MALDI-TOF MS Using a Custom-Made Database, Biomarker Assignment, or Mathematical Classifiers Does Not Differentiate Shigella spp. and Escherichia coli
Abstract
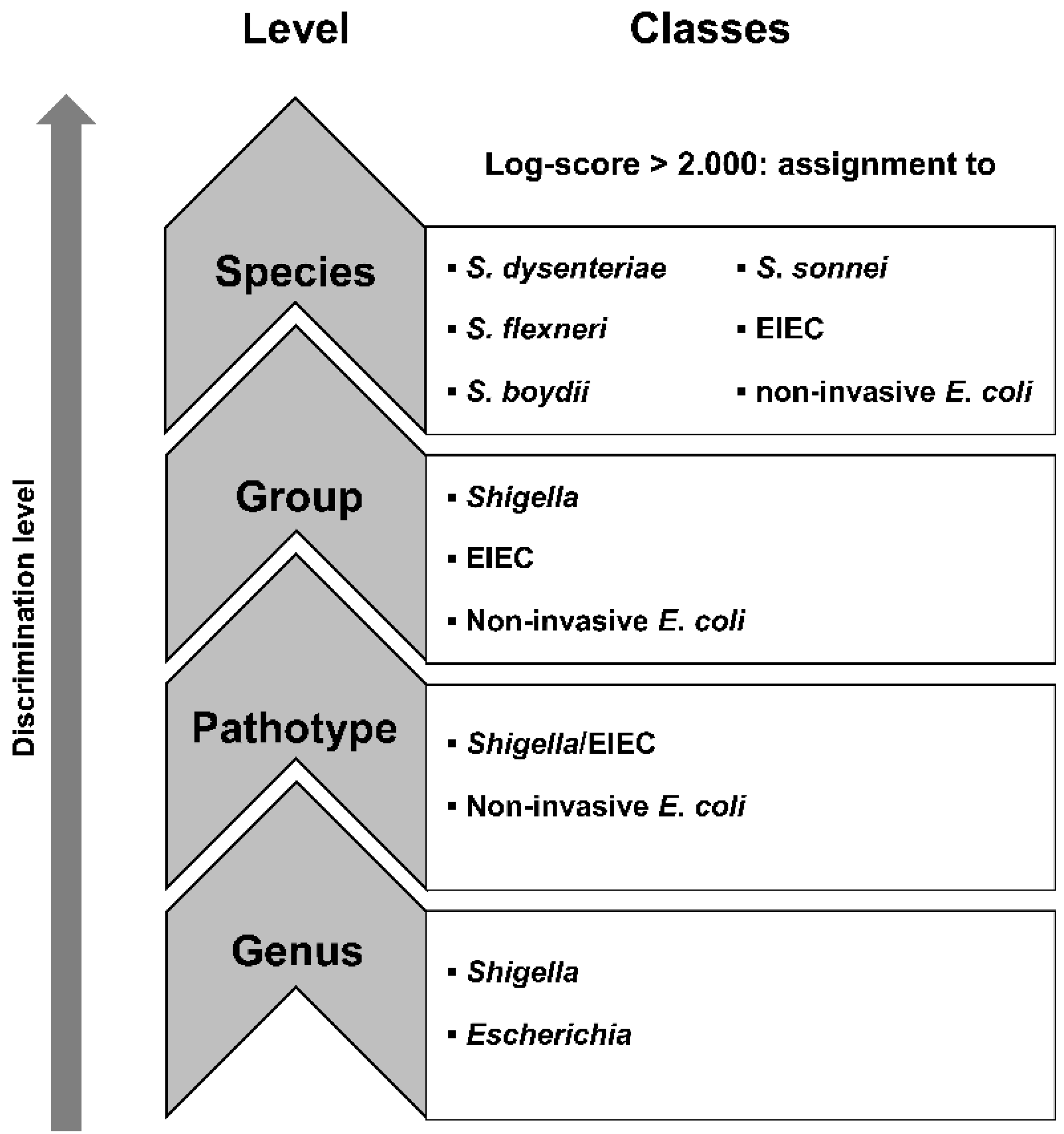
:1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates
2.2. MALDI-TOF MS Preparation of Isolates
2.3. Database Development
2.4. Biomarker Assignment and Principal Component Analysis
2.5. Presence of Biomarkers Identified in Previous Studies
2.6. Classifier Models Based on Machine Learning
3. Results
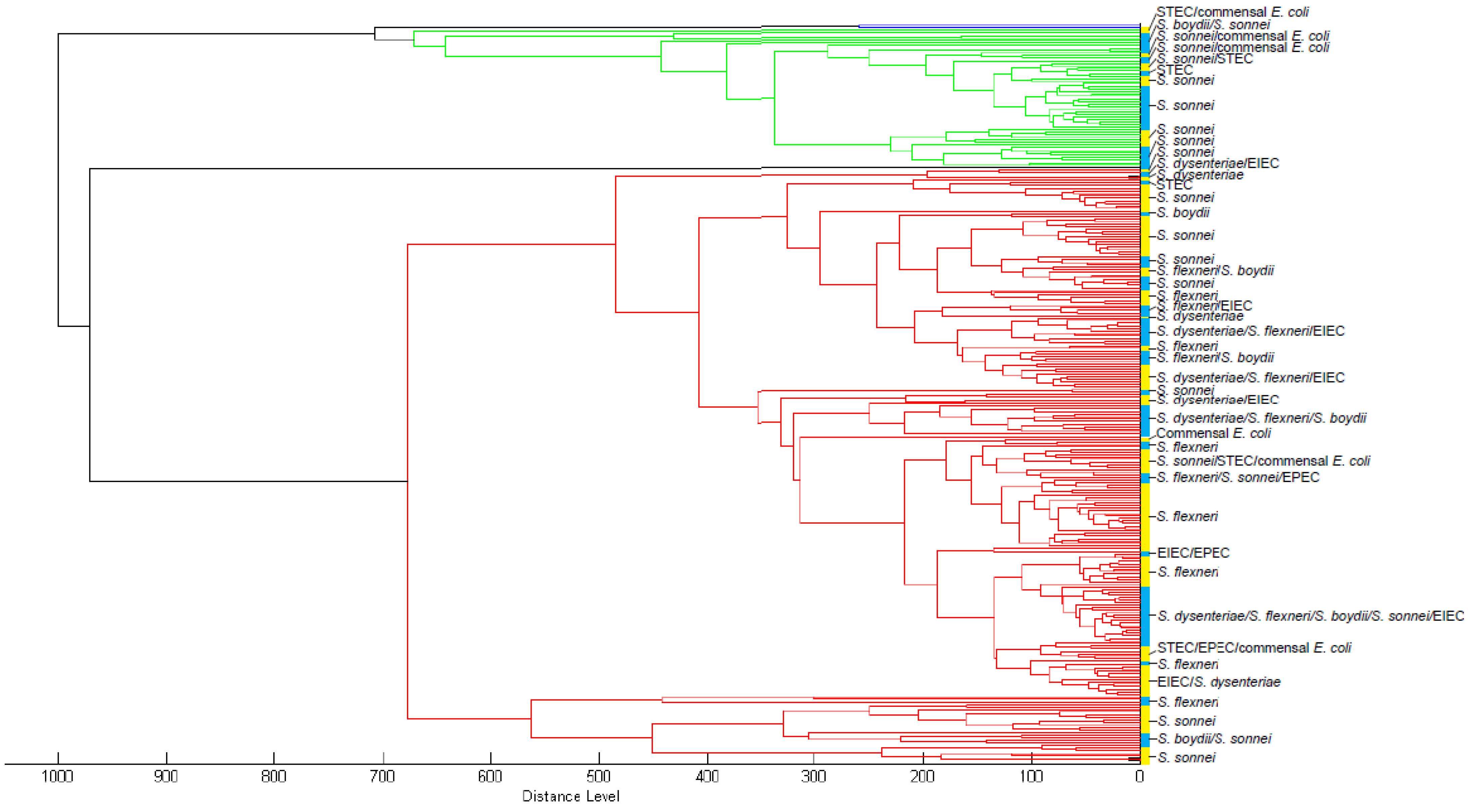
3.1. Database Development
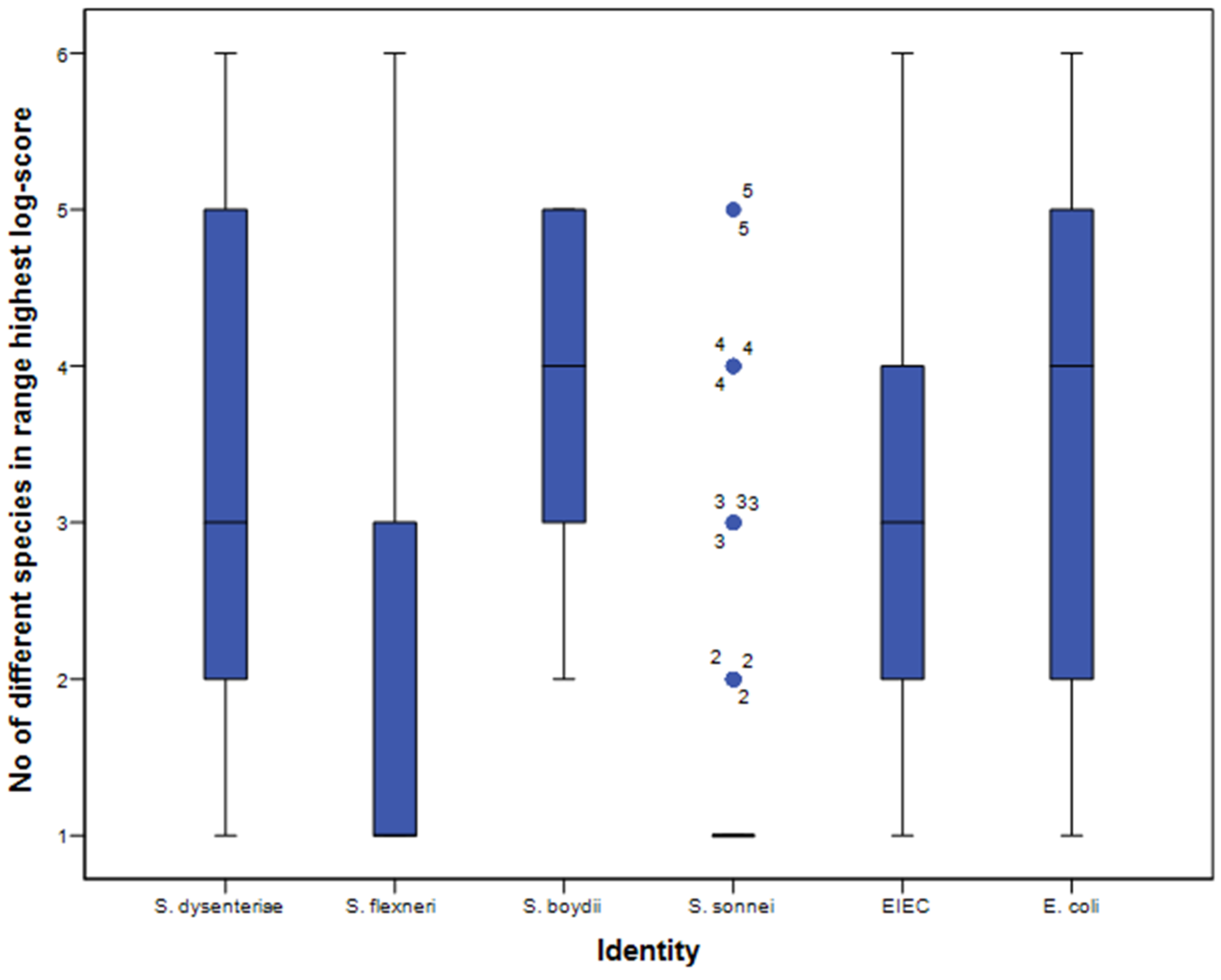
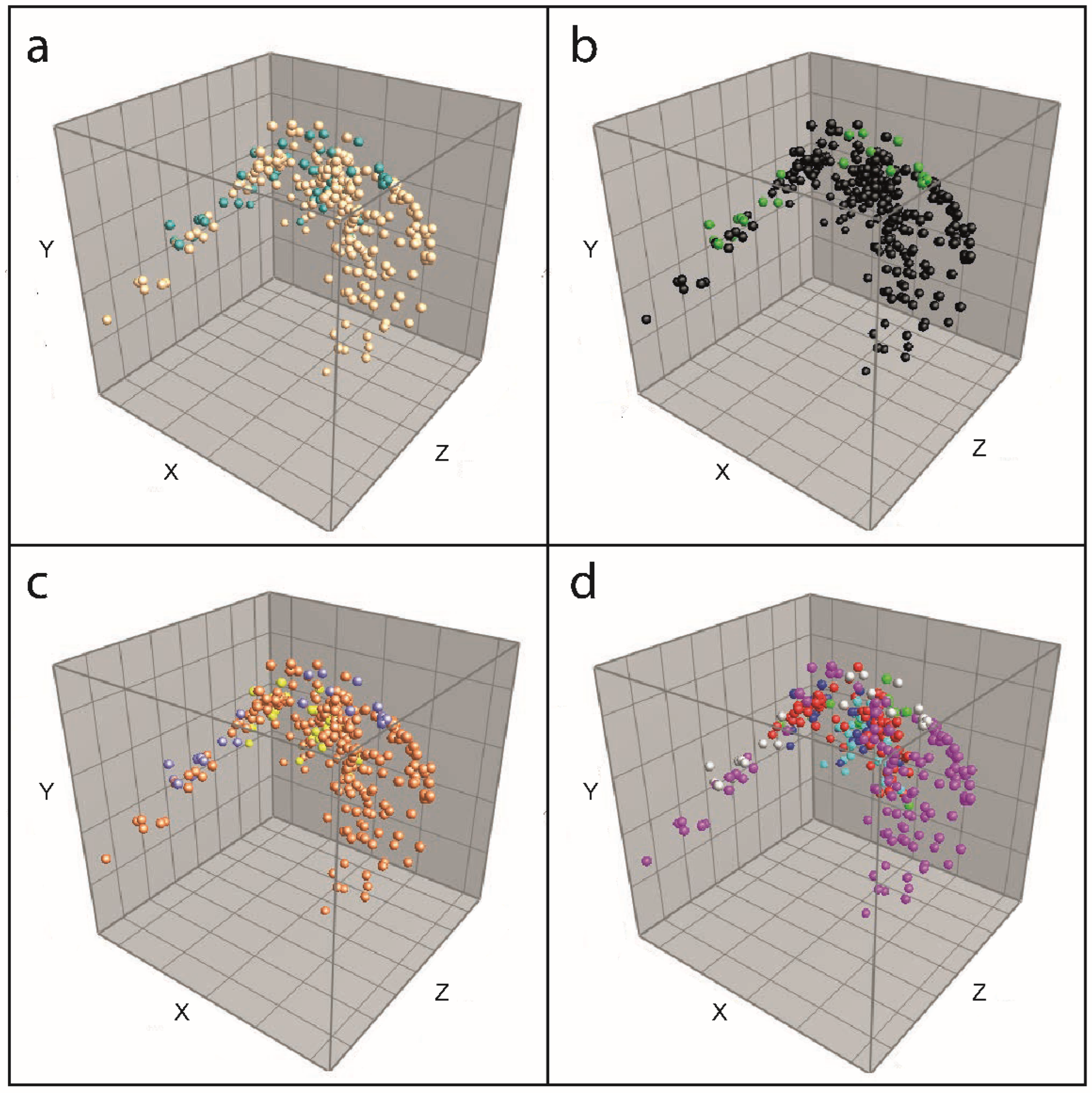
3.2. Biomarker Assignment and Principal Component Analysis
3.3. Presence of Biomarkers Identified in Previous Studies
3.4. Classifier Models Based on Machine Learning
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dupont, H.L.; Formal, S.B.; Hornick, R.B.; Snyder, M.J.; Libonati, J.P.; Sheahan, D.G.; LaBrec, E.H.; Kalas, J.P. Pathogenesis of Escherichia coli Diarrhea. N. Engl. J. Med. 1971, 285, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lan, R.; Alles, M.C.; Donohoe, K.; Martinez, M.B.; Reeves, P.R. Molecular Evolutionary Relationships of Enteroinvasive Escherichia coli and Shigella spp. Infect. Immun. 2004, 72, 5080–5088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettengill, E.A.; Pettengill, J.B.; Binet, R. Phylogenetic Analyses of Shigella and Enteroinvasive Escherichia coli for the Identification of Molecular Epidemiological Markers: Whole-Genome Comparative Analysis Does Not Support Distinct Genera Designation. Front. Microbiol. 2016, 6, 1573. [Google Scholar] [CrossRef] [PubMed]
- Kaper, J.B.; Nataro, J.P.; Mobley, H.L.T. Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2004, 2, 123–140. [Google Scholar] [CrossRef]
- Brenner, D.J.; Family, I. Enterobacteriaceae. In Bergey’s Manual of Systematic Bacteriology; Krieg, N.R., Ed.; 1984; pp. 408–420. [Google Scholar]
- Bopp, C.A.; Brenner, F.W.; Fields, P.I.; Wells, J.G.; Strockbine, N.A. Escherichia, Shigella and Salmonella. In Manual of Clinical Microbiology; Murray, P.R., Ed.; ASM Press: Washington, DC, USA, 2003; pp. 654–671. [Google Scholar]
