Dynamics of Microbial Communities in Phototrophic Polyhydroxyalkanoate Accumulating Cultures
Abstract
:1. Introduction
2. Materials and Methods
2.1. Phototrophic Mixed Cultures Operation
2.2. DNA Sequencing and Taxonomy Assignment
2.3. Fluorescence In Situ Hybridization (FISH) and Morphological Observation
3. Results
3.1. DNA Sequencing and Taxonomic Groups Identification
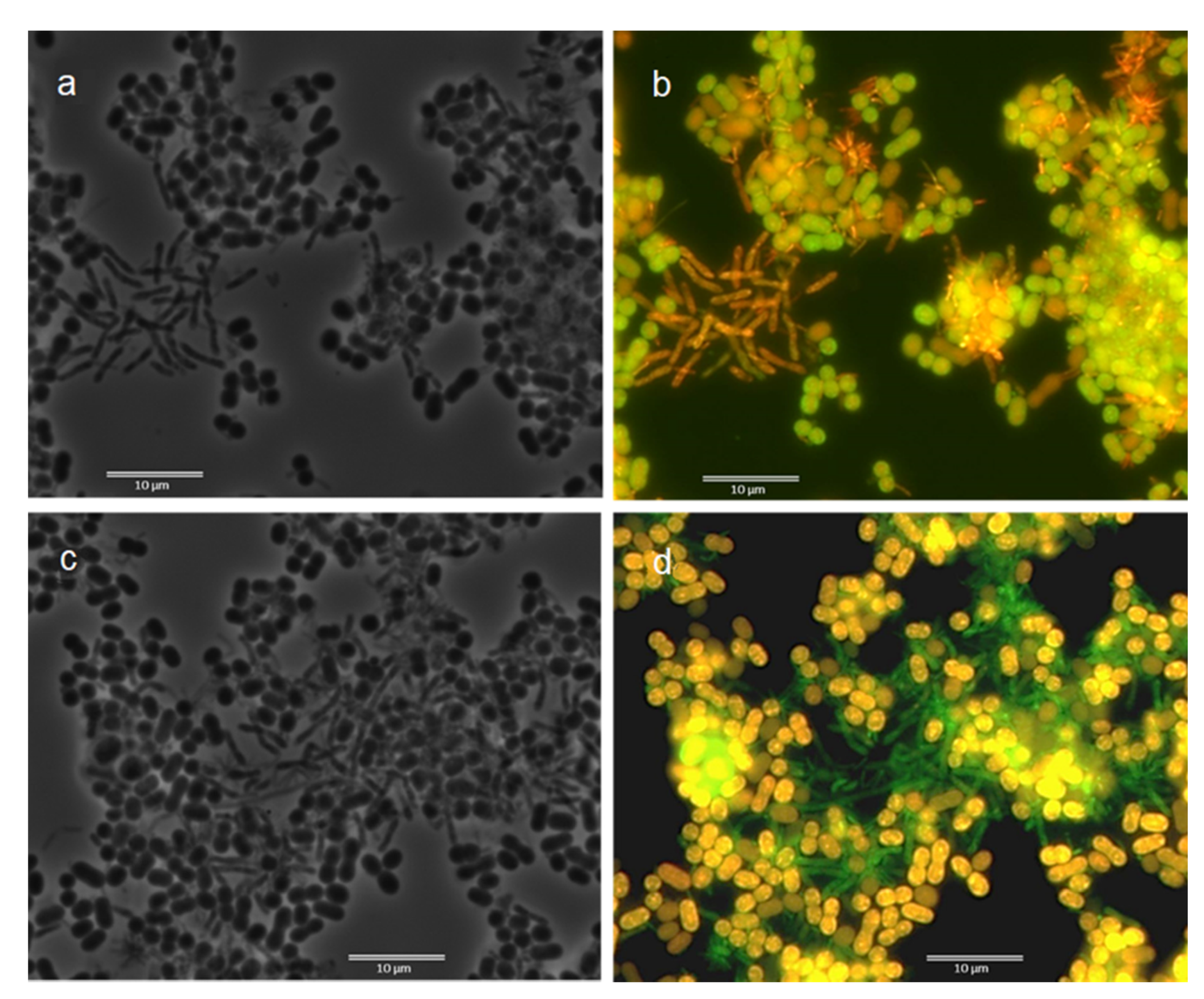
3.2. Fluorescence In Situ Hybridization (FISH)
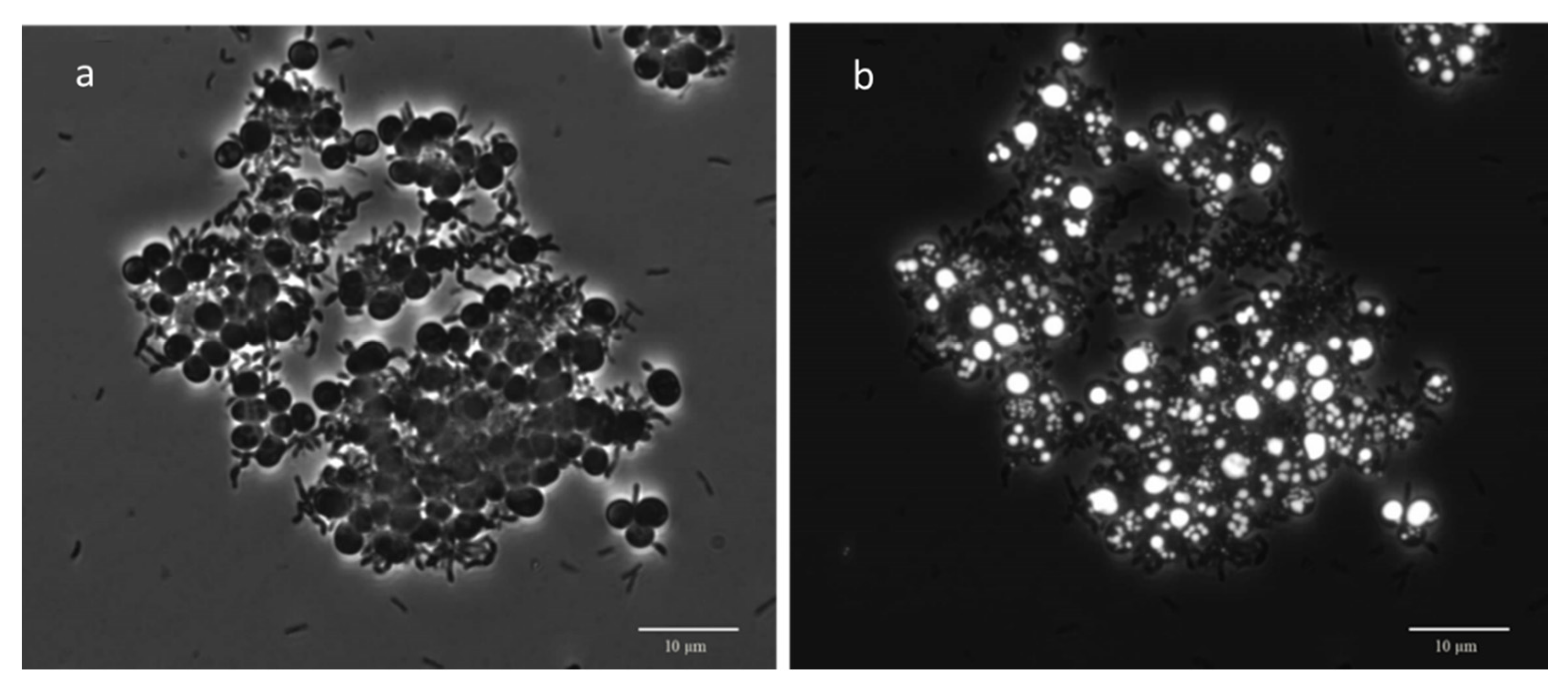
3.3. Microscopic Observation: Cells Morphology through FISH and Nile Blue Staining
4. Discussion
4.1. Synthetic Acetate Feeding (Cultures S1 to S5)
4.2. Fermented Real Waste Feeding (Cultures R1 to R6)
4.2.1. Permanent Feast Operation (R1 to R3)
4.2.2. Feast and Famine Operation (R4 to R6)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shukla, P.R.; Skea, J.; Buendia, E.C.; Masson-Delmotte, V.; Pörtner, H.O.; Roberts, D.; Zhai, P.; Slade, R.; Connors, S.; van Diemen, R.; et al. Summary for Policymakers. In Climate Change and Land: An IPCC Special Report on Climate Change, Desertification, Land Degradation, Sustainable Land Management, Food Security, and Greenhouse Gas Fluxes in Terrestrial Ecosystems; IPCC: Geneva, Switzerland, 2019; 2020 Intergovernmental Panel on Climate Change; ISBN 978-92-9169-154-8. [Google Scholar]
- Costa, C.F.F.A.; Amorim, C.L.; Duque, A.F.; Reis, M.A.M.; Castro, P.M.L. Valorization of wastewater from food industry: Moving to a circular bioeconomy. Rev. Environ. Sci. Bio Technol. 2021, 1–27. [Google Scholar] [CrossRef]
- Yaashikaa, P.; Kumar, P.S.; Varjani, S. Valorization of agro-industrial wastes for biorefinery process and circular bioeconomy: A critical review. Bioresour. Technol. 2021, 343, 126126. [Google Scholar] [CrossRef] [PubMed]
- Capson-Tojo, G.; Batstone, D.J.; Grassino, M.; Vlaeminck, S.E.; Puyol, D.; Verstraete, W.; Kleerebezem, R.; Oehmen, A.; Ghimire, A.; Pikaar, I.; et al. Purple phototrophic bacteria for resource recovery: Challenges and opportunities. Biotechnol. Adv. 2020, 43, 107567. [Google Scholar] [CrossRef] [PubMed]
- Imhoff, J.F. The Chromatiaceae. In The Prokaryotes; Springer: Berlin/Heidelberg, Germany, 2006; Volume 6, pp. 846–873. [Google Scholar]
- Puyol, D.; Batstone, D.J.; Hülsen, T.; Astals, S.; Peces, M.; Krömer, J. Resource Recovery from Wastewater by Biological Technologies: Opportunities, Challenges, and Prospects. Front. Microbiol. 2017, 7, 2106. [Google Scholar] [CrossRef] [Green Version]
