Snf2 Proteins Are Required to Generate Gamete Pronuclei in Tetrahymena thermophila
Abstract
1. Introduction
2. Materials and Methods
2.1. Culture Methods and the Initiation of Mating
2.2. Database Searching and Motif Scanning
2.3. Phylogenetic Analysis
2.4. Construction of C-Terminal EGFP-Tagging and N-Terminal mCherry-Tagging Vectors
2.5. Fluorescence Microscopy of Living Cells
2.6. Indirect Immunofluorescence
2.7. Gene Knockout Constructions for the Snf2 Family Proteins Localizing to the Selected hMIC
2.8. Acetic Orcein Stain
3. Results
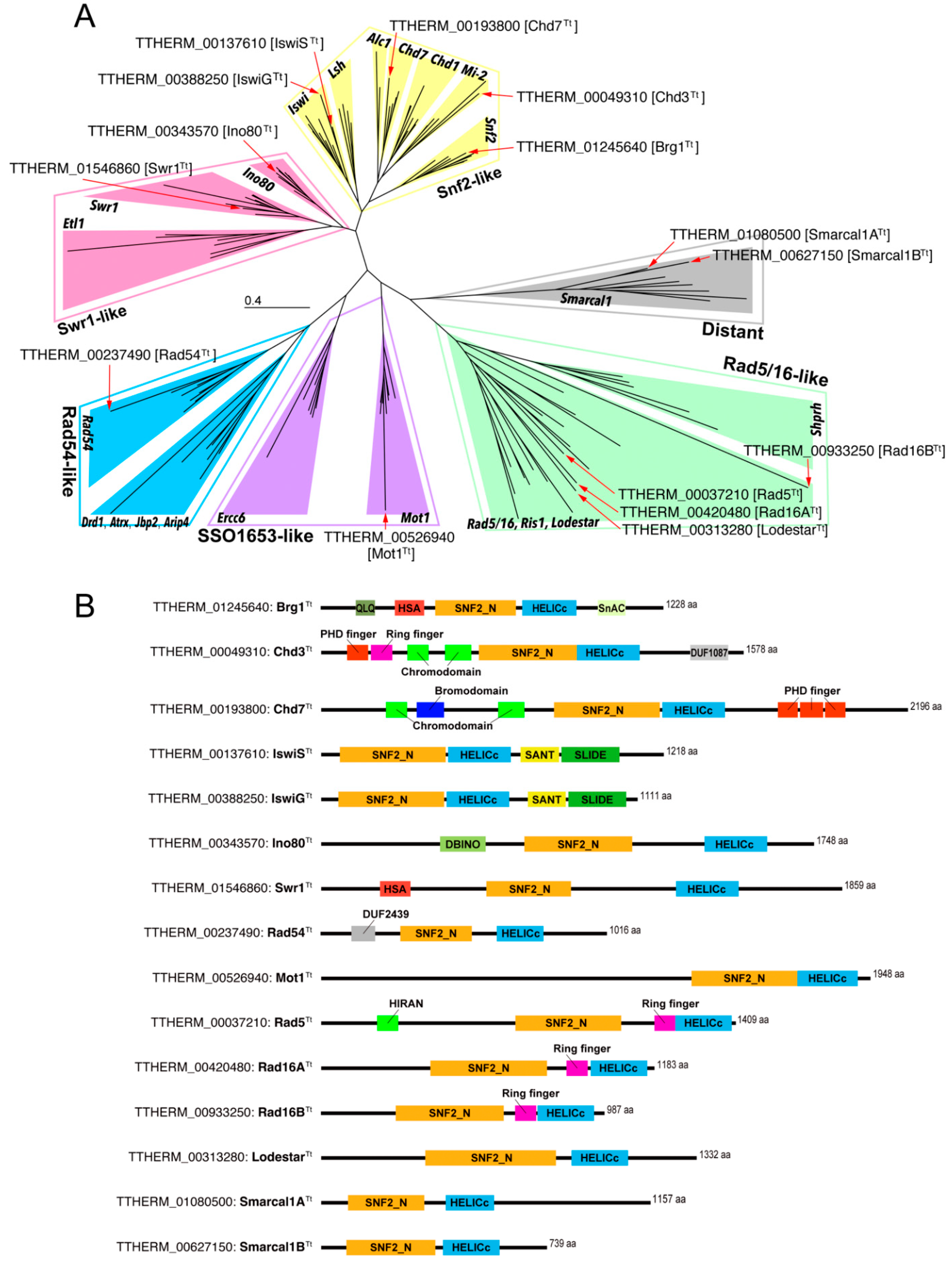
3.1. Tetrahymena Snf2 Family Proteins
3.2. Snf2 Family Proteins Appearing in the Selected hMIC
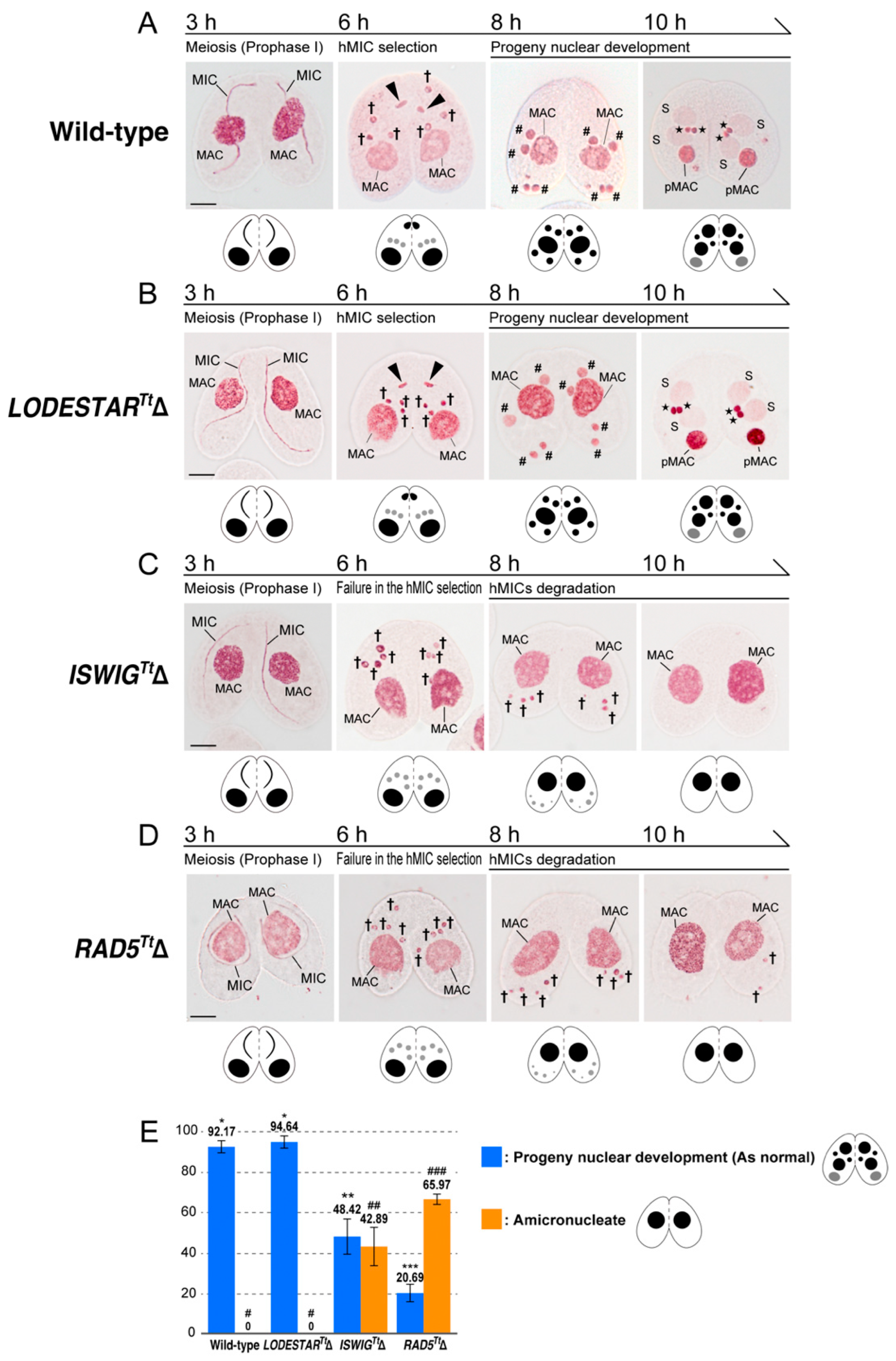
3.3. Loss of ISWIGTt and RAD5Tt Causes a Defect in hMIC Selection
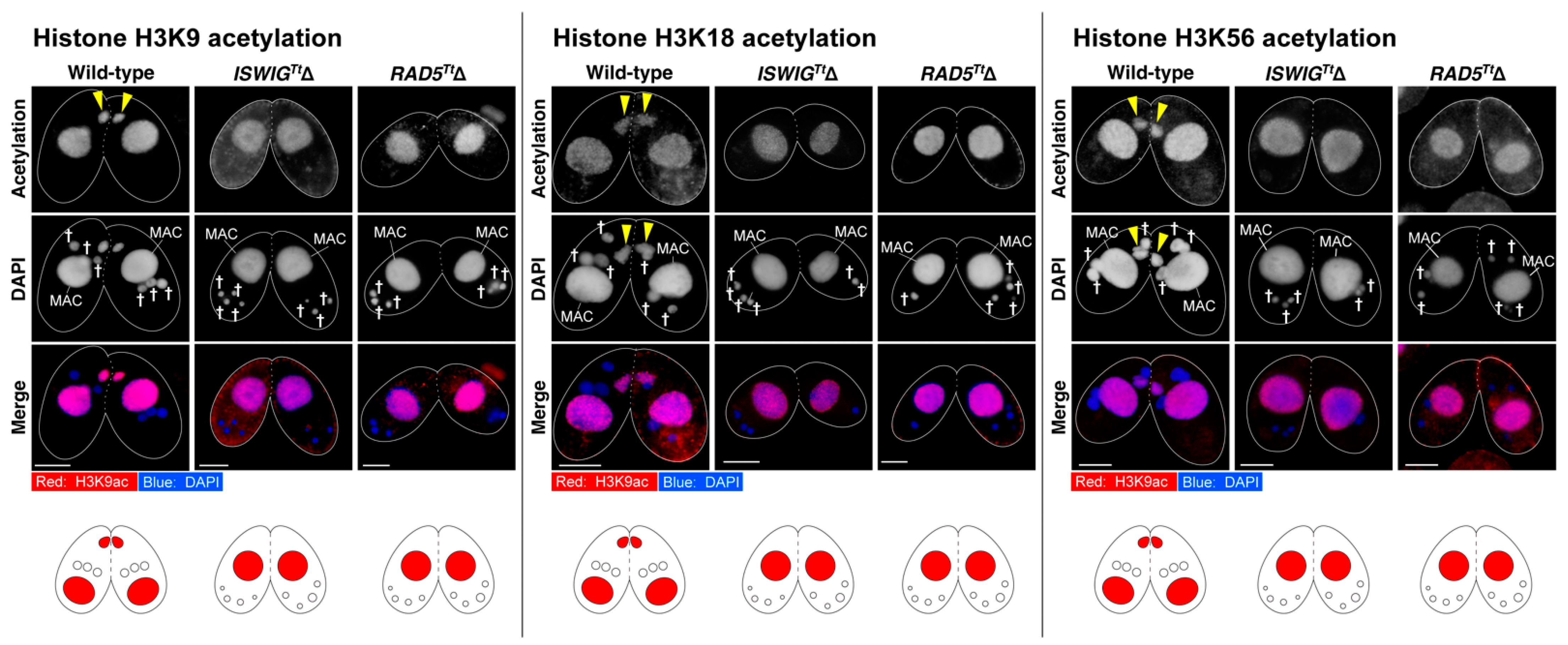
3.4. Depletion of IswiGTt or Rad5Tt Does Not Affect PM-DSB Formation but Does Affect DNA Repair and Euchromatin Formation Which Concomitantly Occur in the Selected hMIC
4. Discussion
4.1. Snf2 Family Proteins Localizing to the MAC
4.2. Rad54
4.3. Lodestar
4.4. Possible Functions of IswiGTt and Rad5Tt for the hMIC Selection
4.5. Tetrahymena Monitors the Appearance of the Selected hMIC in Which DNA Repair and Euchromatin Formation Are Completed
4.6. The Involvement of the Iswi Subfamily in Pronuclear Generation with Euchromatin Formation Is Evolutionarily Conserved
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Iwamoto, M.; Koujin, T.; Osakada, H.; Mori, C.; Kojidani, T.; Matsuda, A.; Asakawa, H.; Hiraoka, Y.; Haraguchi, T. Biased assembly of the nuclear pore complex is required for somatic and germline nuclear differentiation in Tetrahymena. J. Cell Sci. 2015, 128, 1812–1823. [Google Scholar] [CrossRef] [PubMed]
- Orias, E.; Cervantes, M.D.; Hamilton, E.P. Tetrahymena thermophila, a unicellular eukaryote with separate germline and somatic genomes. Res. Microbiol. 2011, 162, 578–586. [Google Scholar] [CrossRef] [PubMed]
- Loidl, J. Tetrahymena meiosis: Simple yet ingenious. PLoS Genet. 2021, 17, e1009627. [Google Scholar] [CrossRef]
- Cole, E.; Sugai, T. Developmental progression of Tetrahymena through the cell cycle and conjugation. Methods Cell Biol. 2012, 109, 177–236. [Google Scholar]