- Beld, M.J.C.V.D.; Reubsaet, F.A.G. Differentiation between Shigella, enteroinvasive Escherichia coli (EIEC) and noninvasive Escherichia coli. Eur. J. Clin. Microbiol. 2012, 31, 899–904. [Google Scholar] [CrossRef] [PubMed]
- Sahl, J.W.; Morris, C.R.; Emberger, J.; Fraser, C.; Ochieng, J.B.; Juma, J.; Fields, B.; Breiman, R.F.; Gilmour, M.; Nataro, J.P.; et al. Defining the Phylogenomics of Shigella Species: A Pathway to Diagnostics. J. Clin. Microbiol. 2015, 53, 951–960. [Google Scholar] [CrossRef] [Green Version]
- Pavlovic, M.; Luze, A.; Konrad, R.; Berger, A.; Sing, A.; Busch, U.; Huber, I. Development of a duplex real-time PCR for differentiation between E. coli and Shigella spp. J. Appl. Microbiol. 2011, 110, 1245–1251. [Google Scholar] [CrossRef]
- Kim, H.-J.; Ryu, J.-O.; Song, J.-Y.; Kim, H.-Y. Multiplex Polymerase Chain Reaction for Identification of Shigellae and Four Shigella Species Using Novel Genetic Markers Screened by Comparative Genomics. Foodborne Pathog. Dis. 2017, 14, 400–406. [Google Scholar] [CrossRef]
- Dhakal, R.; Wang, Q.; Lan, R.; Howard, P.; Sintchenko, V. Novel multiplex PCR assay for identification and subtyping of enteroinvasive Escherichia coli and differentiation from Shigella based on target genes selected by comparative genomics. J. Med. Microbiol. 2018, 67, 1257–1264. [Google Scholar] [CrossRef]
- Martiny, D.; Busson, L.; Wybo, I.; El Haj, R.A.; Dediste, A.; Vandenberg, O. Comparison of the Microflex LT and Vitek MS Systems for Routine Identification of Bacteria by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. J. Clin. Microbiol. 2012, 50, 1313–1325. [Google Scholar] [CrossRef] [Green Version]
- Cameron, M.; Perry, J.; Middleton, J.; Chaffer, M.; Lewis, J.; Keefe, G. Short communication: Evaluation of MALDI-TOF mass spectrometry and a custom reference spectra expanded database for the identification of bovine-associated coagulase-negative staphylococci. J. Dairy Sci. 2018, 101, 590–595. [Google Scholar] [CrossRef] [PubMed]