- Fradinho, J.C.; Carvalho, V.C.F.; Reis, M.A.M. New Phototrophic Factories for Resource Recovery. In Enzymes for Solving Humankind’s Problems; Springer: Singapore, 2021; pp. 413–438. [Google Scholar]
- Cao, K.; Zhi, R.; Zhang, G. Photosynthetic bacteria wastewater treatment with the production of value-added products: A review. Bioresour. Technol. 2020, 299, 122648. [Google Scholar] [CrossRef]
- Sohn, Y.J.; Kim, H.T.; Baritugo, K.; Jo, S.Y.; Song, H.M.; Park, S.Y.; Park, S.K.; Pyo, J.; Gil, C.H.; Kim, H.; et al. Recent Advances in Sustainable Plastic Upcycling and Biopolymers. Biotechnol. J. 2020, 15, e1900489. [Google Scholar] [CrossRef]
- Tan, D.; Wang, Y.; Tong, Y.; Chen, G.-Q. Grand Challenges for Industrializing Polyhydroxyalkanoates (PHAs). Trends Biotechnol. 2021, 39, 953–963. [Google Scholar] [CrossRef]
- Reis, M.; Albuquerque, M.; Villano, M.; Majone, M. Mixed Culture Processes for Polyhydroxyalkanoate Production from Agro-Industrial Surplus/Wastes as Feedstocks. In Comprehensive Biotechnology; Elsevier: Amsterdam, The Netherlands, 2011; pp. 669–683. [Google Scholar]
- Monroy, I.; Buitrón, G. Production of polyhydroxybutyrate by pure and mixed cultures of purple non-sulfur bacteria: A review. J. Biotechnol. 2020, 317, 39–47. [Google Scholar] [CrossRef]
- Sali, S.; Mackey, H.R. The application of purple non-sulfur bacteria for microbial mixed culture polyhydroxyalkanoates production. Rev. Environ. Sci. Bio Technol. 2021, 20, 959–983. [Google Scholar] [CrossRef]
- Fradinho, J.; Allegue, L.; Ventura, M.; Melero, J.; Reis, M.; Puyol, D. Up-scale challenges on biopolymer production from waste streams by Purple Phototrophic Bacteria mixed cultures: A critical review. Bioresour. Technol. 2021, 327, 124820. [Google Scholar] [CrossRef]
- Fradinho, J.; Domingos, J.; Carvalho, G.; Oehmen, A.; Reis, M. Polyhydroxyalkanoates production by a mixed photosynthetic consortium of bacteria and algae. Bioresour. Technol. 2013, 132, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Fradinho, J.; Reis, M.A.; Oehmen, A. Beyond feast and famine: Selecting a PHA accumulating photosynthetic mixed culture in a permanent feast regime. Water Res. 2016, 105, 421–428. [Google Scholar] [CrossRef] [PubMed]