- Akematsu, T.; Fukuda, Y.; Garg, J.; Fillingham, J.S.; Pearlman, R.E.; Loidl, J. Post-meiotic DNA double-strand breaks occur in Tetrahymena, and require Topoisomerase II and Spo11. Elife 2017, 6, e26176. [Google Scholar] [CrossRef]
- Chowdhury, D.; Keogh, M.C.; Ishii, H.; Peterson, C.L.; Buratowski, S.; Lieberman, J. γ-H2AX Dephosphorylation by Protein Phosphatase 2A Facilitates DNA Double-Strand Break Repair. Mol. Cell 2005, 20, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Bo, T.; Kang, Y.; Liu, Y.; Xu, J.; Wang, W. Atg5 Regulates Selective Autophagy of the Parental Macronucleus during Tetrahymena Sexual Reproduction. Cells 2021, 10, 3071. [Google Scholar] [CrossRef]
- Liu, M.L.; Yao, M.C. Role of ATG8 and autophagy in programmed nuclear degradation in Tetrahymena thermophila. Eukaryot. Cell 2012, 11, 494–506. [Google Scholar] [CrossRef]
- Cole, E.S.; Giddings, T.H., Jr.; Ozzello, C.; Winey, M.; O’Toole, E.; Orias, J.; Hamilton, E.; Guerrier, S.; Ballard, A.; Aronstein, T. Membrane dynamics at the nuclear exchange junction during early mating (one to four hours) in the ciliate Tetrahymena thermophila. Eukaryot. Cell 2015, 14, 116–127. [Google Scholar] [CrossRef]
- Akematsu, T.; Sánchez-Fernández, R.; Kosta, F.; Holzer, E.; Loidl, J. The Transmembrane Protein Semi1 Positions Gamete Nuclei for Reciprocal Fertilization in Tetrahymena. iScience 2020, 23, 100749. [Google Scholar] [CrossRef]
- Chen, C.C.; Carson, J.J.; Feser, J.; Tamburini, B.; Zabaronick, S.; Linger, J.; Tyler, J.K. Acetylated Lysine 56 on Histone H3 Drives Chromatin Assembly after Repair and Signals for the Completion of Repair. Cell 2008, 134, 231–243. [Google Scholar] [CrossRef] [PubMed]
- Tie, F.; Banerjee, R.; Stratton, C.A.; Prasad-Sinha, J.; Stepanik, V.; Zlobin, A.; Diaz, M.O.; Scacheri, P.C.; Harte, P.J. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 2009, 136, 3131–3141. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zang, C.; Rosenfeld, J.A.; Schones, D.E.; Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.Y.; Peng, W.; Zhang, M.Q.; et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008, 40, 897–903. [Google Scholar] [CrossRef] [PubMed]