- Seuylemezian, A.; Aronson, H.; Tan, J.; Lin, M.; Schubert, W.; Vaishampayan, P. Development of a Custom MALDI-TOF MS Database for Species-Level Identification of Bacterial Isolates Collected From Spacecraft and Associated Surfaces. Front. Microbiol. 2018, 9, 780. [Google Scholar] [CrossRef] [PubMed]
- Morales, M.P.F.; Hopprich, R.; Wise, R.; Shephard, L.; Weldhagen, G.F. Cost-effective implementation of a custom MALDI-TOF library for the identification of South Australian Nocardia isolates. Pathology 2018, 50, 753–757. [Google Scholar] [CrossRef]
- Paauw, A.; Jonker, D.; Roeselers, G.; Heng, J.M.; Mars-Groenendijk, R.H.; Trip, H.; Molhoek, E.M.; Jansen, H.-J.; van der Plas, J.; de Jong, A.L.; et al. Rapid and reliable discrimination between Shigella species and Escherichia coli using MALDI-TOF mass spectrometry. Int. J. Med. Microbiol. 2015, 305, 446–452. [Google Scholar] [CrossRef] [PubMed]
- Josten, M.; Reif, M.; Szekat, C.; Al-Sabti, N.; Roemer, T.; Sparbier, K.; Kostrzewa, M.; Rohde, H.; Sahl, H.-G.; Bierbaum, G. Analysis of the Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrum of Staphylococcus aureus Identifies Mutations That Allow Differentiation of the Main Clonal Lineages. J. Clin. Microbiol. 2013, 51, 1809–1817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sauget, M.; Valot, B.; Bertrand, X.; Hocquet, D. Can MALDI-TOF Mass Spectrometry Reasonably Type Bacteria? Trends Microbiol. 2017, 25, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Khot, P.D.; Fisher, M.A. Novel Approach for Differentiating Shigella Species and Escherichia coli by Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry. J. Clin. Microbiol. 2013, 51, 3711–3716. [Google Scholar] [CrossRef] [Green Version]
- Everley, R.A.; Mott, T.M.; Wyatt, S.A.; Toney, D.M.; Croley, T.R. Liquid chromatography/mass spectrometry characterization of Escherichia coli and Shigella species. J. Am. Soc. Mass Spectrom. 2008, 19, 1621–1628. [Google Scholar] [CrossRef] [Green Version]
- De Bruyne, K.; Slabbinck, B.; Waegeman, W.; Vauterin, P.; De Baets, B.; Vandamme, P. Bacterial species identification from MALDI-TOF mass spectra through data analysis and machine learning. Syst. Appl. Microbiol. 2011, 34, 20–29. [Google Scholar] [CrossRef]
