Novel Bacteriophages Show Activity against Selected Australian Clinical Strains of Pseudomonas aeruginosa
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Host Strains and Culture Conditions
2.2. Isolation and Enrichment of the Bacteriophages
2.3. Purification and Enumeration of the Bacteriophages
2.4. Phage Morphology by Negative Staining and TEM
2.5. Phage Host Range
2.6. Assessment of Phage Stability at Different Parameters
2.7. Adsorption Assay
2.8. One Step Growth
2.9. Phage Cocktail Killing Action
2.10. Phage Genome Extraction, Sequencing and Analysis
2.10.1. Phage DNA Extraction and Nuclease Treatment
2.10.2. Library Preparation and DNA Sequencing
2.10.3. Bioinformatic Analysis of the Phage Genomes
2.10.4. Phylogenetic Analysis
2.10.5. Comparative Genomics
2.10.6. Nucleotide Sequence Accession Numbers
3. Results
3.1. Isolation and Morphology of the Phages
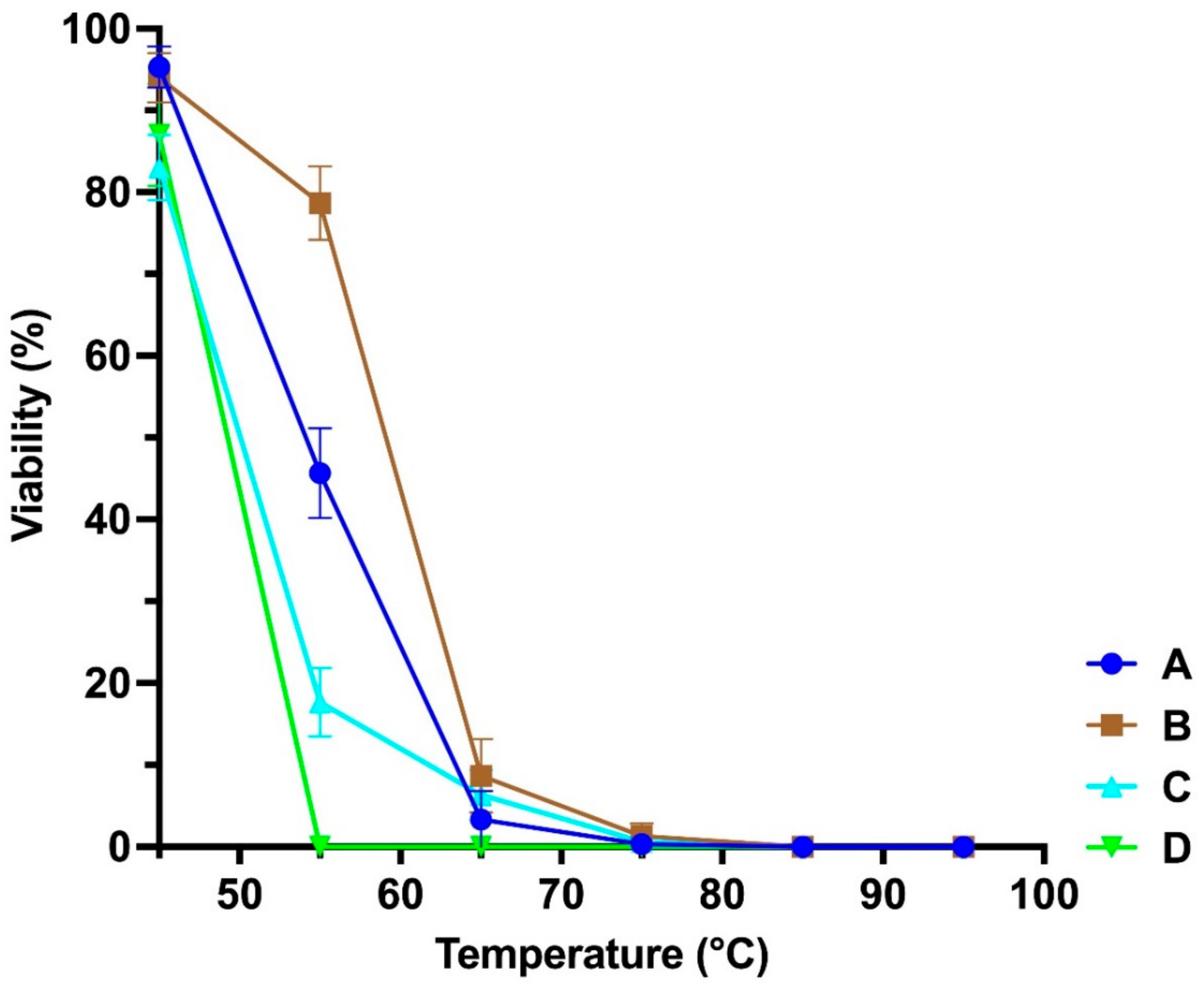
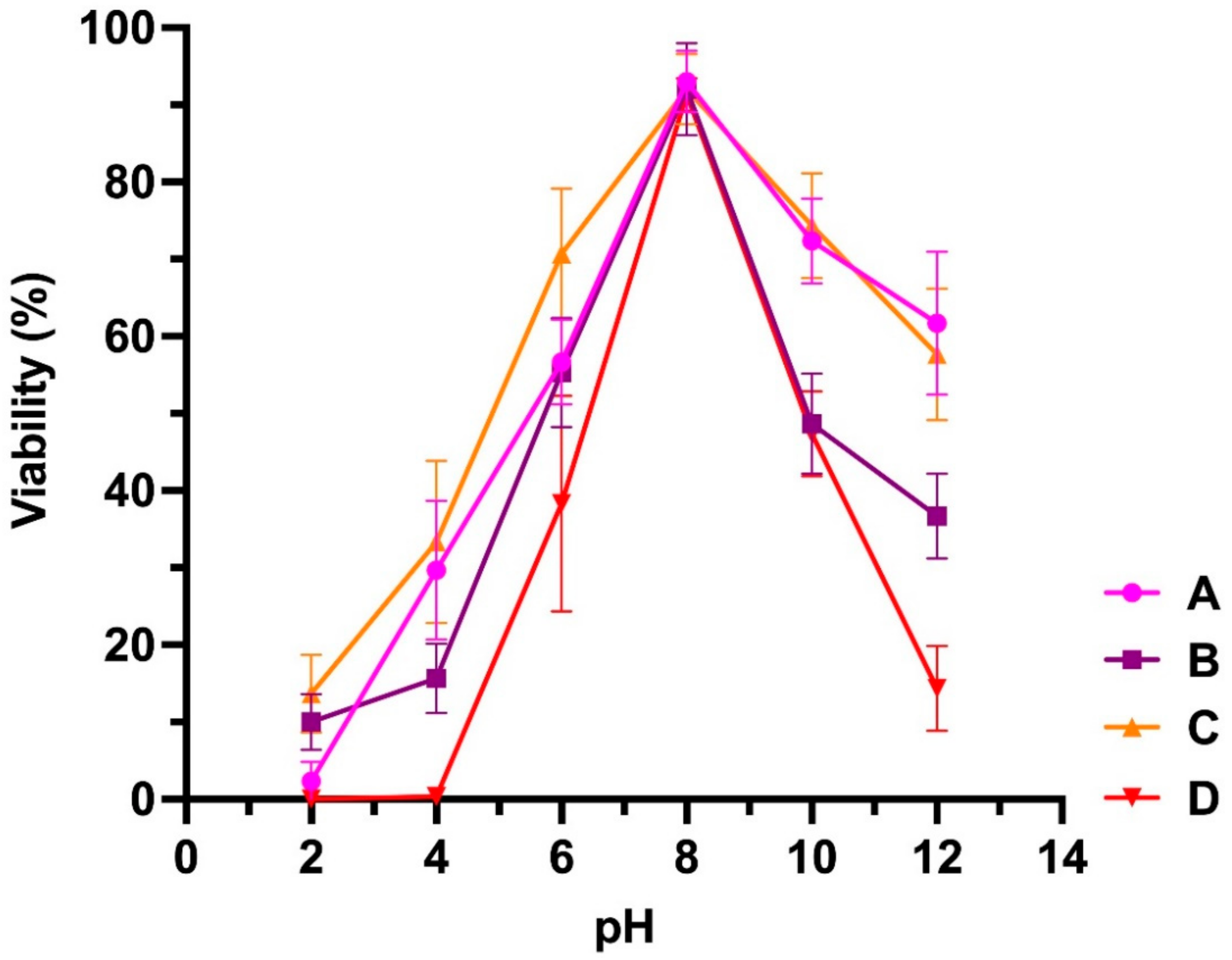
3.2. Stability at Different Temperatures and pH
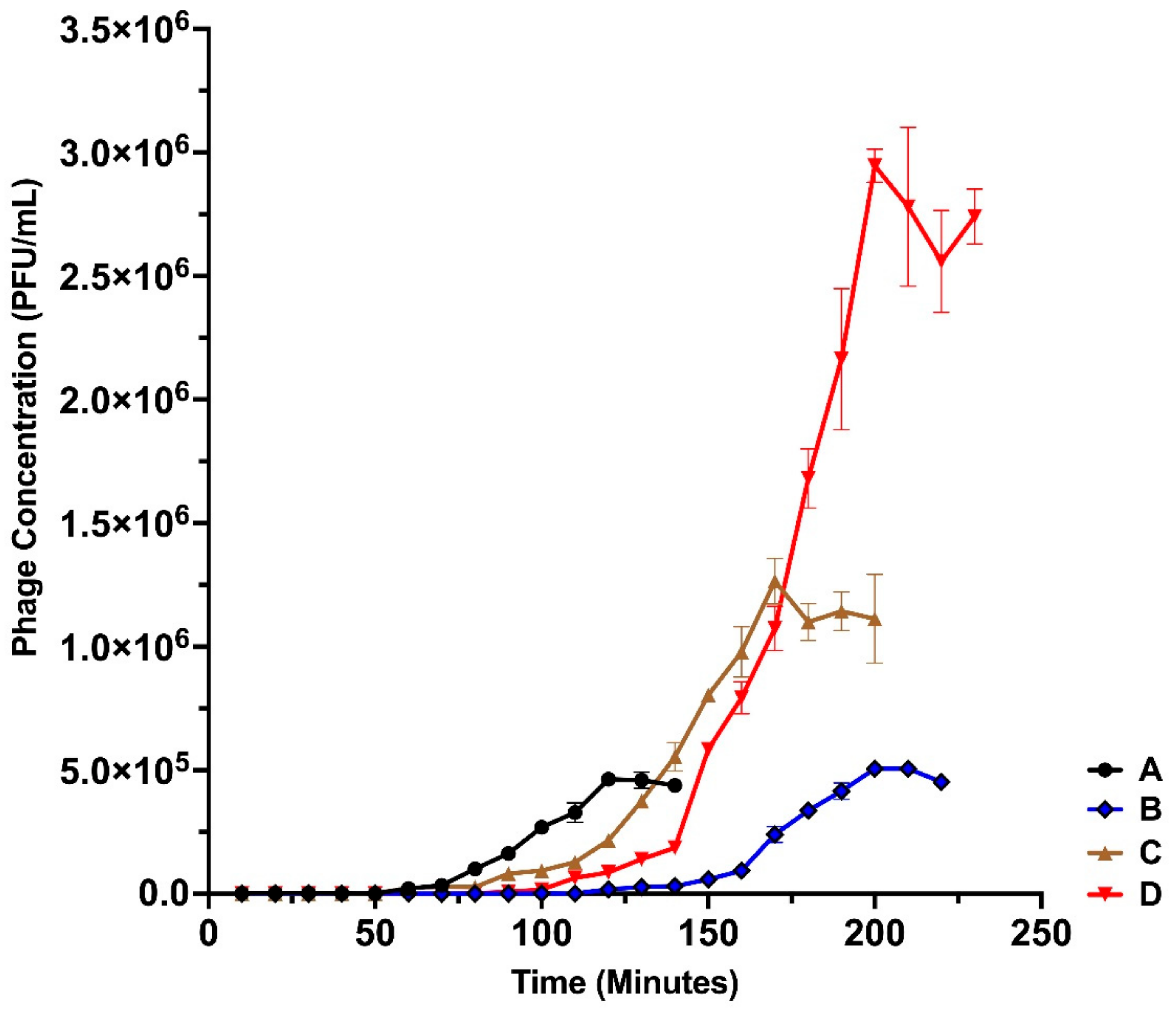
3.3. One-Step Growth Dynamics of the Isolated Bacteriophages
3.4. Host Range Analysis
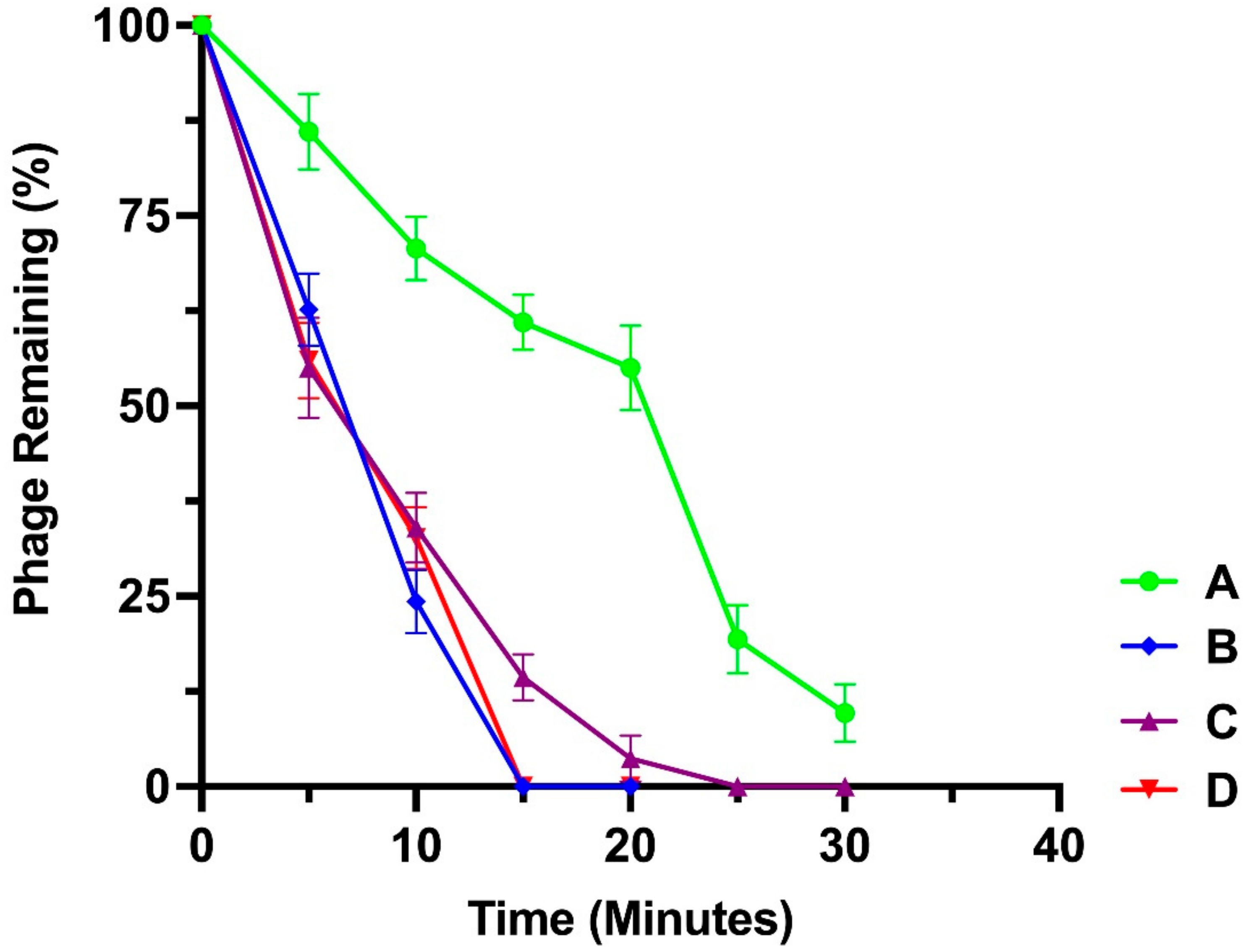
3.5. Adsorption Assay
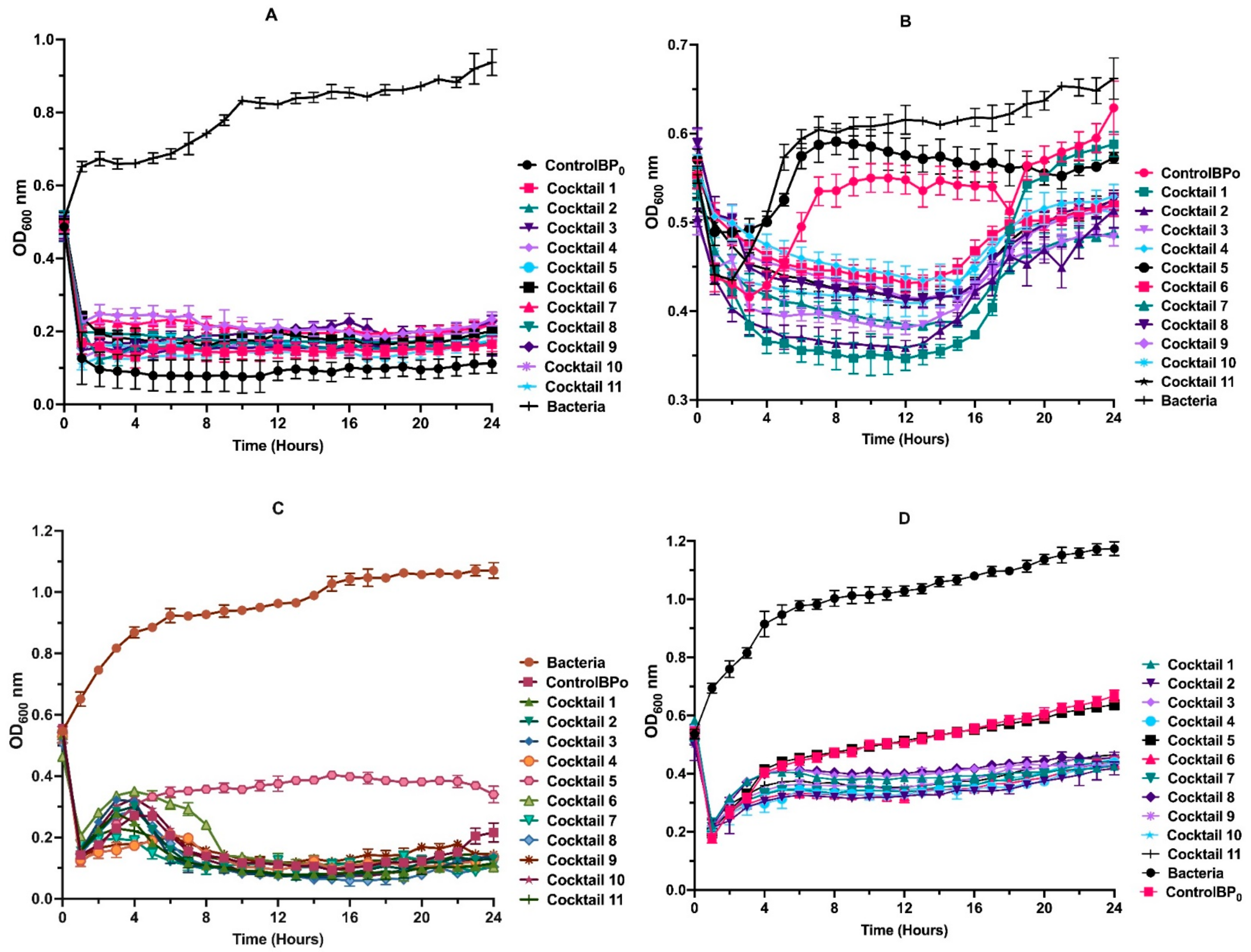
3.6. Phage Cocktail Killing Action
3.7. Analysis of Bacteriophage Genomes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rice, L.B. Federal Funding for the Study of Antimicrobial Resistance in Nosocomial Pathogens: No ESKAPE. J. Infect. Dis. 2008, 197, 1079–1081. [Google Scholar] [CrossRef] [PubMed]
- Pendleton, J.N.; Gorman, S.P.; Gilmore, B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti-Infect. Ther. 2013, 11, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Laxminarayan, R.; Duse, A.; Wattal, C.; Zaidi, A.K.; Wertheim, H.F.; Sumpradit, N.; Vlieghe, E.; Hara, G.L.; Gould, I.M.; Goossens, H.; et al. Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 2013, 13, 1057–1098. [Google Scholar] [CrossRef]