- Catez, F.; Yang, H.; Tracey, K.J.; Reeves, R.; Misteli, T.; Bustin, M. Network of dynamic interactions between histone H1 and high-mobility-group proteins in chromatin. Mol. Cell. Biol. 2004, 24, 4321–4328. [Google Scholar] [CrossRef]
- Agresti, A.; Bianchi, M.E. HMGB proteins and gene expression. Curr. Opin. Genet. Dev. 2003, 13, 170–178. [Google Scholar] [CrossRef]
- Xu, J.; Tian, H.; Liu, X.; Wang, W.; Liang, A. Localization and functional analysis of HmgB3p, a novel protein containing high-mobility-group-box domain from Tetrahymena thermophila. Gene 2013, 526, 87–95. [Google Scholar] [CrossRef]
- Lian, Y.; Hao, H.; Xu, J.; Bo, T.; Liang, A.; Wang, W. The histone chaperone Nrp1 is required for chromatin stability and nuclear division in Tetrahymena thermophila. Epigenet. Chromatin 2021, 14, 34. [Google Scholar] [CrossRef]
- Garg, J.; Lambert, J.P.; Karsou, A.; Marquez, S.; Nabeel-Shah, S.; Bertucci, V.; Retnasothie, D.V.; Radovani, E.; Pawson, T.; Gingras, A.C.; et al. Conserved Asf1-importin β physical interaction in growth and sexual development in the ciliate Tetrahymena thermophila. J. Proteom. 2013, 94, 311–326. [Google Scholar] [CrossRef]
- Park, Y.J.; Luger, K. Histone chaperones in nucleosome eviction and histone exchange. Curr. Opin. Struct. Biol. 2008, 18, 282–289. [Google Scholar] [CrossRef]
- Hota, S.K.; Bruneau, B.G. ATP-dependent chromatin remodeling during mammalian development. Development 2016, 143, 2882–2897. [Google Scholar] [CrossRef]
- Flaus, A.; Owen-Hughes, T. Mechanisms for ATP-dependent chromatin remodelling: The means to the end. FEBS J. 2011, 278, 3579–3595. [Google Scholar] [CrossRef] [PubMed]
- Flaus, A.; Martin, D.M.A.; Barton, G.J.; Owen-Hughes, T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006, 34, 2887–2905. [Google Scholar] [CrossRef] [PubMed]
- DeRango-Adem, E.F. Macromolecular Interactome of Tetrahymena CHD Family Chromatin Remodelers. Master’s Thesis, York University, Toronto, ON, Canada, 2018. Available online: http://hdl.handle.net/10315/34494 (accessed on 29 August 2022).