- Mather, C.A.; Werth, B.J.; Sivagnanam, S.; SenGupta, D.J.; Butler-Wu, S.M. Rapid Detection of Vancomycin-Intermediate Staphylococcus aureus by Matrix-Assisted Laser Desorption Ionization–Time of Flight Mass Spectrometry. J. Clin. Microbiol. 2016, 54, 883–890. [Google Scholar] [CrossRef] [Green Version]
- Ho, P.-L.; Yau, C.-Y.; Ho, L.-Y.; Chen, J.H.K.; Lai, E.L.Y.; Lo, S.W.U.; Tse, C.W.S.; Chow, K.-H. Rapid detection ofcfiAmetallo-β-lactamase-producingBacteroides fragilisby the combination of MALDI-TOF MS and CarbaNP. J. Clin. Pathol. 2017, 70, 868–873. [Google Scholar] [CrossRef] [PubMed]
- Clermont, O.; Bonacorsi, S.; Bingen, E. Rapid and Simple Determination of the Escherichia coli Phylogenetic Group. Appl. Environ. Microbiol. 2000, 66, 4555–4558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beld, M.J.C.V.D.; de Boer, R.F.; Reubsaet, F.A.G.; Rossen, J.; Zhou, K.; Kuiling, S.; Friedrich, A.W.; Kooistra-Smid, M.A.M.D. Evaluation of a Culture-Dependent Algorithm and a Molecular Algorithm for Identification of Shigella spp., Escherichia coli, and Enteroinvasive E. coli. J. Clin. Microbiol. 2018, 56, e00510-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saleeb, P.G.; Drake, S.K.; Murray, P.R.; Zelazny, A.M. Identification of Mycobacteria in Solid-Culture Media by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. J. Clin. Microbiol. 2011, 49, 1790–1794. [Google Scholar] [CrossRef] [Green Version]
| Species and Serotype/O-Type | Training Set | Test Set | ||
|---|---|---|---|---|
| n | Origin | n | Origin | |
| S. dysenteriae serotype 1 | 2 | CIP 57.28T; A1 | 1 | 1 ci 1 |
| S. dysenteriae serotype 2 | 5 | A2, 4 ci 1 | 4 | 4 ci 1 |
| S. dysenteriae serotype 3 | 5 | AMC-43-G-93; 4 ci 1 | 3 | 3 ci 1 |
| S. dysenteriae serotype 4 | 2 | AMC 43-G-86; 1 ci 1 | 0 | |
| S. dysenteriae serotype 5 | 1 | AMC 43-G-84 | 0 | |
| S. dysenteriae serotype 6 | 1 | AMC 43-G-81 | 1 | 1 ci 1 |
| S. dysenteriae serotype 7 | 1 | AMC 43-G-76 | 1 | 1 ci 1 |
| S. dysenteriae serotype 9 | 2 | A58: 1646; 1 ci 1 | 1 | 1 ci 1 |
| S. dysenteriae serotype 10 | 1 | A2050-52 | 0 | |
| S. dysenteriae serotype 12 | 2 | 2 ci 1 | 1 | 1 ci 1 |
