Bacterial Antagonistic Species of the Pathogenic Genus Legionella Isolated from Cooling Tower
Abstract
:1. Introduction
2. Materials and Methods
2.1. Legionella pneumophila Strains
2.2. Isolation of Inhibitory Bacterial Strains of Legionella pneumophila
2.3. Identification of Bacterial Isolates
2.4. Testing Inhibition of Isolates with Different Legionella Strains
2.5. Whole Genome Sequencing of Anti-Legionella Isolates
3. Results
3.1. Inhibition Assay
3.2. Taxonomic Classification of Bacterial Isolates
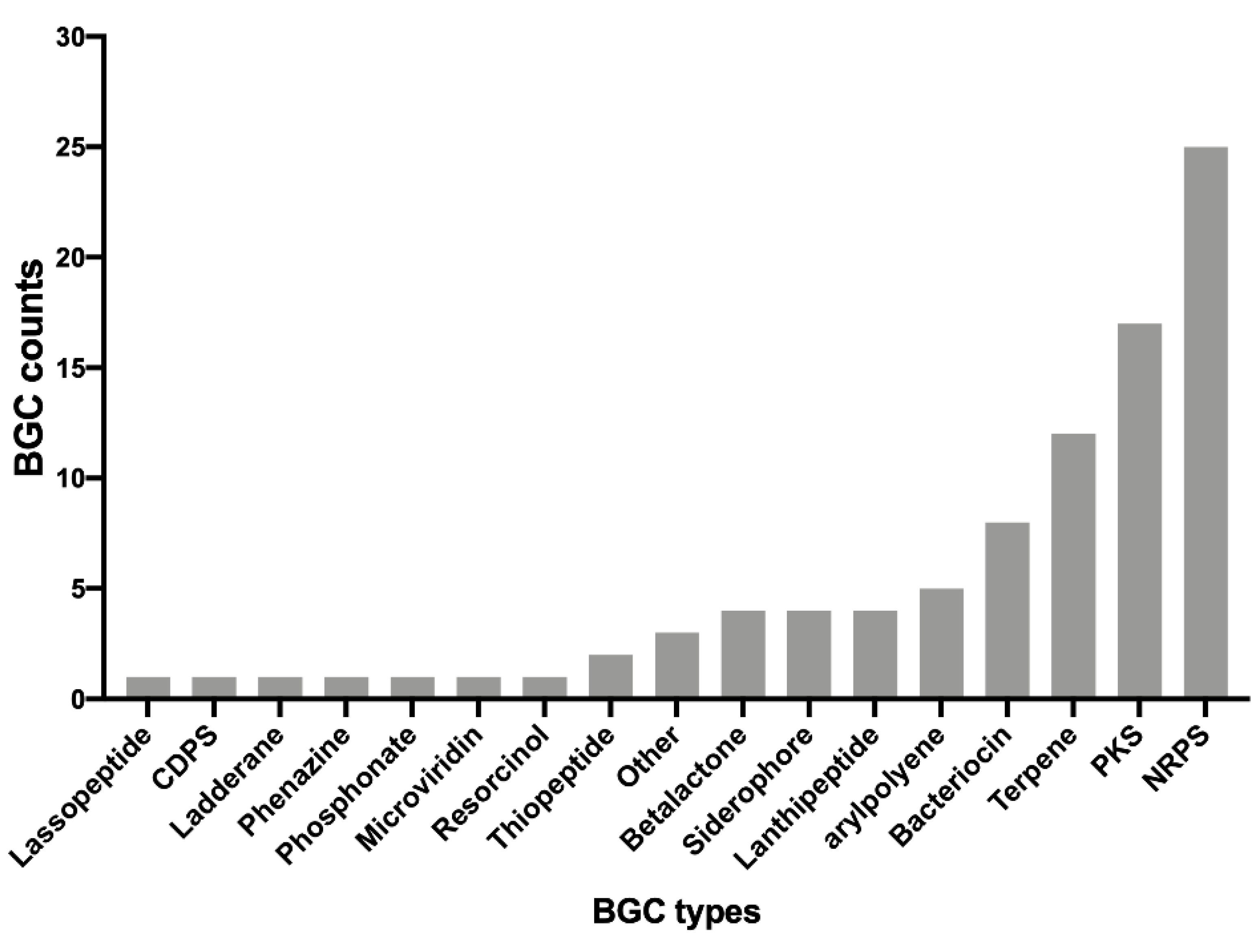
3.3. Identification of Putative Secondary Metabolites
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- van Heijnsbergen, E.; Schalk, J.A.; Euser, S.M.; Brandsema, P.S.; den Boer, J.W.; de Roda Husman, A.M. Confirmed and potential sources of Legionella reviewed. Environ. Sci. Technol. 2015, 49, 4797–4815. [Google Scholar] [CrossRef] [PubMed]
- Benedict, K.M.; Reses, H.; Vigar, M.; Roth, D.M.; Roberts, V.A.; Mattioli, M.; Cooley, L.A.; Hilborn, E.D.; Wade, T.J.; Fullerton, K.E.; et al. Surveillance for Waterborne Disease Outbreaks Associated with Drinking Water—United States, 2013–2014. MMWR Morb. Mortal. Wkly. Rep. 2017, 66, 1216–1221. [Google Scholar] [CrossRef] [PubMed]
- Cassell, K.; Gacek, P.; Rabatsky-Ehr, T.; Petit, S.; Cartter, M.; Weinberger, D.M. Estimating the True Burden of Legionnaires’ Disease. Am. J. Epidemiol. 2019, 188, 1686–1694. [Google Scholar] [CrossRef] [PubMed]
- ECDC. Legionnaires’ Disease. In ECDC Annual Epdimilogical Report for 2017; ECDC: Solna, Sweden, 2019. [Google Scholar]
- National Academies of Sciences, Engineering, and Medicine. Management of Legionella in Water Systems; The National Academies Press: Washington, DC, USA, 2019; 304p. [Google Scholar]
- Phin, N.; Parry-Ford, F.; Harrison, T.; Stagg, H.; Zhang, N.; Kumar, K.; Lortholary, O.; Zumla, P.S.A.; Abubakar, I. Epidemiology and clinical management of Legionnaires’ disease. Lancet Infect. Dis. 2014, 14, 1011–1021. [Google Scholar] [CrossRef]
- Cunha, B.A.; Burillo, A.; Bouza, E. Legionnaires’ disease. Lancet 2016, 387, 376–385. [Google Scholar] [CrossRef]
- Québec, G. Décret 454-2014 Loi sur le Bâtiment. Gaz. Off. Du Québec 2014, 146, 1923–1927. [Google Scholar]
- Standard, A. Standard 188-2015. In Legionellosis: Risk Management for Building Water Systems; ASHRAE: Atlanta, GA, USA, 2015. [Google Scholar]