- Fillingham, J.S.; Garg, J.; Tsao, N.; Vythilingum, N.; Nishikawa, T.; Pearlman, R.E. Molecular Genetic Analysis of an SNF2/brahma-Related Gene in Tetrahymena thermophila Suggests Roles in Growth and Nuclear Development. Eukaryot. Cell 2006, 5, 1347–1359. [Google Scholar] [CrossRef]
- Saettone, A.; Garg, J.; Lambert, J.P.; Nabeel-Shah, S.; Ponce, M.; Burtch, A.; Mudalige, C.T.; Gingras, A.C.; Pearlman, R.E.; Fillingham, J. The bromodomain-containing protein Ibd1 links multiple chromatin-related protein complexes to highly expressed genes in Tetrahymena thermophila. Epigenet. Chromatin 2018, 11, 10. [Google Scholar] [CrossRef] [PubMed]
- Cassidy-Hanley, D.M. Tetrahymena in the laboratory: Strain resources, methods for culture, maintenance, and storage. Methods Cell Biol. 2012, 109, 237–276. [Google Scholar] [PubMed]
- Katoh, K.; Misawa, K.; Kuma, K.I.; Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002, 30, 3059–3066. [Google Scholar] [CrossRef]
- Tanabe, S.A. Kakusan4 and Aminosan: Two programs for comparing nonpartitioned, proportional and separate models for combined molecular phylogenetic analyses of multilocus sequence data. Mol. Ecol. Resour. 2011, 11, 914–921. [Google Scholar] [CrossRef]
- Kozlov, A.M.; Darriba, D.; Flouri, T.; Morel, B.; Stamatakis, A. RAxML-NG: A fast, scalable, and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics 2019, 35, 4453–4455. [Google Scholar] [CrossRef]
- Kataoka, K.; Schoeberl, U.E.; Mochizuki, K. Modules for C-terminal epitope tagging of Tetrahymena genes. J. Microbiol. Methods 2010, 82, 342–346. [Google Scholar] [CrossRef]
- Iwamoto, M.; Mori, C.; Kojidani, T.; Bunai, F.; Hori, T.; Fukagawa, T.; Hiraoka, Y.; Haraguchi, T. Two distinct repeat sequences of Nup98 nucleoporins characterize dual nuclei in the binucleated ciliate Tetrahymena. Curr. Biol. 2009, 19, 843–847. [Google Scholar] [CrossRef]
- Nakagawa, T.; Fujiu, K.; Cole, E.S.; Numata, O. Involvement of a 25 kDa Tetrahymena Ca2+-binding protein in pronuclear exchange. Cell Struct. Funct. 2008, 33, 151–162. [Google Scholar] [CrossRef] [PubMed]
- Merriam, E.V.; Bruns, P.J. Phenotypic assortment in Tetrahymena thermophila: Assortment kinetics of antibiotic-resistance markers, tsA, death, and the highly amplified rDNA locus. Genetics 1988, 120, 389–395. [Google Scholar] [CrossRef]