| S. dysenteriae serotype 14 | 1 | NCTC 11867 | 0 | |
| S. dysenteriae serotype 15 | 1 | NCTC 11868 | 0 | |
| Total number of S. dysenteriae | 24 | 12 | ||
| S. flexneri serotype 1a | 3 | B1A; 2 ci 1 | 0 | |
| S. flexneri serotype 1b | 5 | B1B; 4 ci 1 | 5 | 5 ci 1 |
| S. flexneri serotype 1c | 4 | 4 ci 1 | 3 | 3 ci 1 |
| S. flexneri serotype 2a | 32 | CIP 82.48T; B2A; 30 ci 1 | 32 | 32 ci 1 |
| S. flexneri serotype 2b | 1 | B2B | 3 | 3 ci 1 |
| S. flexneri serotype 3a | 2 | B3A; 1 ci 1 | 14 | 14 ci 1 |
| S. flexneri serotype 3b | 2 | B3B; B3C | 3 | 3 ci 1 |
| S. flexneri serotype 4a | 1 | B4A | 4 | 4 ci 1 |
| S. flexneri serotype 4av | 4 | 5 ci 1 | 0 | |
| S. flexneri serotype 4b | 1 | B4B | 0 | |
| S. flexneri serotype 4c | 3 | 3 ci 1 | 0 | |
| S. flexneri serotype 5b | 1 | B5 | 1 | 1 ci 1 |
| S. flexneri serotype 6 | 10 | B6; 9 ci 1 | 10 | 10 ci 1 |
| S. flexneri serotype X | 1 | |||
| S. flexneri serotype Y | 2 | 2 ci 1 | 2 | 2 ci 1 |
| S. flexneri serotype Yv | 2 | 2 ci 1 | 0 | |
| S. flexneri provisional | 5 | 5 ci 1 | 0 | |
| Total number of S. flexneri | 79 | 77 | ||
| S. boydii serotype 1 | 2 | AMC-43-G-58; 1 ci 1 | 3 | 3 ci 1 |
| S. boydii serotype 2 | 3 | CIP 82.50T; P288; 1 ci 1 | 4 | 4 ci 1 |
| S. boydii serotype 3 | 1 | D1 | 0 | |
| S. boydii serotype 4 | 2 | AMC-43-G-63; 1 ci 1 | 2 | 2 ci 1 |
| S. boydii serotype 5 | 2 | P143; 1 ci 1 | 0 | |
| S. boydii serotype 6 | 1 | CDC 9771 (D19) | 0 | |
| S. boydii serotype 7 | 1 | AMC 4006 (Lavington) | 0 | |
| S. boydii serotype 8 | 0 | 1 | 1 ci 1 | |
| S. boydii serotype 9 | 1 | 1296/7 | 0 | |
| S. boydii serotype 10 | 1 | 430 | 1 | 1 ci 1 |
| S. boydii serotype 11 | 1 | 34 | 0 | |
| S. boydii serotype 12 | 0 | 1 | 1 ci 1 | |
| S. boydii serotype 13 | 0 | 1 | 1 ci 1 | |
| S. boydii serotype 14 | 0 | 1 | 1 ci 1 | |
| S. boydii serotype 15 | 1 | CDC C-703 | 0 | |
| S. boydii serotype 18 | 1 | 1 ci 1 | 1 | 1 ci 1 |
| Total number of S. boydii | 17 | 15 | ||
| S. sonnei | 117 | CIP 82.49T; 116 ci 1 | 115 | 115 ci 1 |
| EIEC | 30 | DSM 9027; DSM 9028; CCUG 11335; CCUG 38080; CCUG 38092; CCUG 38093; EW227; 1624-56; 1184-68; 145/46; L119B-10; 19 ci 1 | 31 | 31 ci 1 |