- Fradinho, J.; Oehmen, A.; Reis, M. Photosynthetic mixed culture polyhydroxyalkanoate (PHA) production from individual and mixed volatile fatty acids (VFAs): Substrate preferences and co-substrate uptake. J. Biotechnol. 2014, 185, 19–27. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Marang, L.; Kleerebezem, R.; Muyzer, G.; Van Loosdrecht, M.C.M. Effect of temperature and cycle length on microbial competition in PHB-producing sequencing batch reactor. ISME J. 2010, 5, 896–907. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, G.; Oehmen, A.; Albuquerque, M.G.; Reis, M.A. The relationship between mixed microbial culture composition and PHA production performance from fermented molasses. New Biotechnol. 2014, 31, 257–263. [Google Scholar] [CrossRef]
- Hülsen, T.; Barry, E.; Lu, Y.; Puyol, D.; Batstone, D. Low temperature treatment of domestic wastewater by purple phototrophic bacteria: Performance, activity, and community. Water Res. 2016, 100, 537–545. [Google Scholar] [CrossRef] [Green Version]
- García, D.; de Godos, I.; Domínguez, C.; Turiel, S.; Bolado, S.; Muñoz, R. A systematic comparison of the potential of microalgae-bacteria and purple phototrophic bacteria consortia for the treatment of piggery wastewater. Bioresour. Technol. 2019, 276, 18–27. [Google Scholar] [CrossRef] [Green Version]
- Cerruti, M.; Stevens, B.; Ebrahimi, S.; Alloul, A.; Vlaeminck, S.E.; Weissbrodt, D.G. Enrichment and Aggregation of Purple Non-sulfur Bacteria in a Mixed-Culture Sequencing-Batch Photobioreactor for Biological Nutrient Removal from Wastewater. Front. Bioeng. Biotechnol. 2020, 8, 8. [Google Scholar] [CrossRef]
- Fradinho, J.; Oehmen, A.; Reis, M.A. Effect of dark/light periods on the polyhydroxyalkanoate production of a photosynthetic mixed culture. Bioresour. Technol. 2013, 148, 474–479. [Google Scholar] [CrossRef]
- Almeida, J.; Serrano, E.; Fernandez, M.; Fradinho, J.; Oehmen, A.; Reis, M. Polyhydroxyalkanoates production from fermented domestic wastewater using phototrophic mixed cultures. Water Res. 2021, 197, 117101. [Google Scholar] [CrossRef]
- Albertsen, M.; Karst, S.M.; Ziegler, A.S.; Kirkegaard, R.H.; Nielsen, P.H. Back to Basics—The Influence of DNA Extraction and Primer Choice on Phylogenetic Analysis of Activated Sludge Communities. PLoS ONE 2015, 10, e0132783. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Huntley, J.; Fierer, N.; Owens, S.M.; Betley, J.; Fraser, L.; Bauer, M.; et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012, 6, 1621–1624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jumpstart Consortium Human Microbiome Project Data Generation Working Group. Evaluation of 16S rDNA-Based Community Profiling for Human Microbiome Research. PLoS ONE 2012, 7, e39315. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Edgar, R.C. UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 2013, 10, 996–998. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high- throughput community sequencing data Intensity normal-ization improves color calling in SOLiD sequencing. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef] [Green Version]
- McIlroy, S.J.; Kirkegaard, R.H.; McIlroy, B.; Nierychlo, M.; Kristensen, J.M.; Karst, S.M.; Albertsen, M.; Nielsen, P.H. MiDAS 2.0: An ecosystem-specific taxonomy and online database for the organisms of wastewater treatment systems expanded for anaerobic digester groups. Database 2017, 2017. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- Bengtsson, S.; Werker, A.; Welander, T. Production of polyhydroxyalkanoates by glycogen accumulating organisms treating a paper mill wastewater. Water Sci. Technol. 2008, 58, 323–330. [Google Scholar] [CrossRef]
- Nielsen, P.H.; Daims, H.; Lemmer, H. FISH Handbook for Biological Wastewater Treatment: Identification and Quantification of Microorganisms in Activated Sludge and Biofilms by FISH, 1st ed.; IWA Publishing: London, UK, 2009. [Google Scholar]
- Greuter, D.; Loy, A.; Horn, M.; Rattei, T. ProbeBase—An online resource for rRNA-targeted oligonucleotide probes and primers: New features 2016. Nucleic Acids Res. 2016, 44, D586–D589. [Google Scholar] [CrossRef] [PubMed]
- Imhoff, J.F. Blastochloris. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2020; pp. 1–8. [Google Scholar]
- Carvalho, V.; Kessler, M.; Fradinho, J.; Oehmen, A.; Reis, M. Achieving nitrogen and phosphorus removal at low C/N ratios without aeration through a novel phototrophic process. Sci. Total. Environ. 2021, 793, 148501. [Google Scholar] [CrossRef] [PubMed]
- Kolakovic, S.; Santos, J.M.; Reis, M.A. Phosphorus and carbon solubilization strategies for wastewater sludge valorisation. J. Environ. Chem. Eng. 2021, 9, 106261. [Google Scholar] [CrossRef]
- Dionisi, D.; Majone, M.; Vallini, G.; Di Gregorio, S.; Beccari, M. Effect of the applied organic load rate on biodegradable polymer production by mixed microbial cultures in a sequencing batch reactor. Biotechnol. Bioeng. 2006, 93, 76–88. [Google Scholar] [CrossRef]