- Luepke, K.H.; Suda, K.J.; Boucher, H.; Russo, R.L.; Bonney, M.W.; Hunt, T.D.; Mohr, J.F., 3rd. Past, Present, and Future of Antibacterial Economics: Increasing Bacterial Resistance, Limited Antibiotic Pipeline, and Societal Implications. Pharmacotherapy 2017, 37, 71–84. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, J. Tackling Drug Resistant Infections Globally: Final Report and Recommendations; Government of the United Kingdom: London, UK, 2016.
- Tacconelli, E.; Carrara, E.; Savoldi, A.; Harbarth, S.; Mendelson, M.; Monnet, D.L.; Pulcini, C.; Kahlmeter, G.; Kluytmans, J.; Carmeli, Y.; et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018, 18, 318–327. [Google Scholar] [CrossRef]
- Mena, K.D.; Gerba, C.P. Risk assessment of Pseudomonas aeruginosa in water. Rev. Environ. Contam. Toxicol. 2009, 201, 71–115. [Google Scholar] [PubMed]
- Aendekerk, S.; Diggle, S.P.; Song, Z.; Høiby, N.; Cornelis, P.; Williams, P.; Cámara, M. The MexGHI-OpmD multidrug efflux pump controls growth, antibiotic susceptibility and virulence in Pseudomonas aeruginosa via 4-quinolone-dependent cell-to-cell communication. Microbiology 2005, 151, 1113–1125. [Google Scholar] [CrossRef]
- Driscoll, J.A.; Brody, S.L.; Kollef, M.H. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 2007, 67, 351–368. [Google Scholar] [CrossRef]
- Gellatly, S.L.; Hancock, R.E. Pseudomonas aeruginosa: New insights into pathogenesis and host defenses. Pathog. Dis. 2013, 67, 159–173. [Google Scholar] [CrossRef]
- Moradali, M.F.; Donati, I.; Sims, I.M.; Ghods, S.; Rehm, B.H.A.; Nitz, M.; Pier, G.B. Alginate Polymerization and Modification Are Linked in Pseudomonas aeruginosa. mBio 2015, 6, e00453-15. [Google Scholar] [CrossRef]
- Breidenstein, E.B.; de la Fuente-Núñez, C.; Hancock, R.E. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011, 19, 419–426. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.P.; Boas, D.V.; Sillankorva, S.; Azeredo, J.; Goff, S.P. Phage Therapy: A Step Forward in the Treatment of Pseudomonas aeruginosa Infections. J. Virol. 2015, 89, 7449–7456. [Google Scholar] [CrossRef] [PubMed]