| Other E. coli pathotypes (human) | 11 | 7 STEC ci 1, 4 EPEC ci 1 | 11 | 8 STEC ci 1; 3 EPEC ci 1 |
| Other E. coli pathotypes (animal) 2 | 10 | 5 mussel, 3 pigeon, 2 turkey | 10 | 4 mussel, 3 pigeon, 2 turkey, 1 oyster |
| Biomarkers (m/z) | 2691 | 2877 | 3129 | 3636 | 3647 | 3930 | 3939 | 4163 | 4189 | 4368 | 4501 | 4769 | 4775 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S. dysenteriae | (n = 24) | 92 | 4 | 100 | 0 | 0 | 0 | 100 | 0 | 100 | 100 | 100 | 0 | 100 |
| S. flexneri | (n = 46) | 100 | 0 | 100 | 0 | 63 | 53 | 18 | 1 | 94 | 97 | 99 | 22 | 97 |
| S. boydii | (n = 17) | 88 | 0 | 100 | 18 | 0 | 0 | 94 | 0 | 100 | 88 | 100 | 18 | 100 |
| S. sonnei | (n = 117) | 56 | 49 | 59 | 22 | 0 | 1 | 56 | 17 | 89 | 68 | 56 | 23 | 98 |
| EIEC | (n = 31) | 100 | 0 | 100 | 3 | 6 | 0 | 97 | 0 | 97 | 100 | 94 | 26 | 97 |
| Other E. coli | (n = 21) | 52 | 24 | 52 | 71 | 0 | 5 | 62 | 38 | 90 | 67 | 57 | 67 | 100 |
| Biomarkers (m/z) | 4784 | 5156 | 5239 | 5386 | 5415 | 6262 | 6322 | 6412 | 6488 | 7275 | 7295 | 7715 | 7868 | 7879 |
| S. dysenteriae | 100 | 100 | 8 | 92 | 52 | 100 | 100 | 0 | 0 | 0 | 0 | 0 | 0 | 100 |
| S. flexneri | 76 | 97 | 55 | 99 | 45 | 99 | 99 | 4 | 42 | 1 | 83 | 0 | 41 | 23 |
| S. boydii | 76 | 94 | 18 | 88 | 59 | 100 | 88 | 18 | 6 | 18 | 0 | 6 | 12 | 88 |
| S. sonnei | 69 | 86 | 27 | 73 | 0 | 74 | 62 | 13 | 1 | 17 | 0 | 28 | 18 | 75 |
| EIEC | 71 | 100 | 39 | 100 | 42 | 94 | 97 | 19 | 0 | 23 | 6 | 3 | 16 | 84 |
| Other E. coli | 19 | 86 | 0 | 67 | 0 | 57 | 67 | 48 | 10 | 52 | 0 | 24 | 43 | 33 |
| Biomarkers (m/z) | 8326 | 8370 | 8379 | 9002 | 9227 | 9535 | 9546 | 9563 | 9739 | 10,300 | 10,310 | 10,488 | 10,934 | |
| S. dysenteriae | 0 | 0 | 100 | 100 | 0 | 0 | 100 | 100 | 0 | 0 | 100 | 0 | 0 | |
| S. flexneri | 5 | 4 | 90 | 94 | 4 | 17 | 82 | 92 | 8 | 9 | 86 | 0 | 0 | |
| S. boydii | 0 | 18 | 82 | 88 | 12 | 18 | 82 | 88 | 6 | 18 | 82 | 0 | 12 | |
| S. sonnei | 15 | 15 | 82 | 38 | 12 | 16 | 85 | 54 | 15 | 13 | 70 | 34 | 36 | |
| EIEC | 6 | 16 | 77 | 87 | 13 | 23 | 77 | 81 | 6 | 19 | 77 | 0 | 0 | |
| Other E. coli | 43 | 38 | 38 | 24 | 48 | 48 | 62 | 38 | 43 | 48 | 48 | 0 | 5 | |