- Rainey, F.A. Trichococcus. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; pp. 1–7. [Google Scholar]
- Li, J.; Li, Y.; Zhou, Y.; Wang, C.; Wu, B.; Wan, J. Actinomycesand Alimentary Tract Diseases: A Review of Its Biological Functions and Pathology. BioMed Res. Int. 2018, 2018, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Maszenan, A.M.; Seviour, R.J.; Patel, B.K.C. Amaricoccus. In Bergey’s Manual of Systematics of Archaea and Bacteria; Trujillo, M.E., Dedysh, S., DeVos, P., Hedlund, B., Kämpfer, P., Rainey, F.A., Whitman, W.B., Eds.; Wiley: Hoboken, NJ, USA, 2015; pp. 1–7. [Google Scholar] [CrossRef]
- Ueki, A.; Akasaka, H.; Ueki, K.; Genus, X. Propionicimonas. In Bergey’s Manual of Systematic Bacteriology; Goodfellow, M., Kämpfer, P., Busse, H.J., Trujillo, M.E., Suzuki, K.I., Ludwig, W., Whitman, W.B., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; p. 1175. [Google Scholar] [CrossRef]
- Morgan-Sagastume, F. Characterisation of open, mixed microbial cultures for polyhydroxyalkanoate (PHA) production. Rev. Environ. Sci. Bio Technol. 2016, 15, 593–625. [Google Scholar] [CrossRef]
- Kuever, J.; Rainey, F.A.; Widdel, F. Class IV. Deltaproteobacteria class nov. In Bergey’s Manual® of Systematic Bacteriology; Springer Science and Business Media LLC: Berlin/Heidelberg, Germany, 2005; Volume 478, pp. 922–1144. [Google Scholar]
- Shimkets, L. Development, Prokaryotic: Variety and Versatility. In Reference Module in Biomedical Sciences; Elsevier BV: Amsterdam, The Netherlands, 2015. [Google Scholar]
- Lorenzen, W.; Ahrendt, T.; Bozhüyük, K.; Bode, H. A multifunctional enzyme is involved in bacterial ether lipid biosynthesis. Nat. Chem. Biol. 2014, 10, 425–427. [Google Scholar] [CrossRef]
| ID | Reactor | Feed | Selection | Operating Conditions | Culture Performance | Ref. | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Light | Cycle | SRT | OLR | VSS | SBR | Accumulation Test | |||||||
| (W/L) | (Days) | (Cmmol/L·d) | (g/L) | PHA Content a | qPHA b | PHA Content a | qPHA b | ||||||
| S1 | Closed, argon sparging, 30 °C | Synthetic acetate | FF | 1.3 | 24 h light | 6 | 10 | 1.1 c | 2–4 (4–8; 100 HB) | 0.13 ± 0.03 | n.d. | n.d. | - |
| S2 | 8 h light | 6 | 10 | 1.4 | 0.5–2 (0.5–4; 100 HB) | 1.2 ± 0.2 | ≈18 d (20; 100 HB) | 1.1 ± 0.1 | [15] | ||||