- Mochizuki, K. High efficiency transformation of Tetrahymena using a codon-optimized neomycin resistance gene. Gene 2008, 425, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Sheng, Y.; Duan, L.; Cheng, T.; Qiao, Y.; Stover, N.A.; Gao, S. The completed macronuclear genome of a model ciliate Tetrahymena thermophila and its application in genome scrambling and copy number analyses. Sci. China Life Sci. 2020, 63, 1534–1542. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Lu, X.; Zhou, Z.; Chang, Y.; Yuan, D.; Tian, M.; Zhou, Z.; Wang, L.; Fu, C.; Orias, E.; et al. Transcriptome analysis of the model protozoan, Tetrahymena thermophila, using deep RNA sequencing. PLoS ONE 2012, 7, e30630. [Google Scholar] [CrossRef]
- Coyne, R.S.; Thiagarajan, M.; Jones, K.M.; Wortman, J.R.; Tallon, L.J.; Haas, B.J.; Cassidy-Hanley, D.M.; Wiley, E.A.; Smith, J.J.; Collins, K.; et al. Refined annotation and assembly of the Tetrahymena thermophila genome sequence through EST analysis; comparative genomic hybridization; and targeted gap closure. BMC Genom. 2008, 9, 562–579. [Google Scholar] [CrossRef]
- Eisen, J.A.; Coyne, R.S.; Wu, M.; Wu, D.; Thiagarajan, M.; Wortman, J.R.; Badger, J.H.; Ren, Q.; Amedeo, P.; Jones, K.M.; et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006, 4, e286. [Google Scholar] [CrossRef]
- Hamilton, E.P.; Dear, P.H.; Rowland, T.; Saks, K.; Eisen, J.A.; Orias, E. Use of HAPPY mapping for the higher order assembly of the Tetrahymena genome. Genomics 2006, 88, 443–451. [Google Scholar] [CrossRef][Green Version]
- Stover, N.A.; Krieger, C.J.; Binkley, G.; Dong, Q.; Fisk, D.G.; Nash, R.; Sethuraman, A.; Weng, S.; Cherry, J.M. Tetrahymena Genome Database (TGD): A new genomic resource for Tetrahymena thermophila research. Nucleic Acids Res. 2006, 34, 500–503. [Google Scholar] [CrossRef]
- Pinskaya, M.; Nair, A.; Clynes, D.; Morillon, A.; Mellor, J. Nucleosome Remodeling and Transcriptional Repression Are Distinct Functions of Isw1 in Saccharomyces cerevisiae. Mol. Cell. Biol. 2009, 29, 2419–2430. [Google Scholar] [CrossRef]