- Parr, A.; Whitney, E.A.; Berkelman, R.L. Legionellosis on the Rise: A Review of Guidelines for Prevention in the United States. J. Public Health Manag. Pract. 2015, 21, E17–E26. [Google Scholar] [CrossRef] [Green Version]
- Gaia, V.; Fry, N.K.; Afshar, B.; Lück, P.C.; Meugnier, H.; Etienne, J.; Peduzzi, R.; Harrison, T.G. Consensus sequence-based scheme for epidemiological typing of clinical and environmental isolates of Legionella pneumophila. J. Clin. Microbiol. 2005, 43, 2047–2052. [Google Scholar] [CrossRef] [Green Version]
- Ratzow, S.; Gaia, V.; Helbig, J.H.; Fry, N.K.; Lück, P.C. Addition of neuA, the gene encoding N-acylneuraminate cytidylyl transferase, increases the discriminatory ability of the consensus sequence-based scheme for typing Legionella pneumophila serogroup 1 strains. J. Clin. Microbiol. 2007, 45, 1965–1968. [Google Scholar] [CrossRef] [Green Version]
- Kozak-Muiznieks, N.A.; Lucas, C.E.; Brown, E.; Pondo, T.; Taylor, T.H., Jr.; Frace, M.; Miskowski, D.; Winchell, J.M. Prevalence of sequence types among clinical and environmental isolates of Legionella pneumophila serogroup 1 in the United States from 1982 to 2012. J. Clin. Microbiol. 2014, 52, 201–211. [Google Scholar] [CrossRef] [Green Version]
- Benson, R.F.; Thacker, W.L.; Waters, R.P.; Quinlivan, P.A.; Mayberry, W.R.; Brenner, D.J.; Wilkinson, H.W. Legionella quinlivanii sp. nov. isolated from water. Curr. Microbiol. 1989, 18, 195–197. [Google Scholar] [CrossRef]
- Lalancette, C.; Leduc, J.-M.; Malo, J.; Fournier, É.; Saoud, J.; Faucher, S.P.; Pacheco, A.L.; Bernard, K.; Martineau, C.; Lévesque, S. Legionella quinlivanii strain isolated from a human: A case report and whole genome sequencing analysis. Off. J. Assoc. Med. Microbiol. Infect. Dis. Can. 2020, 5, 112–114. [Google Scholar] [CrossRef] [Green Version]
- Boamah, D.K.; Zhou, G.; Ensminger, A.W.; O’Connor, T.J. From many hosts, one accidental pathogen: The diverse protozoan hosts of Legionella. Front. Cell. Infect. Microbiol. 2017, 7, 477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borella, P.; Guerrieri, E.; Marchesi, I.; Bondi, M.; Messi, P. Water ecology of Legionella and protozoan: Environmental and public health perspectives. Biotechnol. Annu. Rev. 2005, 11, 355–380. [Google Scholar]