| 8326 | 8370 | 8379 | 9002 | 9227 | 9535 | 9546 | 9563 | 9739 | 10,300 | 10,310 | 10,488 | 10,934 |
| Correct Identification with MALDI-TOF, Direct Smear | Correct Identification with MALDI-TOF, Ethanol | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bruker Databases 1, n (%) | Custom Databases 1, n (%) | Bruker Databases 1 + Custom, n (%) | Biomarker Assignment, n (%) | Classifier Models, n (%) | Bruker Databases 1, n (%) | Custom Databases 1, n (%) | Bruker Databases 1 + Custom, n (%) | Biomarker Assignment, n (%) | Classifier Models, n (%) | |||||||||||
| Genus | ||||||||||||||||||||
| Shigella (n = 217) | 19 | (9) | 205 | (94) | 205 | (94) | 10 | (5) | 209 | (96) | 12 | (6) | 207 | (95) | 205 | (94) | 15 | (7) | 217 | (100) |
| E. coli (n = 52) | 49 | (94) | 26 | (50) | 29 | (56) | NA | 11 | (21) | 47 | (90) | 35 | (67) | 37 | (71) | NA | 4 | (8) | ||
| Unassigned 2 | 2 | (1) | 1 | (0.4) | 3 | (1) | 257 | (96) | 0 | (0) | 1 | (0.4) | 0 | (0) | 3 | (1) | 250 | (93) | 0 | (0) |
| Pathotype | ||||||||||||||||||||
| Shigella/EIEC (n = 248) | NA | 233 | (94) | 241 | (97) | 217 | (88) | 145 | (58) | NA | 245 | (99) | 242 | (98) | 225 | (92) | 147 | (59) | ||
| Other E. coli (n = 21) | 21 | (100) | 6 | (29) | 10 | (48) | NA | 14 | (67) | 21 | (100) | 11 | (52) | 13 | (62) | NA | 6 | (29) | ||
| Unassigned 2 | 2 | 1 | (0.4) | 3 | (1) | 46 | (17) | 0 | (0) | 0 | (0) | 0 | (0) | 3 | (1) | 27 | (10) | 0 | (0) | |
| Group | ||||||||||||||||||||
| Shigella (n = 217) | 19 | (9) | 205 | (94) | 205 | (94) | 193 | (89) | 131 | (60) | 12 | (6) | 207 | (95) | 205 | (94) | 195 | (90) | 134 | (62) |
| EIEC (n = 31) | NA | 9 | (29) | 8 | (26) | NA | 2 | (6) | NA | 19 | (61) | 19 | (61) | NA | 0 | (0) | ||||
| Other E. coli (n = 21) | 21 | (100) | 6 | (29) | 10 | (48) | NA | 13 | (62) | 21 | (100) | 11 | (52) | 13 | (62) | NA | 7 | (33) | ||
| Unassigned 2 | 2 | (1) | 1 | (0.4) | 3 | (1) | 49 | (23) | 0 | (0) | 1 | (0.4) | 0 | (0) | 3 | (1) | 36 | (13) | 0 | (0) |
| Species | ||||||||||||||||||||
| S. dysenteriae (n = 11) | 5 | (45) | 5 | (45) | 5 | (45) | 0 | (0) | 0 | (0) | 4 | (36) | 7 | (64) | 6 | (55) | 0 | (0) | 0 | (0) |