| S3 | 4 h light + 4 h dark | 6 | 10 | 0.7 c | 4–7 (12–17; 100 HB) | 1.6 ± 0.1 | 15 e (30; 100 HB) | 2.2 ± 0.1 | [23] | ||||
| S4 | 8 h light | 6 | 20 | 2.0 c | 8–14 (8–12; 100 HB) | 1.5 ± 0.1 | 22 f (18; 76 HB:24 HV) | n.d. | [17] | ||||
| S5 | PF | 1.8 | 24 h light | 3 | 40 | 1.2 c | ≈2 (4 -8; 100 HB) | 0.05 ± 0.04 | 75 g (60; 100 HB) | 0.73 ± 0.13 | [16] | ||
| R0 | Inoculum from high-rate algae ponds used for domestic wastewater treatment | [24] | |||||||||||
| R1 | Open, 25 °C | Fermented mixture molasses + domestic wastewater | PF | 1.1 | 12 h light + 12 h dark | 6 | 12 | 1.7 ± 0.2 | 14 (20; 86 HB:14 HV) | 0.13 | n.d. | n.d. | |
| * R2 | 6 | 12.5 | 0.2 | 0.5–2 (4–6; ≈65 HB:35 HV) | n.d. | n.d. | n.d. | ||||||
| R3 | 6 | 2–5 | 0.6 ± 0.2 | 7 (18; 60 HB:40 HV) | 0.33 | n.d. | n.d. | ||||||
| R4 | FF | 1.9 | 12 h light + 12 h dark | 6 | 2–5 | 0.9 ± 0.2 | 0.1–1.5 (0.5–6; ≈80 HB:20 HV) | 0.42 | n.d. | n.d. | |||
| * R5 | 6 | 7 | 1.0 ± 0.2 | 0.3–6 (0.5–13; ≈40 HB:60 HV) | n.d. | n.d. | n.d. | ||||||
| R6 | 6 | 7–10 | 1.6 ± 0.2 | 1–6 (4–9; 63 HB:37 HV) | 0.62 | 32 h (31; 85 HB:15 HV) | 1.84 | ||||||
 less abundant to more abundant.
less abundant to more abundant.
 less abundant to more abundant.
less abundant to more abundant.| S1 | S2 | S3 | S4 | S5 | |||||
|---|---|---|---|---|---|---|---|---|---|
| Phylum | Class | Order | Family | Genus | % | ||||
| Actinobacteria | - | - | - | - | 0.0 | 2.1 | 0.0 | 0.1 | 1.8 |
| Bacteroidetes | - | - | - | - | 5.8 | 0.1 | 0.2 | 0.1 | 7.4 |
| Chloroflexi | - | - | - | - | 2.2 | 0.1 | 0.4 | 0.1 | 0.0 |
| Cyanobacteria | - | - | - | - | 5.1 | 4.6 | 6.6 | 1.7 | 0.4 |
| Proteobacteria | Alpha proteobacteria | Rhizobiales | Bradyrhizobiaceae | - | 37.5 | 6.5 | 1.6 | 5.1 | 58.6 |
| Phyllobacteriaceae | - | 0.0 | 1.0 | 0.0 | 2.0 | 0.0 | |||
| Rhizobiaceae | - | 0.1 | 0.5 | 72.5 | 0.0 | 0.1 | |||
| Hyphomicrobiaceae | Blastochloris | 30.9 | 60.0 | 12.7 | 74.7 | 0.2 | |||
| Rhodobacterales | Rhodobacteraceae | - | 7.7 | 5.2 | 0.2 | 0.2 | 0.0 | ||
| Rickettsiales | - | - | 0.0 | 2.7 | 0.0 | 0.3 | 0.0 | ||
| Gamma proteobacteria | Chromatiales | Chromatiaceae | - | 2.2 | 0.0 | 0.3 | 5.1 | 26.0 | |
| Xanthomonadales | Xanthomonadaceae | - | 0.0 | 10.1 | 0.5 | 0.6 | 0.0 | ||
| Other phyla | - | - | - | - | 3.5 | 0.1 | 0.6 | 0.8 | 0.1 |
| Total | 95.0 | 93.1 | 95.6 | 90.9 | 94.6 | ||||
 less abundant to more abundant.
