Trophic State Drives the Diversity of Protists in a Tropical River (New River, Belize)
Abstract
1. Introduction
2. Materials and Methods
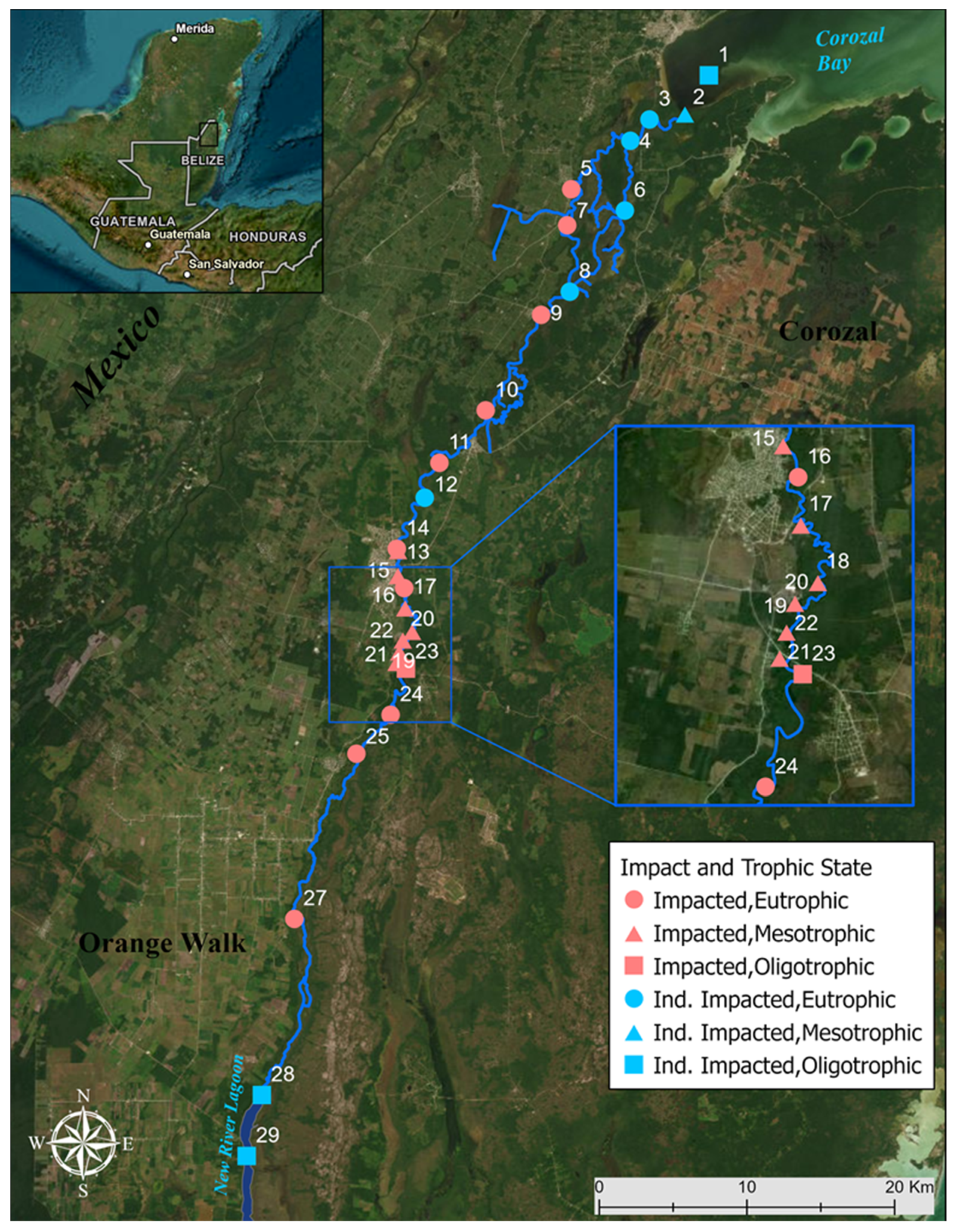