- Holloway, B.W.; Egan, J.B.; Monk, M. Lysogeny in Pseudomonas aeruginosa. Aust. J. Exp. Biol. Med. Sci. 1960, 38, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Lindberg, R.B.; Latta, R.L. Phage Typing of Pseudomonas aeruginosa: Clinical and Epidemiologic Considerations. J. Infect. Dis. 1974, 130, S33–S42. [Google Scholar] [CrossRef] [PubMed]
- Krylov, V.; Shaburova, O.; Pleteneva, E.; Bourkaltseva, M.; Krylov, S.; Kaplan, A.; Chesnokova, E.; Kulakov, L.; Magill, D.; Polygach, O. Modular Approach to Select Bacteriophages Targeting Pseudomonas aeruginosa for Their Application to Children Suffering with Cystic Fibrosis. Front. Microbiol. 2016, 7, 1631. [Google Scholar] [CrossRef]
- Shabalova, I.; Karpanov, N.; Krylov, V.; Sharibjanova, T.; Akhverdijan, V. Pseudomonas aeruginosa bacteriophage in treatment of P. aeruginosa infection in cystic fibrosis patients. In Proceedings of the IX International Cystic Fibrosis Congress, Brighton, UK, 9–15 June 1984; Volume 443, pp. 377–383. [Google Scholar]
- Verbeken, G.; Vos, D.D.; Vaneechoutte, M.; Merabishvili, M.; Zizi, M.; Pirnay, J.-P. European regulatory conundrum of phage therapy. Future Microbiol. 2007, 2, 485–491. [Google Scholar] [CrossRef]
- Anomaly, J. The Future of Phage: Ethical Challenges of Using Phage Therapy to Treat Bacterial Infections. Public Health Ethics 2020, 13, 82–88. [Google Scholar] [CrossRef]
- Rossitto, M.; Fiscarelli, E.V.; Rosati, P. Challenges and Promises for Planning Future Clinical Research into Bacteriophage Therapy Against Pseudomonas aeruginosa in Cystic Fibrosis. An Argumentative Review. Front. Microbiol. 2018, 9, 775. [Google Scholar] [CrossRef] [PubMed]
- Merabishvili, M.; Pirnay, J.P.; Verbeken, G.; Chanishvili, N.; Tediashvili, M.; Lashkhi, N.; Glonti, T.; Krylov, V.; Mast, J.; Van Parys, L.; et al. Quality-controlled small-scale production of a well-defined bacteriophage cocktail for use in human clinical trials. PLoS ONE 2009, 4, e4944. [Google Scholar] [CrossRef]