| S. flexneri (n = 77) | NA | 70 | (91) | 70 | (91) | 24 | (31) | 6 | (8) | NA | 73 | (95) | 73 | (95) | 30 | (39) | 3 | (4) | ||
| S. boydii (n = 14) | NA | 1 | (7) | 0 | (0) | 0 | (0) | 0 | (0) | NA | 0 | (0) | 0 | (0) | 0 | (0) | 0 | (0) | ||
| S. sonnei (n = 115) | NA | 110 | (96) | 110 | (96) | 113 | (98) | 92 | (80) | NA | 112 | (97) | 112 | (97) | 108 | (94) | 101 | (88) | ||
| EIEC (n = 31) | NA | 9 | (29) | 8 | (26) | 0 | (0) | 1 | (3) | NA | 19 | (61) | 19 | (61) | 1 | (3) | 3 | (10) | ||
| Other E. coli (n = 21) | 21 | (100) | 6 | (29) | 10 | (48) | 0 | (0) | 12 | (57) | 21 | (100) | 11 | (52) | 13 | (62) | 0 | (0) | 4 | (19) |
| Unassigned 2 | 2 | (1) | 1 | (0.4) | 3 | (1) | 85 | (32) | 0 | (0) | 1 | (0.4) | 0 | (0) | 3 | (1) | 97 | (36) | 0 | (0) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
van den Beld, M.J.C.; Rossen, J.W.A.; Evers, N.; Kooistra-Smid, M.A.M.D.; Reubsaet, F.A.G. MALDI-TOF MS Using a Custom-Made Database, Biomarker Assignment, or Mathematical Classifiers Does Not Differentiate Shigella spp. and Escherichia coli. Microorganisms 2022, 10, 435. https://doi.org/10.3390/microorganisms10020435
van den Beld MJC, Rossen JWA, Evers N, Kooistra-Smid MAMD, Reubsaet FAG. MALDI-TOF MS Using a Custom-Made Database, Biomarker Assignment, or Mathematical Classifiers Does Not Differentiate Shigella spp. and Escherichia coli. Microorganisms. 2022; 10(2):435. https://doi.org/10.3390/microorganisms10020435
Chicago/Turabian Stylevan den Beld, Maaike J. C., John W. A. Rossen, Noah Evers, Mirjam A. M. D. Kooistra-Smid, and Frans A. G. Reubsaet. 2022. "MALDI-TOF MS Using a Custom-Made Database, Biomarker Assignment, or Mathematical Classifiers Does Not Differentiate Shigella spp. and Escherichia coli" Microorganisms 10, no. 2: 435. https://doi.org/10.3390/microorganisms10020435
APA Stylevan den Beld, M. J. C., Rossen, J. W. A., Evers, N., Kooistra-Smid, M. A. M. D., & Reubsaet, F. A. G. (2022). MALDI-TOF MS Using a Custom-Made Database, Biomarker Assignment, or Mathematical Classifiers Does Not Differentiate Shigella spp. and Escherichia coli. Microorganisms, 10(2), 435. https://doi.org/10.3390/microorganisms10020435