- Llewellyn, A.C.; Lucas, C.E.; Roberts, S.E.; Brown, E.W.; Nayak, B.S.; Raphael, B.H.; Winchell, J.M. Distribution of Legionella and bacterial community composition among regionally diverse US cooling towers. PLoS ONE 2017, 12, e0189937. [Google Scholar] [CrossRef] [Green Version]
- Paranjape, K.; Bédard, É.; Shetty, D.; Hu, M.; Choon, F.C.P.; Prévost, M.; Faucher, S.P. Unravelling the importance of the eukaryotic and bacterial communities and their relationship with Legionella spp. ecology in cooling towers: A complex network. Microbiome 2020, 8, 1–19. [Google Scholar] [CrossRef]
- Paranjape, K.; Bédard, É.; Whyte, L.G.; Ronholm, J.; Prévost, M.; Faucher, S.P. Presence of Legionella spp. in cooling towers: The role of microbial diversity, Pseudomonas, and continuous chlorine application. Water Res. 2020, 169, 115252. [Google Scholar] [CrossRef]
- Marchand, A.; Verdon, J.; Lacombe, C.; Crapart, S.; Hechard, Y.; Berjeaud, J. Anti-Legionella activity of staphylococcal hemolytic peptides. Peptides 2011, 32, 845–851. [Google Scholar] [CrossRef]
- Loiseau, C.; Schlusselhuber, M.; Bigot, R.; Bertaux, J.; Berjeaud, J.-M.; Verdon, J. Surfactin from Bacillus subtilis displays an unexpected anti-Legionella activity. Appl. Microbiol. Biotechnol. 2015, 99, 5083–5093. [Google Scholar] [CrossRef]
- Abd, H.; Wretlind, B.; Saeed, A.; Idsund, E.; Hultenby, K.; Sandström, G. Pseudomonas aeruginosa utilises its type III secretion system to kill the free-living amoeba Acanthamoeba castellanii. J. Eukaryot. Microbiol. 2008, 55, 235–243. [Google Scholar] [CrossRef]
- Matz, C.; Moreno, A.M.; Alhede, M.; Manefield, M.; Hauser, A.R.; Givskov, M.; Kjelleberg, S. Pseudomonas aeruginosa uses type III secretion system to kill biofilm-associated amoebae. ISME J. 2008, 2, 843. [Google Scholar] [CrossRef] [PubMed]
- Faucher, S.P.; Matthews, S.; Nickzad, A.; Vounba, P.; Shetty, D.; Bedard, E.; Prévost, M.; Déziel, E.; Paranjape, K. Toxoflavin secreted by Pseudomonas alcaliphila inhibits growth of Legionella pneumophila and its host Vermamoeba vermiformis. bioRxiv 2022, 1–40. [Google Scholar]
- Tison, D.; Pope, D.; Cherry, W.; Fliermans, C. Growth of Legionella pneumophila in association with blue-green algae (cyanobacteria). Appl. Environ. Microbiol. 1980, 39, 456–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wadowsky, R.M.; Yee, R.B. Satellite growth of Legionella pneumophila with an environmental isolate of Flavobacterium breve. Appl. Environ. Microbiol. 1983, 46, 1447–1449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, H.; Ashbolt, N. The role of biofilms and protozoa in Legionella pathogenesis: Implications for drinking water. J. Appl. Microbiol. 2009, 107, 368–378. [Google Scholar] [CrossRef] [PubMed]