- Al-Wrafy, F.; Brzozowska, E.; Górska, S.; Gamian, A. Pathogenic factors of Pseudomonas aeruginosa—the role of biofilm in pathogenicity and as a target for phage therapy. Postepy Hig. Med. Dosw. 2017, 71, 78–91. [Google Scholar] [CrossRef] [PubMed]
- Sacher, J.C.; Zheng, J.; McCallin, S. Sourcing phages for compassionate use. Microbiol. Aust. 2019, 40, 24–27. [Google Scholar] [CrossRef]
- Corporation, A.B. AmpliPhi to Collaborate with Western Sydney Local Health District and Westmead Institute for Medical Research on Expanded Access for Investigational Bacteriophage Therapeutics AB-SA01 and AB-PA01; Business Wire: San Diego, CA, USA, 2018; Available online: https://investor.ampliphibio.com/press-release/featured/ampliphi-collaborate-western-sydney-local-health-district-and-westmead (accessed on 15 September 2021).
- Khawaldeh, A.; Morales, S.; Dillon, B.; Alavidze, Z.; Ginn, A.N.; Thomas, L.; Chapman, S.J.; Dublanchet, A.; Smithyman, A.; Iredell, J.R. Bacteriophage therapy for refractory Pseudomonas aeruginosa urinary tract infection. J. Med. Microbiol. 2011, 60, 169–1700. [Google Scholar] [CrossRef]
- Donovan, P. Access to unregistered drugs in Australia. Aust. Prescr. 2017, 40, 194–196. [Google Scholar] [CrossRef] [PubMed]
- Moore, M.P.; Lamont, I.L.; Williams, D.; Paterson, S.; Kukavica-Ibrulj, I.; Tucker, N.P.; Kenna, D.; Turton, J.F.; Jeukens, J.; Freschi, L.; et al. Transmission, adaptation and geographical spread of the Pseudomonas aeruginosa Liverpool epidemic strain. Microb. Genom. 2021, 7, 000511. [Google Scholar] [CrossRef]
- Wiehlmann, L.; Wagner, G.; Cramer, N.; Siebert, B.; Gudowius, P.; Morales, G.; Köhler, T.; van Delden, C.; Weinel, C.; Slickers, P.; et al. Population structure of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2007, 104, 8101–8106. [Google Scholar] [CrossRef] [PubMed]
- LaBauve, A.E.; Wargo, M.J. Growth and Laboratory Maintenance of Pseudomonas aeruginosa. Curr. Protoc. Microbiol. 2012, 25, 6E.1.1–6E.1.8. [Google Scholar] [CrossRef] [PubMed]