less abundant to more abundant.
 less abundant to more abundant.
less abundant to more abundant.| R0 | R1 | R2 | R3 | R4 | R5 | R6 | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Phylum | Class | Order | Family | Genus | % | ||||||
| Actinobacteria | Actinobacteridae | Actinomycetales | - | - | 0.5 | 0.0 | 55.2 | 0.1 | 0.0 | 0.2 | 0.1 |
| unknown | Corynebacteriales | - | - | 4.3 | 0.0 | 0.0 | 0.0 | 0.2 | 0.0 | 0.0 | |
| Propionibacteriales | Propionibacteriaceae | Propionicimonas | 0.0 | 0.0 | 0.8 | 0.2 | 0.1 | 11.1 | 0.4 | ||
| Micrococcales | - | - | 2.1 | 0.0 | 0.1 | 0.0 | 0.6 | 0.3 | 0.9 | ||
| PeM15 | - | - | 10.9 | 0.0 | 0.0 | 0.0 | 0.1 | 0.1 | 0.0 | ||
| Chloroflexi | Caldilineae | Caldilineales | - | - | 11.7 | 0.0 | 0.0 | 0.0 | 0.8 | 0.3 | 0.0 |
| Cyanobacteria | - | - | - | - | 0.3 | 0.0 | 0.4 | 1.0 | 0.4 | 1.4 | 2.4 |
| Firmicutes | Bacilli | Lactobacillales | Carnobacteriaceae | Trichococcus | 0.6 | 68.8 | 3.5 | 0.0 | 0.0 | 0.0 | 0.0 |
| Clostridia | Clostridiales | Clostridiaceae | Clostridium | 2.0 | 0.3 | 1.2 | 0.0 | 0.3 | 0.1 | 0.0 | |
| Peptostreptococcaceae | - | 6.7 | 10.0 | 2.0 | 0.0 | 0.3 | 0.1 | 0.0 | |||
| Proteobacteria | Alpha proteobacteria | Rhizobiales | Bradyrhizobiaceae | Rhodopseudomonas | 7.2 | 2.7 | 8.7 | 84.4 | 1.2 | 1.2 | 10.9 |
| Rhizobiaceae | Rhizobium | 0.6 | 0.0 | 0.0 | 11.2 | 1.0 | 1.9 | 32.9 | |||
| Hyphomicrobiaceae | - | 3.0 | 0.1 | 0.8 | 0.1 | 1.8 | 0.5 | 22.5 | |||
| Rhodobacterales | Rhodobacteraceae | Rhodobacter | 5.7 | 6.7 | 1.5 | 1.3 | 15.4 | 0.5 | 5.7 | ||
| Paracoccus | 0.5 | 0.0 | 0.1 | 0.0 | 53.7 | 9.5 | 0.7 | ||||
| Caulobacterales | - | - | 0.8 | 0.4 | 0.0 | 0.0 | 1.0 | 3.7 | 0.0 | ||
| Beta proteobacteria | Burkholderiales | Comamonadaceae | - | 3.6 | 0.0 | 0.0 | 0.3 | 0.5 | 0.2 | 0.1 | |
| Rhodocyclales | Rhodocyclaceae | - | 1.6 | 0.0 | 0.1 | 0.1 | 4.4 | 62.9 | 0.1 | ||
| Gamma proteobacteria | Chromatiales | Chromatiaceae | - | 0.4 | 5.0 | 2.1 | 0.0 | 2.9 | 0.1 | 3.9 | |
| Xanthomonadales | - | - | 1.7 | 0.1 | 0.0 | 0.0 | 5.9 | 0.3 | 0.5 | ||
| Other phyla | - | - | - | - | 4.1 | 0.2 | 2.0 | 0.1 | 1.8 | 1.0 | 1.8 |
| Total | 68.3 | 94.3 | 78.5 | 98.8 | 92.4 | 95.4 | 82.9 | ||||
| Samples | ALF969 | BET42a | GAM42a | Delta495a | ARC915 | LGC0354 | GRb | RHC439 | Rhodo-2 | Rhodopseud | DSBAC357 | DSB706 | DSV687 | PAR651 | Azo644 | Thau832 | ZRA23a | AMAR839 | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| S1 | +++ | ± | ++ | n.a. | n.a. | n.a. | n.a. | n.a. | n.a. | ++ | n.a | n.a | n.a | n.a | n.a | n.a | n.a | n.a | * |
| S2 | ++ | - | +++ | - | - | n.a | + | - | + | + | n.a. | n.a. | n.a. | - | - | - | - | - | [15] |
| S3 | ++ | - | ++ | - | n.a | n.a | n.a | n.a | n.a | - | n.a | n.a | n.a | n.a | n.a | n.a | n.a | n.a | [23] |