- Guerrieri, E.; Bondi, M.; Sabia, C.; de Niederhäusern, S.; Borella, P.; Messi, P. Effect of Bacterial Interference on Biofilm Development by Legionella pneumophila. Curr. Microbiol. 2008, 57, 532–536. [Google Scholar] [CrossRef]
- Corre, M.-H.; Delafont, V.; Legrand, A.; Berjeaud, J.-M.; Verdon, J. Exploiting the Richness of Environmental Waterborne Bacterial Species to Find Natural Legionella pneumophila Competitors. Front. Microbiol. 2019, 9, 3360. [Google Scholar] [CrossRef]
- Lévesque, S.; Plante, P.-L.; Mendis, N.; Cantin, P.; Marchand, G.; Charest, H.; Raymond, F.; Huot, C.; Goupil-Sormany, I.; Desbiens, F. Genomic characterization of a large outbreak of Legionella pneumophila serogroup 1 strains in Quebec City, 2012. PLoS ONE 2014, 9, e103852. [Google Scholar] [CrossRef]
- Lévesque, S.; Lalancette, C.; Bernard, K.; Pacheco, A.L.; Dion, R.; Longtin, J.; Tremblay, C. Molecular Typing of Legionella pneumophila Isolates in the Province of Quebec from 2005 to 2015. PLoS ONE 2016, 11, e0163818. [Google Scholar] [CrossRef]
- Paniagua, A.T.; Paranjape, K.; Hu, M.; Bédard, É.; Faucher, S. Impact of temperature on Legionella pneumophila, its protozoan host cells, and the microbial diversity of the biofilm community of a pilot cooling tower. Sci. Total Environ. 2019, 712, 136131. [Google Scholar] [CrossRef]
- Johnson, M.; Zaretskaya, I.; Raytselis, Y.; Merezhuk, Y.; McGinnis, S.; Madden, T.L. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008, 36, W5–W9. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.; Fast, Q.C. A Quality Control Tool for High Throughput Sequence Data; Babraham Bioinformatics, Babraham Institute: Cambridge, UK, 2010. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nurk, S.; Bankevich, A.; Antipov, D.; Gurevich, A.A.; Korobeynikov, A.; Lapidus, A.; Prjibelski, A.D.; Pyshkin, A.; Sirotkin, A.; Sirotkin, Y.; et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 2013, 20, 714–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodriguez-R, L.M.; Gunturu, S.; Harvey, W.T.; Rosselló-Mora, R.; Tiedje, J.M.; Cole, J.R.; Konstantinidis, K.T. The Microbial Genomes Atlas (MiGA) webserver: Taxonomic and gene diversity analysis of Archaea and Bacteria at the whole genome level. Nucleic Acids Res. 2018, 46, W282–W288. [Google Scholar] [CrossRef]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M. Twelve years of SAMtools and BCFtools. Gigascience 2021, 10, giab008. [Google Scholar] [CrossRef]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:13033997. [Google Scholar]
- Blin, K.; Shaw, S.; Steinke, K.; Villebro, R.; Ziemert, N.; Lee, S.Y.; Medema, M.H.; Weber, T. antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res. 2019, 47, W81–W87. [Google Scholar] [CrossRef] [Green Version]
- Ciufo, S.; Kannan, S.; Sharma, S.; Badretdin, A.; Clark, K.; Turner, S.; Brover, S.; Schoch, C.L.; Kimchi, A.; DiCuccio, M. Using average nucleotide identity to improve taxonomic assignments in prokaryotic genomes at the NCBI. Int. J. Syst. Evol. Microbiol. 2018, 68, 2386. [Google Scholar] [CrossRef]
- Caboche, S.; Leclère, V.; Pupin, M.; Kucherov, G.; Jacques, P. Diversity of monomers in nonribosomal peptides: Towards the prediction of origin and biological activity. J. Bacteriol. 2010, 192, 5143–5150. [Google Scholar] [CrossRef] [Green Version]
- Rappé, M.S.; Giovannoni, S.J. The uncultured microbial majority. Annu. Rev. Microbiol. 2003, 57, 369–394. [Google Scholar] [CrossRef] [Green Version]
- Epstein, S.S. The phenomenon of microbial uncultivability. Curr. Opin. Microbiol. 2013, 16, 636–642. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Du, L.; Liu, F.; Xu, F.; Hu, B.; Venturi, V.; Qian, G. Involvement of both PKS and NRPS in antibacterial activity in Lysobacter enzymogenes OH11. FEMS Microbiol. Lett. 2014, 355, 170–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathur, H.; Field, D.; Rea, M.C.; Cotter, P.D.; Hill, C.; Ross, R.P. Bacteriocin-antimicrobial synergy: A medical and food perspective. Front. Microbiol. 2017, 8, 1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, K.; Thilakan, B.; Raola, V.K.; Joy, M. Antibacterial polyketides from Bacillus amyloliquefaciens associated with edible red seaweed Laurenciae papillosa. Food Chem. 2017, 218, 427–434. [Google Scholar] [CrossRef] [PubMed]
- Patil, P.S.; Kumbhar, T.S. Antioxidant, antibacterial and cytotoxic potential of silver nanoparticles synthesized using terpenes rich extract of Lantana camara L. leaves. Biochem. Biophys. Rep. 2017, 10, 76–81. [Google Scholar]