2.1. Study Area
2.2. Sampling Methods
2.3. Laboratory Methods
2.4. Trophic State Index
2.5. Anthropogenic Impact Classification
2.6. DNA Extraction and Amplification
2.7. Library Preparation
2.8. Sequence and Phylogenetic Analysis
2.9. Statistical Analysis
2.10. Protist Functional Diversity Analysis
3. Results
3.1. Water Quality Parameters
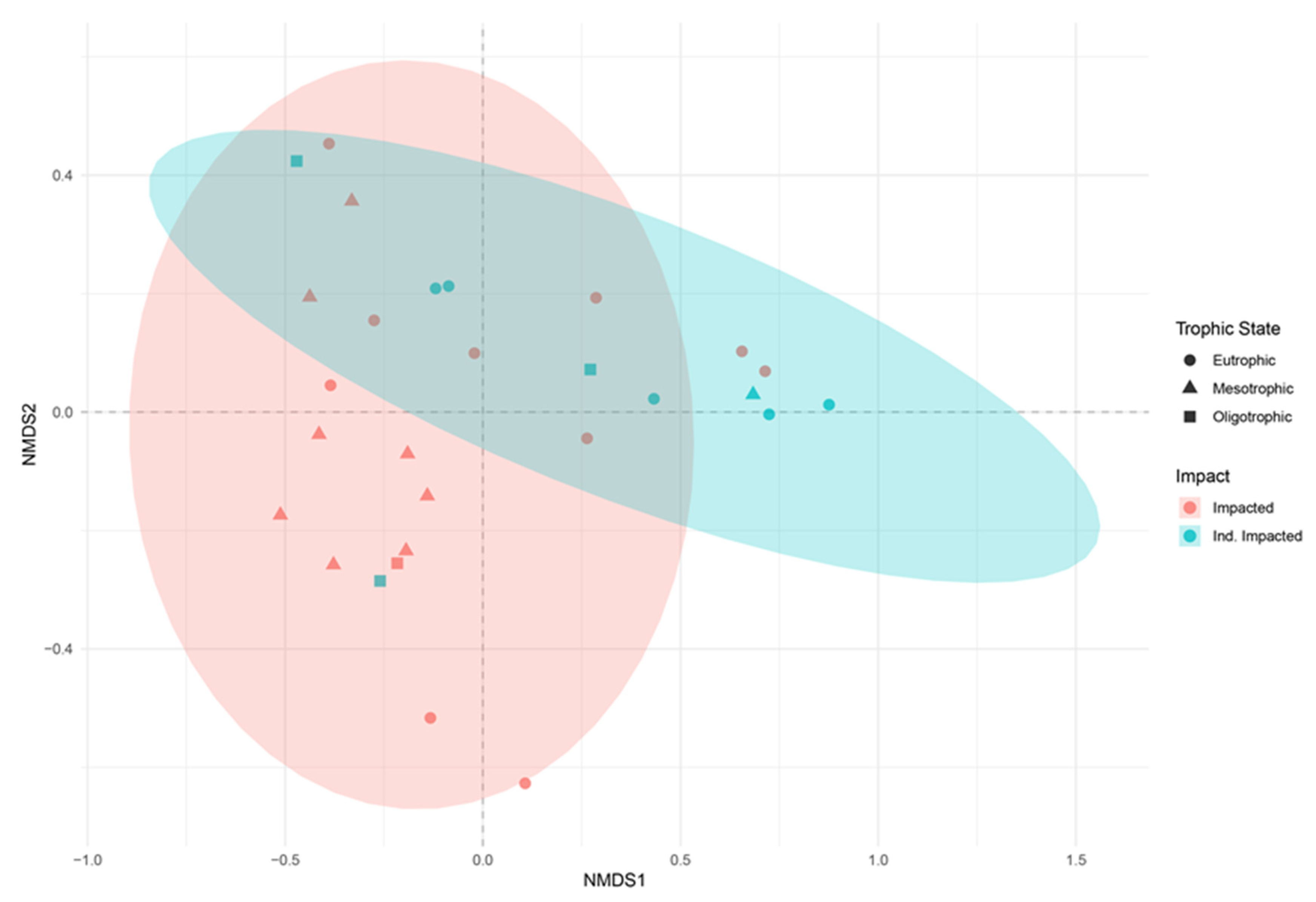
