; Righi, E.; et al. Management of KPC-producing Klebsiella pneumoniae infections. Clin. Microbiol. Infect. 2018, 24, 133–144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navon-Venezia, S.; Kondratyeva, K.; Carattoli, A. Klebsiella pneumoniae: A major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol. Rev. 2017, 41, 252–275. [Google Scholar] [CrossRef] [PubMed]
- Tokunou, Y.; Hashimoto, K.; Okamoto, A. Acceleration of Extracellular Electron Transfer by Alternative Redox-Active Molecules to Riboflavin for Outer-Membrane Cytochrome c of Shewanella oneidensis MR-1. J. Phys. Chem. C 2016, 120, 16168–16173. [Google Scholar] [CrossRef]
- Okamoto, A.; Hashimoto, K.; Nealson, K.H.; Nakamura, R. Rate enhancement of bacterial extracellular electron transport involves bound flavin semiquinones. Proc. Natl. Acad. Sci. USA 2013, 110, 7856. [Google Scholar] [CrossRef] [Green Version]
- Tokunou, Y.; Hashimoto, K.; Okamoto, A. Electrochemical Detection of Deuterium Kinetic Isotope Effect on Extracellular Electron Transport in Shewanella oneidensis MR-1. J. Vis. Exp. 2018, 134, e57584. [Google Scholar] [CrossRef] [PubMed]
- Zeraik, A.; Nitschke, M. Influence of growth media and temperature on bacterial adhesion to polystyrene surfaces. Braz. Arch. Biol. Technol. 2012, 55, 569–576. [Google Scholar] [CrossRef] [Green Version]
- Cichocki, N.; Hübschmann, T.; Schattenberg, F.; Kerckhof, F.-M.; Overmann, J.; Müller, S. Bacterial mock communities as standards for reproducible cytometric microbiome analysis. Nat. Protoc. 2020, 15, 2788–2812. [Google Scholar] [CrossRef] [PubMed]
- Lindqvist, R.; Barmark, G. Specific growth rate determines the sensitivity of Escherichia coli to lactic acid stress: Implications for predictive microbiology. Biomed. Res. Int. 2014, 2014, 471317. [Google Scholar] [CrossRef]
- Chen, Y.; Pouillot, R.; Burall, S.L.; Strain, E.A.; Van Doren, J.M.; De Jesus, A.J.; Laasri, A.; Wang, H.; Ali, L.; Tatavarthy, A.; et al. Comparative evaluation of direct plating and most probable number for enumeration of low levels of Listeria monocytogenes in naturally contaminated ice cream products. Int. J. Food Microbiol. 2017, 241, 15–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fairchild, G.; Tasseff, B.; Khalsa, H.; Generous, N.; Daughton, A.R.; Velappan, N.; Priedhorsky, R.; Deshpande, A. Epidemiological Data Challenges: Planning for a More Robust Future Through Data Standards. Front. Public Health 2018, 6, 336. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miran, W.; Long, X.; Huang, W.; Okamoto, A. Current Production Capability of Drug-Resistant Pathogen Enables Its Rapid Label-Free Detection Applicable to Wastewater-Based Epidemiology. Microorganisms 2022, 10, 472. https://doi.org/10.3390/microorganisms10020472
Miran W, Long X, Huang W, Okamoto A. Current Production Capability of Drug-Resistant Pathogen Enables Its Rapid Label-Free Detection Applicable to Wastewater-Based Epidemiology. Microorganisms. 2022; 10(2):472. https://doi.org/10.3390/microorganisms10020472
Chicago/Turabian StyleMiran, Waheed, Xizi Long, Wenyuan Huang, and Akihiro Okamoto. 2022. "Current Production Capability of Drug-Resistant Pathogen Enables Its Rapid Label-Free Detection Applicable to Wastewater-Based Epidemiology" Microorganisms 10, no. 2: 472. https://doi.org/10.3390/microorganisms10020472
APA StyleMiran, W., Long, X., Huang, W., & Okamoto, A. (2022). Current Production Capability of Drug-Resistant Pathogen Enables Its Rapid Label-Free Detection Applicable to Wastewater-Based Epidemiology. Microorganisms, 10(2), 472. https://doi.org/10.3390/microorganisms10020472