- Bakshi, R.; Prakash, T.; Dash, D.; Brahmachari, V. In silico characterization of the INO80 subfamily of SWI2/SNF2 chromatin remodeling proteins. Biochem. Biophys. Res. Commun. 2004, 320, 197–204. [Google Scholar] [CrossRef] [PubMed]
- Narlikar, G.J.; Sundaramoorthy, R.; Owen-Hughes, T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell 2013, 154, 490–503. [Google Scholar] [CrossRef] [PubMed]
- Chen, S. The RING finger ATPase Rad5p of Saccharomyces cerevisiae contributes to DNA double-strand break repair in a ubiquitin-independent manner. Nucleic Acids Res. 2005, 33, 5878–5886. [Google Scholar] [CrossRef]
- Elserafy, M.; Abugable, A.A.; Atteya, R.; El-Khamisy, S. Rad5, HLTF, and SHPRH: A Fresh View of an Old Story. Trends Genet. 2018, 34, 574–577. [Google Scholar] [CrossRef]
- Shang, Y.; Song, X.; Bowen, J.; Corstanje, R.; Gao, Y.; Gaertig, J.; Gorovsky, M.A. A robust inducible-repressible promoter greatly facilitates gene knockouts, conditional expression, and overexpression of homologous and heterologous genes in Tetrahymena thermophila. Proc. Natl. Acad. Sci. USA 2002, 99, 3734–3739. [Google Scholar] [CrossRef]
- Fukuda, Y.; Akematsu, T.; Attiq, R.; Tada, C.; Nakai, Y.; Pearlman, R.E. Role of the Cytosolic Heat Shock Protein 70 Ssa5 in the Ciliate Protozoan Tetrahymena thermophila. J. Eukaryot. Microbiol. 2015, 62, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Gates, L.A.; Shi, J.; Rohira, A.D.; Feng, Q.; Zhu, B.; Bedford, M.T.; Sagum, C.A.; Jung, S.Y.; Qin, J.; Tsai, M.J.; et al. Acetylation on histone H3 lysine 9 mediates a switch from transcription initiation to elongation. J. Biol. Chem. 2017, 292, 14456–14472. [Google Scholar] [CrossRef]
- Aslam, M.; Fakher, B.; Jakada, B.H.; Cao, S.; Qin, Y. SWR1 Chromatin remodeling complex: A key transcriptional regulator in plants. Cells 2019, 8, 1621. [Google Scholar] [CrossRef]
- Tyagi, M.; Imam, N.; Verma, K.; Patel, A.K. Chromatin remodelers: We are the drivers! Nucleus 2016, 7, 388–404. [Google Scholar] [CrossRef]
- Xue, Y.; Van, C.; Pradhan, S.K.; Su, T.; Gehrke, J.; Kuryan, B.G.; Kitada, T.; Vashisht, A.; Tran, N.; Wohlschlegel, J.; et al. The Ino80 complex prevents invasion of euchromatin into silent chromatin. Genes Dev. 2015, 29, 350–355. [Google Scholar] [CrossRef][Green Version]