- Van Twest, R.; Kropinski, A.M. Bacteriophage enrichment from water and soil. Methods Mol. Biol. 2009, 501, 15–21. [Google Scholar]
- Kropinski, A.M.; Mazzocco, A.; Waddell, T.E.; Lingohr, E.; Johnson, R.P. Enumeration of bacteriophages by double agar overlay plaque assay. Methods Mol. Biol. 2009, 501, 69–76. [Google Scholar]
- Baer, A.; Kehn-Hall, K. Viral Concentration Determination Through Plaque Assays: Using Traditional and Novel Overlay Systems. J. Vis. Exp. 2014, 93, e52065. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Kutter, E. Phage host range and efficiency of plating. Methods Mol. Biol. 2009, 501, 141–149. [Google Scholar]
- Verma, V.; Harjai, K.; Chhibber, S. Characterization of a T7-like lytic bacteriophage of Klebsiella pneumoniae B5055: A potential therapeutic agent. Curr. Microbiol. 2009, 59, 274–281. [Google Scholar] [CrossRef]
- Gallet, R.; Shao, Y.; Wang, I.N. High adsorption rate is detrimental to bacteriophage fitness in a biofilm-like environment. BMC Evol. Biol. 2009, 9, 241. [Google Scholar] [CrossRef]
- Kropinski, A.M. Measurement of the Rate of Attachment of Bacteriophage to Cells. In Bacteriophages: Methods and Protocols; Clokie, M.R.J., Kropinski, A.M., Eds.; Humana Press: Totowa, NJ, USA, 2009; Volume 1, pp. 151–155. [Google Scholar]
- Kropinski, A.M. Practical Advice on the One-Step Growth Curve. In Bacteriophages: Methods and Protocols; Clokie, M.R.J., Kropinski, A.M., Lavigne, R., Eds.; Springer: New York, NY, USA, 2018; Volume 3, pp. 41–47. [Google Scholar]
- Chen, L.; Yuan, S.; Liu, Q.; Mai, G.; Yang, J.; Deng, D.; Zhang, B.; Liu, C.; Ma, Y. In Vitro Design and Evaluation of Phage Cocktails Against Aeromonas salmonicida. Front. Microbiol. 2018, 9, 1476. [Google Scholar] [CrossRef]
- Andrews, S. FastQC, version 0.11.9; A Quality Control Tool for High Throughput Sequence Data; 2010. Available online: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 14 December 2021).