| S4 | ++ | - | +++ | n.a | n.a | n.a | - | n.a | - | + | n.a | n.a | n.a | n.a | n.a | n.a | n.a | n.a | [17] |
| S5 | ++ | - | ++ | n.a | n.a | n.a | n.a | n.a | n.a | - | n.a | n.a | n.a | n.a | n.a | n.a | n.a | n.a | [16] |
| R1 | inc. | ± | ++ | ± | ++ | ± | + | - | - | ++ | - | ± | - | ± | + | + | ± | ± | * |
| R2 | inc. | - | ++ | + | + | ± | inc. | - | - | + | ± | ± | - | ± | - | - | ± | + | |
| R3 | +++ | ++ | + | ++ | ++ | ± | +++ | ± | - | ++ | - | + | - | ± | - | - | - | + | |
| R4 | +++ | ++ | ++ | ++ | ± | ± | +++ | ± | - | ± | ± | + | ± | ++ | - | + | ++ | ± | |
| R5 | ++ | ++ | + | +++ | ± | ± | ++ | ± | - | ± | ± | ± | ± | ++ | ± | + | ++ | - | |
| R6 | ++ | + | + | ++ | - | - | ++ | ± | - | - | - | + | ± | ++ | - | ± | - | ± |
| Morphology from Nile Blue Observation | Morphology from FISH Observation | ||||||
|---|---|---|---|---|---|---|---|
| ALF969 | BET42a | GAM42a | Delta495a | ||||
| R1 | Coccus (large) | a | Rods (short) | Rods (thin) | Coccus | Not present | |
| R2 | Coccus (aggregated in rectangular sets) Rods (thin); Rods (thick) | a | Rods (short) | Not present | Rods (thick) Coccus | Rods (thin) | |
| R3 | Rods (long and thick) | Rods (short and thick) Rods (thin) | Rods (short and thick) | Rods (long and thick) | Rods (thin) Ovoid | ||
| R4 | Rods (aggregated in swarms) * | Rods Ovoid | Rods (short and thin) | Rods (long and thick) | Rods (swarms) | ||
| R5 | Coccus Rods (aggregated in swarms) * | Rods (short and thick) | b | Coccus (large) | Rods (short) | Rods (swarms) | |
| R6 | Coccus (large) Rods (short) | Coccus | Rods | Rods | Rods | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Almeida, J.R.; Fradinho, J.C.; Carvalho, G.; Oehmen, A.; Reis, M.A.M. Dynamics of Microbial Communities in Phototrophic Polyhydroxyalkanoate Accumulating Cultures. Microorganisms 2022, 10, 351. https://doi.org/10.3390/microorganisms10020351
Almeida JR, Fradinho JC, Carvalho G, Oehmen A, Reis MAM. Dynamics of Microbial Communities in Phototrophic Polyhydroxyalkanoate Accumulating Cultures. Microorganisms. 2022; 10(2):351. https://doi.org/10.3390/microorganisms10020351
Chicago/Turabian StyleAlmeida, Juliana R., Joana C. Fradinho, Gilda Carvalho, Adrian Oehmen, and Maria A. M. Reis. 2022. "Dynamics of Microbial Communities in Phototrophic Polyhydroxyalkanoate Accumulating Cultures" Microorganisms 10, no. 2: 351. https://doi.org/10.3390/microorganisms10020351
APA StyleAlmeida, J. R., Fradinho, J. C., Carvalho, G., Oehmen, A., & Reis, M. A. M. (2022). Dynamics of Microbial Communities in Phototrophic Polyhydroxyalkanoate Accumulating Cultures. Microorganisms, 10(2), 351. https://doi.org/10.3390/microorganisms10020351