- Ryan, D.P.; Owen-Hughes, T. Snf2-family proteins: Chromatin remodellers for any occasion. Curr. Opin. Chem. Biol. 2011, 15, 649–656. [Google Scholar] [CrossRef] [PubMed]
- Dasgupta, A.; Darst, R.P.; Martin, K.J.; Afshari, C.A.; Auble, D.T. Mot1 activates and represses transcription by direct, ATPase-dependent mechanisms. Proc. Natl. Acad. Sci. USA 2002, 99, 2666–2671. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Liu, H.; Gill, H.W.; Yu, Y.; Waters, R.; Reed, S.H. Saccharomyces cerevisiae Rad16 mediates ultraviolet-dependent histone H3 acetylation required for efficient global genome nucleotide-excision repair. EMBO Rep. 2008, 9, 97–102. [Google Scholar] [CrossRef]
- Wolner, B.; Peterson, C.L. ATP-dependent and ATP-independent Roles for the Rad54 Chromatin Remodeling Enzyme during Recombinational Repair of a DNA Double Strand Break. J. Biol. Chem. 2005, 280, 10855–10860. [Google Scholar] [CrossRef]
- Smirnova, M.; Van Komen, S.; Sung, P.; Klein, H.L. Effects of tumor-associated mutations on Rad54 functions. J. Biol. Chem. 2004, 279, 24081–24088. [Google Scholar] [CrossRef] [PubMed]
- Mazin, A.V.; Mazina, O.M.; Bugreev, D.V.; Rossi, M.J. Rad54, the motor of homologous recombination. DNA Repair 2010, 9, 286–302. [Google Scholar] [CrossRef]
- Szalontai, T.; Gaspar, I.; Belecz, I.; Kerekes, I.; Erdelyi, M.; Boros, I.; Szabad, J. Horka D, a Chromosome Instability-Causing Mutation in Drosophila, Is a Dominant-Negative Allele of lodestar. Genetics 2009, 181, 367–377. [Google Scholar] [CrossRef][Green Version]
- Girdham, C.H.; Glover, D.M. Chromosome tangling and breakage at anaphase result from mutations in lodestar, a Drosophila gene encoding a putative nucleoside triphosphate-binding protein. Genes Dev. 1991, 5, 1786–1799. [Google Scholar] [CrossRef]
- Fujiu, K.; Numata, O. Localization of Microtubules during Macronuclear Division in Tetrahymena and Possible Involvement of Macronuclear Microtubules in ‘Amitotic’ Chromatin Distribution. Cell Struct. Funct. 1999, 24, 401–404. [Google Scholar] [CrossRef][Green Version]