- Konetschny-Rapp, S.; Jung, G.; Meiwes, J.; Zähner, H. Staphyloferrin A: A structurally new siderophore from staphylococci. Eur. J. Biochem. 1990, 191, 65–74. [Google Scholar] [CrossRef]
- Miethke, M.; Klotz, O.; Linne, U.; May, J.J.; Beckering, C.L.; Marahiel, M.A. Ferri-bacillibactin uptake and hydrolysis in Bacillus subtilis. Mol. Microbiol. 2006, 61, 1413–1427. [Google Scholar] [CrossRef]
- Rajavel, M.; Mitra, A.; Gopal, B. Role of Bacillus subtilis BacB in the synthesis of bacilysin. J. Biol. Chem. 2009, 284, 31882–31892. [Google Scholar] [CrossRef] [Green Version]
- Yakimov, M.M.; Timmis, K.N.; Wray, V.; Fredrickson, H.L. Characterization of a new lipopeptide surfactant produced by thermotolerant and halotolerant subsurface Bacillus licheniformis BAS50. Appl. Environ. Microbiol. 1995, 61, 1706–1713. [Google Scholar] [CrossRef] [Green Version]
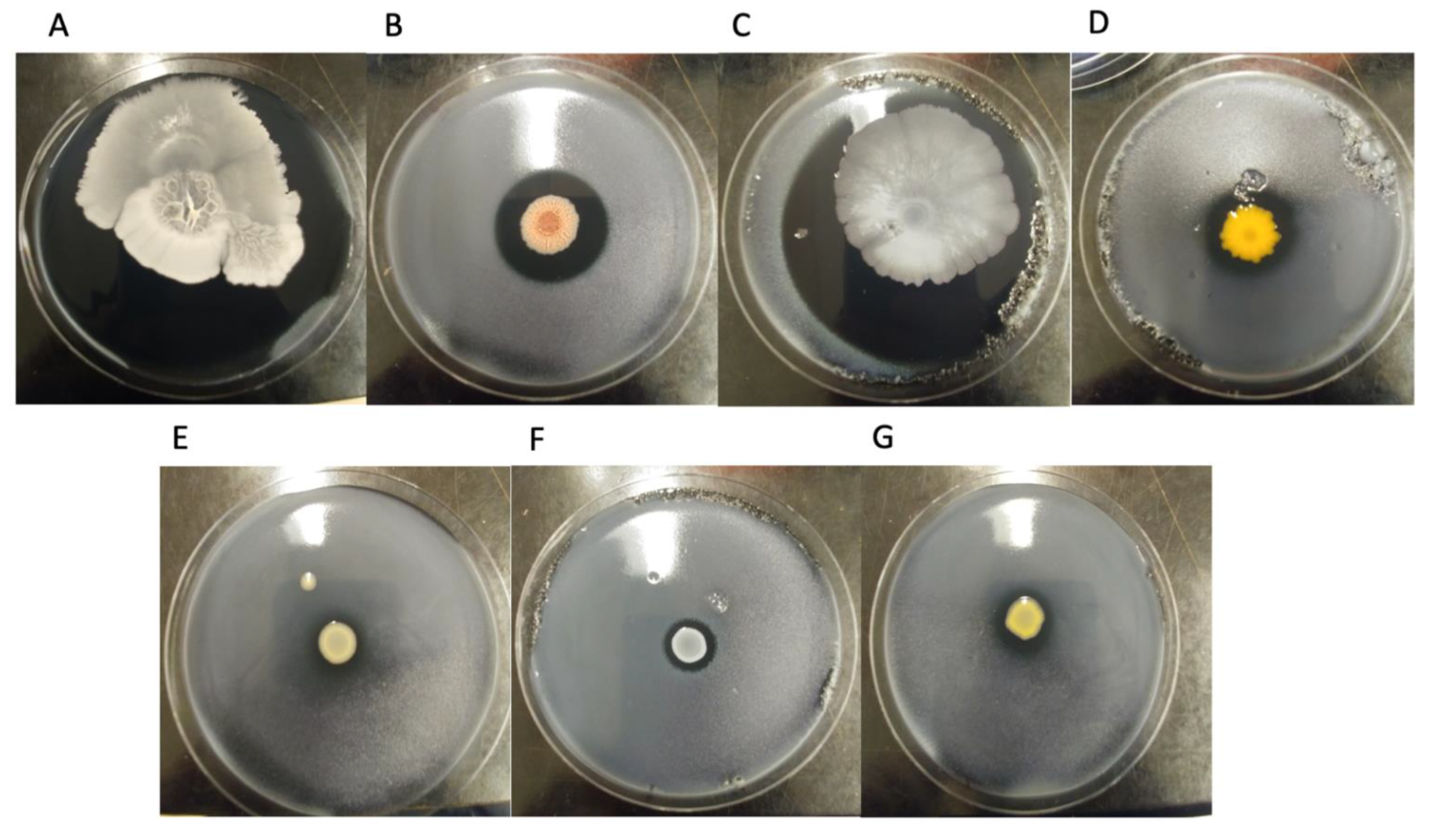

| Strain Name * | Strain Number ** | Species | Sequence Base Type (SBT) | Source |
|---|---|---|---|---|
| LpPhili | ATCC33152 | L. pneumophila philadelphia 1 (ATCC33152) | 37 | Patient |
| LpS62P | ID143016 | L. pneumophila | 62 | Patient |
| LpS62E | ID120292 | L. pneumophila (2012 Quebec City Outbreak) | 62 | Environmental |
| LpS1P | ID126851 | L. pneumophila | 1 | Patient |
| LpS1E | ID142903 | L. pneumophila | 1 | Environmental |
| LpS256P | ID128014 | L. pneumophila | 256 | Patient |
| LpS256E | ID128471 | L. pneumophila | 256 | Environmental |
| LpS213P | ID120882 | L. pneumophila | 213 | Patient |
| Lq | ID143958 | L. quinlivanii | NA | Patient |
| Strain Number | Closest Species (% ANI) | p-Value at Species Level | p-Value at Genus Level | Predicted Number of Proteins | Genome Size (bp) | Source |
|---|---|---|---|---|---|---|
| SPF474 | Bacillus amyloliquefaciens LL3 NC 017190 (99.98%) | 8.02 × 10−5 | 8.02 × 10−5 | 3638 | 3,522,847 | Cooling tower MTL3 [20] |
| SPF497 | Bacillus paralicheniformis NZ CP033389 (100%) | 8.02 × 10−5 | 8.02 × 10−5 | 4407 | 4,415,689 | Cooling tower MTL3 [20] |
| SPF498 | Stenotrophomonas sp. MYb57 NZ CP023271 (95.67%) | 0.124 | 0.0063 | 4091 | 4,581,475 | Cooling tower MTL5 [20] |
| SPF499 | Cupriviadus pauculus NZ CP033969 (86.84%) | 0.468 | 0.0338 | 6397 | 6,854,167 | Cooling tower MTL3 [20] |
| SPF475 | Chryseobacterium indologenes NZ CP018786 (90.65% ANI) | 0.296 | 0.0085 | 4971 | 5,302,653 | Cooling tower MTL5 [20] |