- Bolger, A.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [PubMed]
- Roux, S.; Enault, F.; Hurwitz, B.L.; Sullivan, M.B. VirSorter: Mining viral signal from microbial genomic data. PeerJ 2015, 3, e985. [Google Scholar] [CrossRef] [PubMed]
- Solovyev, V.V.; Salamov, A.A. INFOGENE: A database of known gene structures and predicted genes and proteins in sequences of genome sequencing projects. Nucleic Acids Res. 1999, 27, 248–250. [Google Scholar] [CrossRef] [PubMed]
- Besemer, J.; Lomsadze, A.; Borodovsky, M. GeneMarkS: A self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 2001, 29, 2607–2618. [Google Scholar] [CrossRef] [PubMed]
- Brettin, T.; Davis, J.J.; Disz, T.; Edwards, R.A.; Gerdes, S.; Olsen, G.J.; Olson, R.; Overbeek, R.; Parrello, B.; Pusch, G.D.; et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015, 5, 8365. [Google Scholar] [CrossRef] [PubMed]
- Hyatt, D.; Chen, G.-L.; LoCascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [PubMed]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Biomatters Ltd. Geneious, version 6.0.5. 2012.
- Schattner, P.; Brooks, A.N.; Lowe, T.M. The tRNAscan-SE, snoscan and snoGPS web servers for the detection of tRNAs and snoRNAs. Nucleic Acids Res. 2005, 33, W686–W689. [Google Scholar] [CrossRef]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Göker, M. VICTOR: Genome-based phylogeny and classification of prokaryotic viruses. Bioinformatics 2017, 33, 3396–3404. [Google Scholar] [CrossRef]
- Rambaut, A. FigTree, version 1.4.4; A graphical viewer of phylogenetic trees and a program for producing publication-ready figures; Ashworth Laboratories, Institute of Evolutionary Biology, University of Edinburgh: Edinburgh, UK, 2018. [Google Scholar]
- Chevenet, F.; Brun, C.; Bañuls, A.-L.; Jacq, B.; Christen, R. TreeDyn: Towards dynamic graphics and annotations for analyses of trees. BMC Bioinform. 2006, 7, 439. [Google Scholar] [CrossRef]
- Carver, T.; Harris, S.R.; Berriman, M.; Parkhill, J.; McQuillan, J.A. Artemis: An integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 2012, 28, 464–469. [Google Scholar] [CrossRef] [PubMed]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef]
- Angelis, A.; Kanavos, P.; López-Bastida, J.; Linertová, R.; Nicod, E.; Serrano-Aguilar, P.; Network, B.-R.R. Social and economic costs and health-related quality of life in non-institutionalised patients with cystic fibrosis in the United Kingdom. BMC Health Serv. Res. 2015, 15, 428. [Google Scholar] [CrossRef]
- Chevreul, K.; Michel, M.; Brigham, K.B.; López-Bastida, J.; Linertová, R.; Oliva-Moreno, J.; Serrano-Aguilar, P.; Posada-de-la-Paz, M.; Taruscio, D.; Schieppati, A.; et al. Social/economic costs and health-related quality of life in patients with cystic fibrosis in Europe. Eur. J. Health Econ. 2016, 17, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Czaplewski, L.; Bax, R.; Clokie, M.; Dawson, M.; Fairhead, H.; Fischetti, V.A.; Foster, S.; Gilmore, B.F.; Hancock, R.E.; Harper, D.; et al. Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect. Dis. 2016, 16, 239–251. [Google Scholar] [CrossRef]
- Furfaro, L.L.; Payne, M.S.; Chang, B.J. Bacteriophage Therapy: Clinical Trials and Regulatory Hurdles. Front. Cell. Infect. Microbiol. 2018, 8, 376. [Google Scholar] [CrossRef] [PubMed]
- Chegini, Z.; Khoshbayan, A.; Taati Moghadam, M.; Farahani, I.; Jazireian, P.; Shariati, A. Bacteriophage therapy against Pseudomonas aeruginosa biofilms: A review. Ann. Clin. Microbiol. Antimicrob. 2020, 19, 45. [Google Scholar] [CrossRef]
- Brives, C.; Pourraz, J. Phage therapy as a potential solution in the fight against AMR: Obstacles and possible futures. Palgrave Commun. 2020, 6, 100. [Google Scholar] [CrossRef]
- Hutchison, J.S.; Ward, R.E.; Lacroix, J.; Hébert, P.C.; Barnes, M.A.; Bohn, D.J.; Dirks, P.B.; Doucette, S.; Fergusson, D.; Gottesman, R.; et al. Hypothermia Therapy after Traumatic Brain Injury in Children. N. Engl. J. Med. 2008, 358, 2447–2456. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, S.; Joo, N.S.; Reitz, B.; Wine, J.J.; Verkman, A.S. Submucosal gland secretions in airways from cystic fibrosis patients have normal [Na(+)] and pH but elevated viscosity. Proc. Natl. Acad. Sci. USA 2001, 98, 8119–8123. [Google Scholar] [CrossRef] [PubMed]
- Kellum, J.A. Determinants of blood pH in health and disease. Crit. Care 2000, 4, 6. [Google Scholar] [CrossRef][Green Version]
- Cummings, N.A.; Nordby, G.L. Measurement of synovial fluid pH in normal and arthritic knees. Arthritis Rheum. 1966, 9, 47–56. [Google Scholar] [CrossRef]
- Khan Mirzaei, M.; Nilsson, A.S. Isolation of Phages for Phage Therapy: A Comparison of Spot Tests and Efficiency of Plating Analyses for Determination of Host Range and Efficacy. PLoS ONE 2015, 10, e0118557. [Google Scholar] [CrossRef]