- Aydin, Ö.Z.; Vermeulen, W.; Lans, H. ISWI chromatin remodeling complexes in the DNA damage response. Cell Cycle 2014, 13, 3016–3025. [Google Scholar] [CrossRef]
- Unk, I.; Hajdú, I.; Blastyák, A.; Haracska, L. Role of yeast Rad5 and its human orthologs, HLTF and SHPRH in DNA damage tolerance. DNA Repair 2010, 9, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Ortiz-Bazán, M.A.; Gallo-Fernández, M.; Saugar, I.; Jiménez-Martín, A.; Vázquez, M.A.; Tercero, J.A. Rad5 plays a major role in the cellular response to DNA damage during chromosome replication. Cell Rep. 2014, 9, 460–468. [Google Scholar] [CrossRef] [PubMed]
- Lewis, J.D.; Song, Y.; de Jong, M.E.; Bagha, S.M.; Ausió, J. A walk though vertebrate and invertebrate protamines. Chromosoma 2003, 111, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Poccia, D.; Collas, P. Transforming sperm nuclei into male pronuclei in vivo and in vitro. Curr. Top. Dev. Biol. 1996, 34, 25–88. [Google Scholar] [CrossRef] [PubMed]
- Doyen, C.M.; Chalkley, G.E.; Voets, O.; Bezstarosti, K.; Demmers, J.A.; Moshkin, Y.M.; Verrijzer, C.P. A Testis-Specific Chaperone and the Chromatin Remodeler ISWI Mediate Repackaging of the Paternal Genome. Cell Rep. 2015, 13, 1310–1318. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fukuda, Y.; Akematsu, T.; Bando, H.; Kato, K. Snf2 Proteins Are Required to Generate Gamete Pronuclei in Tetrahymena thermophila. Microorganisms 2022, 10, 2426. https://doi.org/10.3390/microorganisms10122426
Fukuda Y, Akematsu T, Bando H, Kato K. Snf2 Proteins Are Required to Generate Gamete Pronuclei in Tetrahymena thermophila. Microorganisms. 2022; 10(12):2426. https://doi.org/10.3390/microorganisms10122426
Chicago/Turabian StyleFukuda, Yasuhiro, Takahiko Akematsu, Hironori Bando, and Kentaro Kato. 2022. "Snf2 Proteins Are Required to Generate Gamete Pronuclei in Tetrahymena thermophila" Microorganisms 10, no. 12: 2426. https://doi.org/10.3390/microorganisms10122426
APA StyleFukuda, Y., Akematsu, T., Bando, H., & Kato, K. (2022). Snf2 Proteins Are Required to Generate Gamete Pronuclei in Tetrahymena thermophila. Microorganisms, 10(12), 2426. https://doi.org/10.3390/microorganisms10122426