| SPF476 | Staphylococcus epidermidis ATCC 12228 NC 004461 (99.59%) | 0.0086 | 0.0014 | 2422 | 2,530,472 | Cooling tower MTL3 [20] |
| SPF437 | Bacillus subtilis subsp inaquosorum NZ_CP013984 (99.98% ANI) | 8.02 × 10−5 | 8.02 × 10−5 | 4123 | 4,195,215 | Cooling tower model [33] |
| Strain Name | Number of BGCs | High Similarity Clusters (>70% Similarity) | Low Similarity Clusters (<70% Similarity) | Number of Unassigned BGCs |
|---|---|---|---|---|
| Bacillus amyloliquefaciens | 10 | Bacillaene (100%), Bacillibactin (100%), Bacilysin (100%), Fengycin (93%) | Butirosin A/B (7%), Bacillomycin (60%), Surfactin (39%) | 3 |
| Bacillus paralicheniformis | 14 | Fengycin (73%), Lichenysin (100%), Bacitracin (88%), | Bacilibactin (53%), Fengycin (23%), Butirosin (7%), Haloduracin (40%), Fengycin (20%) | 6 |
| Stenotrophomonas sp. | 3 | 0 | Myxochelin (25%), APE Vf (35%) | 1 |
| Cupriavidus sp. | 9 | 0 | Desferrioxamine (50%), APEVf (40%), WS9326 (12%) | 6 |
| Chryseobacterium sp. | 12 | 0 | Desferrioxamine(50%), Flexirubin (52%), Flexirubin (22%), Caratenoid (28%) | 8 |
| Staphylococcus epidermidis | 3 | Staphyloferrin (100%) | 0 | 2 |
| Bacillus subtilis | 16 | Subtilosin A (100%), Bacilysin (100%), Surfactin (82%), Bacillibactin (100%), Fengycin (80%), Sublancin (100%), | Plipastatin (53%), Zwittermycin (18%), Aurantinins (21%), Aurantinins (39%), Plipastatin (23%), Aurantinins (28%) | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paranjape, K.; Lévesque, S.; Faucher, S.P. Bacterial Antagonistic Species of the Pathogenic Genus Legionella Isolated from Cooling Tower. Microorganisms 2022, 10, 392. https://doi.org/10.3390/microorganisms10020392
Paranjape K, Lévesque S, Faucher SP. Bacterial Antagonistic Species of the Pathogenic Genus Legionella Isolated from Cooling Tower. Microorganisms. 2022; 10(2):392. https://doi.org/10.3390/microorganisms10020392
Chicago/Turabian StyleParanjape, Kiran, Simon Lévesque, and Sébastien P. Faucher. 2022. "Bacterial Antagonistic Species of the Pathogenic Genus Legionella Isolated from Cooling Tower" Microorganisms 10, no. 2: 392. https://doi.org/10.3390/microorganisms10020392
APA StyleParanjape, K., Lévesque, S., & Faucher, S. P. (2022). Bacterial Antagonistic Species of the Pathogenic Genus Legionella Isolated from Cooling Tower. Microorganisms, 10(2), 392. https://doi.org/10.3390/microorganisms10020392