- Hyman, P. Phages for Phage Therapy: Isolation, Characterization, and Host Range Breadth. Pharmaceuticals 2019, 12, 35. [Google Scholar] [CrossRef]
- Anderson, G.G. Pseudomonas Aeruginosa Biofilm Formation in the CF Lung and its Implications for Therapy. In Cystic Fibrosis-Renewed Hopes Through Research; Sriramulu, D., Ed.; InTech: London, UK, 2012; pp. 153–180. [Google Scholar]
- Hraiech, S.; Brégeon, F.; Rolain, J.-M. Bacteriophage-based therapy in cystic fibrosis-associated Pseudomonas aeruginosa infections: Rationale and current status. Drug Des. Devel. Ther. 2015, 9, 3653–3663. [Google Scholar] [PubMed]
- Merabishvili, M.; Pirnay, J.-P.; De Vos, D. Guidelines to Compose an Ideal Bacteriophage Cocktail. In Bacteriophage Therapy: From Lab to Clinical Practice; Azeredo, J., Sillankorva, S., Eds.; Springer: New York, NY, USA, 2018; pp. 99–110. [Google Scholar]
- Alves, D.R.; Perez-Esteban, P.; Kot, W.; Bean, J.E.; Arnot, T.; Hansen, L.H.; Enright, M.C.; Jenkins, A.T.A. A novel bacteriophage cocktail reduces and disperses Pseudomonas aeruginosa biofilms under static and flow conditions. Microb. Biotechnol. 2016, 9, 61–74. [Google Scholar] [CrossRef]
- Dion, M.B.; Oechslin, F.; Moineau, S. Phage diversity, genomics and phylogeny. Nat. Rev. Microbiol. 2020, 18, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Gordillo Altamirano, F.L.; Barr, J.J. Unlocking the next generation of phage therapy: The key is in the receptors. Curr. Opin. Biotechnol. 2021, 68, 115–123. [Google Scholar] [CrossRef]
- Tabassum, R.; Shafique, M.; Khawaja, K.A.; Alvi, I.A.; Rehman, Y.; Sheik, C.S.; Abbas, Z.; Rehman, S.U. Complete genome analysis of a Siphoviridae phage TSK1 showing biofilm removal potential against Klebsiella pneumoniae. Sci. Rep. 2018, 8, 17904. [Google Scholar] [CrossRef] [PubMed]
- Loc-Carrillo, C.; Abedon, S.T. Pros and cons of phage therapy. Bacteriophage 2011, 1, 111–114. [Google Scholar] [CrossRef]
- Abedon, S.T.; García, P.; Mullany, P.; Aminov, R. Editorial: Phage Therapy: Past, Present and Future. Front. Microbiol. 2017, 8, 981. [Google Scholar] [CrossRef] [PubMed]
- Łobocka, M.; Hejnowicz, M.; Gągała, U.; Weber-Dąbrowska, B.; Węgrzyn, G.; Dadlez, M. Phage Therapy: Current Research and Applications; Borysowski, J., Miedzybrodzki, R., Górski, A., Eds.; Caister Academic Press: Norfolk, UK, 2014; p. 460. [Google Scholar]
- Bailly-Bechet, M.; Vergassola, M.; Rocha, E. Causes for the intriguing presence of tRNAs in phages. Genome Res. 2007, 17, 1486–1495. [Google Scholar] [CrossRef]
- Bishal, A.K.; Saha, S.; Sau, K. Synonymous codon usage in forty staphylococcal phages identifies the factors controlling codon usage variation and the phages suitable for phage therapy. Bioinformation 2012, 8, 1187–1194. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.-Z.; Liu, Q.P.; Fan, L.J.; Cui, X.F.; Zhou, X.P. Analysis of synonymous codon usage and evolution of begomoviruses. J. Zhejiang Univ. Sci. B 2008, 9, 667–674. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Batra, H.; Dong, J.; Chen, C.; Rao, V.B.; Tao, P. Genetic Engineering of Bacteriophages Against Infectious Diseases. Front. Microbiol. 2019, 10, 954. [Google Scholar] [CrossRef] [PubMed]
- Gibb, B.; Hyman, P.; Schneider, C.L. The Many Applications of Engineered Bacteriophages-An Overview. Pharmaceuticals 2021, 14, 634. [Google Scholar] [CrossRef] [PubMed]
- Tao, P.; Wu, X.; Rao, V. Unexpected evolutionary benefit to phages imparted by bacterial CRISPR-Cas9. Sci. Adv. 2018, 4, eaar4134. [Google Scholar] [CrossRef]
- Pawluk, A.; Davidson, A.R.; Maxwell, K.L. Anti-CRISPR: Discovery, mechanism and function. Nat. Rev. Microbiol. 2018, 16, 12–17. [Google Scholar] [CrossRef]
- Adams, M.H. Bacteriophages; InterScience: New York, NY, USA, 1959; p. 592. [Google Scholar]



| Clinical Strain | Isolation Source |
|---|---|
| P. aeruginosa AUS34 | Sputum |
| P. aeruginosa AUS260 | Sputum |
| P. aeruginosa AUS301 | Blood |
| P. aeruginosa AUS391 | Joint aspirate |
| Bacteria | ID (PubMLST) or Accession Number (ACCN) | Source |
|---|---|---|
| P. aeruginosa AUS432 | ID: 1249 | Biobank at CFS-QCH Brisbane, Australia |
| P. aeruginosa AUS247 | ID: 1065 | Biobank at CFS-QCH Brisbane, Australia |
| P. aeruginosa AUS391 | ID: 1209 | Biobank at CFS-QCH Brisbane, Australia |
| P. aeruginosa AUS34 | ID: 580 | Biobank at CFS-QCH Brisbane, Australia |
| P. aeruginosa AUS291 | ID: 1109 | Biobank at CFS-QCH Brisbane, Australia |
| P. aeruginosa AUS23 | ID: 569 | Biobank at CFS-QCH Brisbane, Australia |
| P. aeruginosa AUS455 | ID: 1272 | Biobank at CFS-QCH Brisbane, Australia |
| P. aeruginosa AUS260 | ID: 1078 | Biobank at CFS-QCH Brisbane, Australia |
| P. aeruginosa AUS301 | ID: 1119 | Biobank at CFS-QCH Brisbane, Australia |
| P. aeruginosa AUS403 | ID: 1220 | Biobank at CFS-QCH Brisbane, Australia |
| P. aeruginosa PAO1 | ACCN: NC 002516 | Lab strain (ACWEB) |
| Pseudomonas putida KT2440 (RP4 plasmid) | ACCN: AE015451 | Lab strain (ACWEB) |
| Cocktail | Phage Constituents of the Cocktails |
|---|---|
| Cocktail 1 | Pseudomonas Phage_AUS034 + Pseudomonas Phage_AUS260 |
| Cocktail 2 | Pseudomonas Phage_AUS034 + Pseudomonas Phage_AUS301 |
| Cocktail 3 | Pseudomonas Phage_AUS034 + Pseudomonas Phage_AUS391 |
| Cocktail 4 | Pseudomonas Phage_AUS260 + Pseudomonas Phage_AUS301 |
| Cocktail 5 | Pseudomonas Phage_AUS260 + Pseudomonas Phage_AUS391 |
| Cocktail 6 | Pseudomonas Phage_AUS301 + Pseudomonas Phage_AUS391 |
| Cocktail 7 | Pseudomonas Phage_AUS034 + Pseudomonas Phage_AUS260 + Pseudomonas Phage_AUS301 |
| Cocktail 8 | Pseudomonas Phage_AUS034 + Pseudomonas Phage_AUS260 + Pseudomonas Phage_AUS391 |
| Cocktail 9 | Pseudomonas Phage_AUS260 + Pseudomonas Phage_AUS301 + Pseudomonas Phage_AUS391 |
| Cocktail 10 | Pseudomonas Phage_AUS034 + Pseudomonas Phage_AUS301 + Pseudomonas Phage_AUS391 |
| Cocktail 11 | Pseudomonas Phage_AUS034 + Pseudomonas Phage_AUS260 + Pseudomonas Phage_AUS301 + Pseudomonas Phage_AUS391 |
| Isolated Phage | Bacteria | Plaque Diameter (mm) | Phage Size (nm) | |
|---|---|---|---|---|
| Head | Tail | |||
| Pseudomonas Phage_AUS034 | P. aeruginosa AUS34 | 3 | 56 ± 0.5 | 45 ± 2 |
| Pseudomonas Phage_AUS260 | P. aeruginosa AUS260 | 1.5 | 58 ± 0.5 | 190 ± 1 |
| Pseudomonas Phage_AUS301 | P. aeruginosa AUS301 | 0.6 | 73 ± 1 | 70 ± 2 (Contracted) 132 ± 2 (Non-contracted) |
| Pseudomonas Phage_AUS391 | P. aeruginosa AUS391 | 0.6 | 75 ± 2 | 81 ± 2 (Contracted) 121 ± 3 (Non-contracted) |
| Isolated Phage | Latent Period (Minutes) | Burst Size (PFU/Cell) |
|---|---|---|
| Pseudomonas Phage_AUS034 | 50 | 143 ± 16 |
| Pseudomonas Phage_AUS260 | 60 | 86 ± 7 |
| Pseudomonas Phage_AUS301 | 80 | 121 ± 11 |
| Pseudomonas Phage_AUS391 | 90 | 51 ± 4 |
| Bacteria | Lytic Activity of Phage | |||
|---|---|---|---|---|
| Pseudomonas Phage_AUS034 | Pseudomonas Phage_AUS260 | Pseudomonas Phage_AUS301 | Pseudomonas Phage_AUS391 | |
| P. aeruginosa AUS432 | + + | − − | + − | − − |
| P. aeruginosa AUS247 | − − | − − | − − | − − |
| P. aeruginosa AUS391 | − − | + + | − − | + + |
| P. aeruginosa AUS034 | + + | − − | + + | + + |
| P. aeruginosa AUS291 | − − | + − | + − | + + |
| P. aeruginosa AUS023 | − − | − − | − − | − − |
| P. aeruginosa AUS455 | + − | + − | + − | + + |
| P. aeruginosa AUS260 | + + | + + | + + | + − |
| P. aeruginosa AUS301 | + + | + − | + + | + − |
| P. aeruginosa AUS403 | − − | − − | − − | − − |
| P. aeruginosa PAO1 | + + | − − | + + | + + |
| Pseudomonas putida KT2440 (RP4 plasmid) | − − | − − | − − | − − |
| Phage | Adsorption (%) | k (mL/min) |
|---|---|---|
| Pseudomonas Phage_AUS034 | 81 | 1.26 × 10−8 |
| Pseudomonas Phage_AUS260 | 100 | 3.36 × 10−9 |
| Pseudomonas Phage_AUS301 | 97 | 4.76 × 10−9 |
| Pseudomonas Phage_AUS391 | 100 | 4.33 × 10−9 |
| Bacteriophage Name | Genome Size (bp) | GC Content (%) | Number of ORFs | Number of CDSs | CDSs with Putative Functions | Number of tRNAs |
|---|---|---|---|---|---|---|
| Pseudomonas Phage_AUS034 | 44,167 | 52.2 | 75 | 64 | 19 | 4 |
| Pseudomonas Phage_AUS260 | 42,020 | 54.7 | 78 | 54 | 28 | 1 |
| Pseudomonas Phage_AUS301 | 49,455 | 55.8 | 98 | 62 | 27 | 0 |
| Pseudomonas Phage_AUS391 | 65,515 | 55.5 | 125 | 89 | 29 | 0 |
| Isolated Phage | Most Similar to | Identity (%) | Query Cover (%) |
|---|---|---|---|
| Pseudomonas Phage_AUS034 | Pseudomonas phage oldone | 97.10 | 98 |
| Pseudomonas Phage_AUS260 | Xanthomonas phage Samson | 95.45 | 99 |
| Pseudomonas Phage_AUS301 | Pseudomonas phage S50 | 98.75 | 100 |
| Pseudomonas Phage_AUS391 | Pseudomonas virus Pa193 | 97.59 | 98 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Namonyo, S.; Carvalho, G.; Guo, J.; Weynberg, K.D. Novel Bacteriophages Show Activity against Selected Australian Clinical Strains of Pseudomonas aeruginosa. Microorganisms 2022, 10, 210. https://doi.org/10.3390/microorganisms10020210
Namonyo S, Carvalho G, Guo J, Weynberg KD. Novel Bacteriophages Show Activity against Selected Australian Clinical Strains of Pseudomonas aeruginosa. Microorganisms. 2022; 10(2):210. https://doi.org/10.3390/microorganisms10020210
Chicago/Turabian StyleNamonyo, Samuel, Gilda Carvalho, Jianhua Guo, and Karen D. Weynberg. 2022. "Novel Bacteriophages Show Activity against Selected Australian Clinical Strains of Pseudomonas aeruginosa" Microorganisms 10, no. 2: 210. https://doi.org/10.3390/microorganisms10020210
APA StyleNamonyo, S., Carvalho, G., Guo, J., & Weynberg, K. D. (2022). Novel Bacteriophages Show Activity against Selected Australian Clinical Strains of Pseudomonas aeruginosa. Microorganisms, 10(2), 210. https://doi.org/10.3390/microorganisms10020210