3.2. Trophic State Indices and Anthropogenic Impact
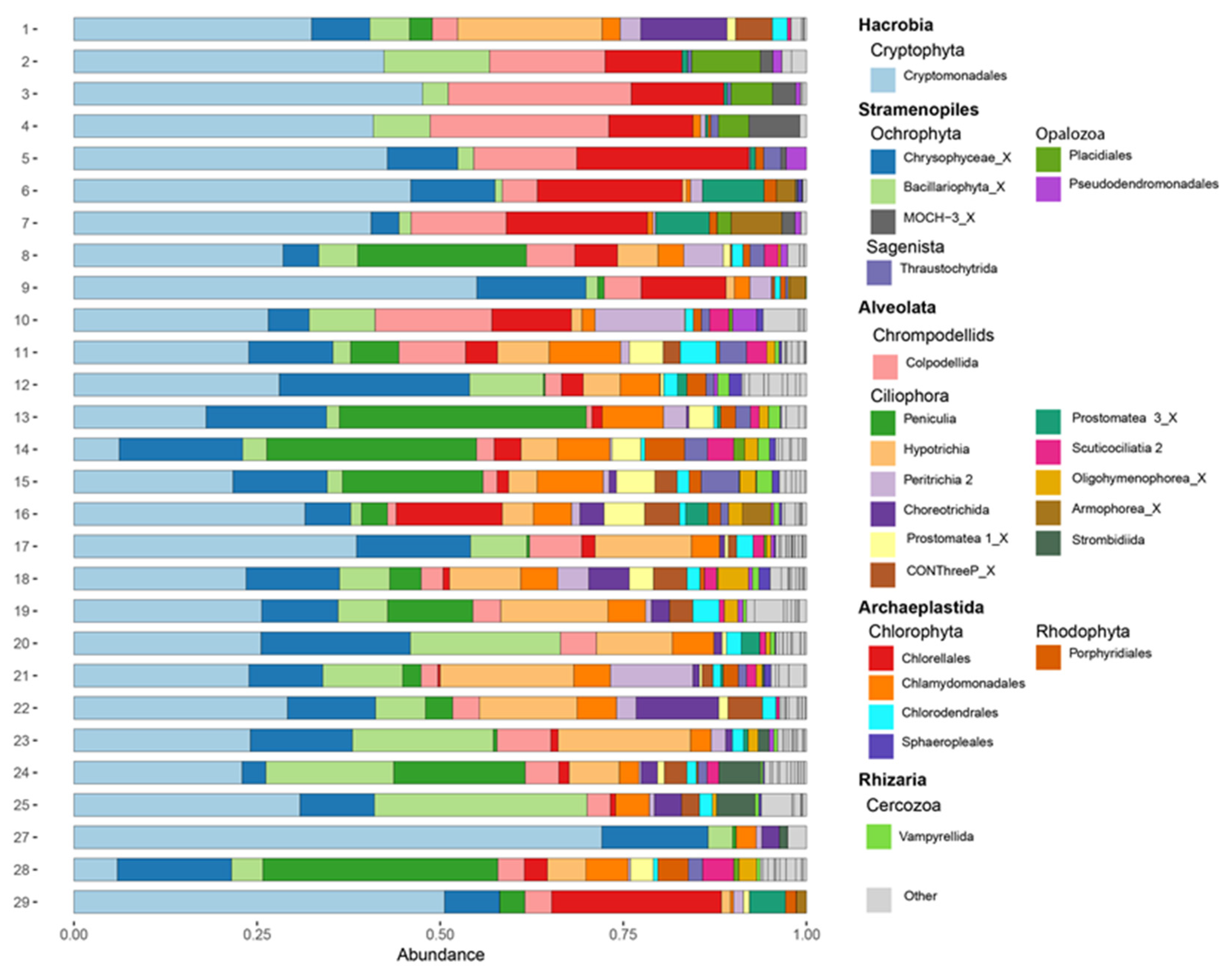
3.3. Protist Community Composition
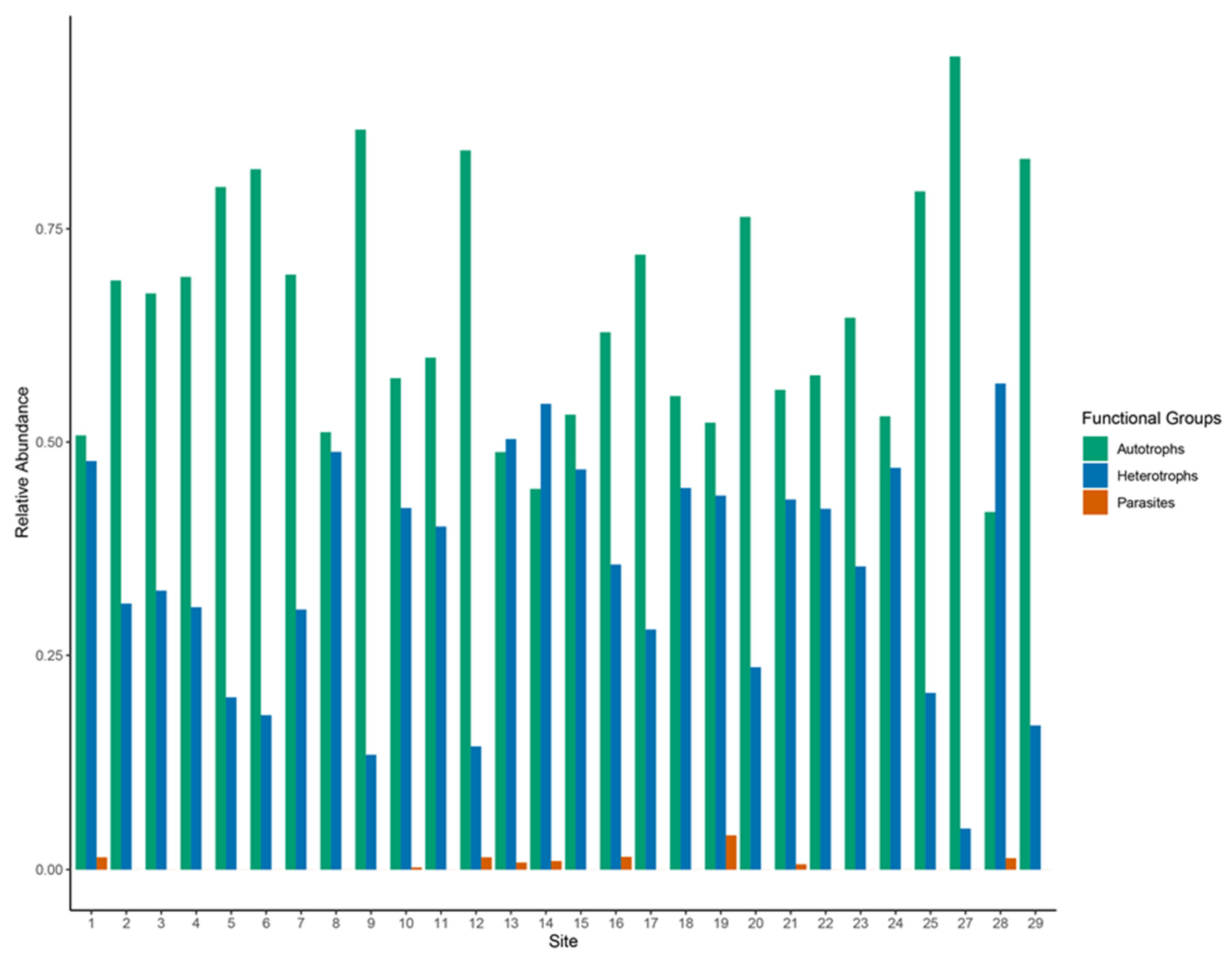
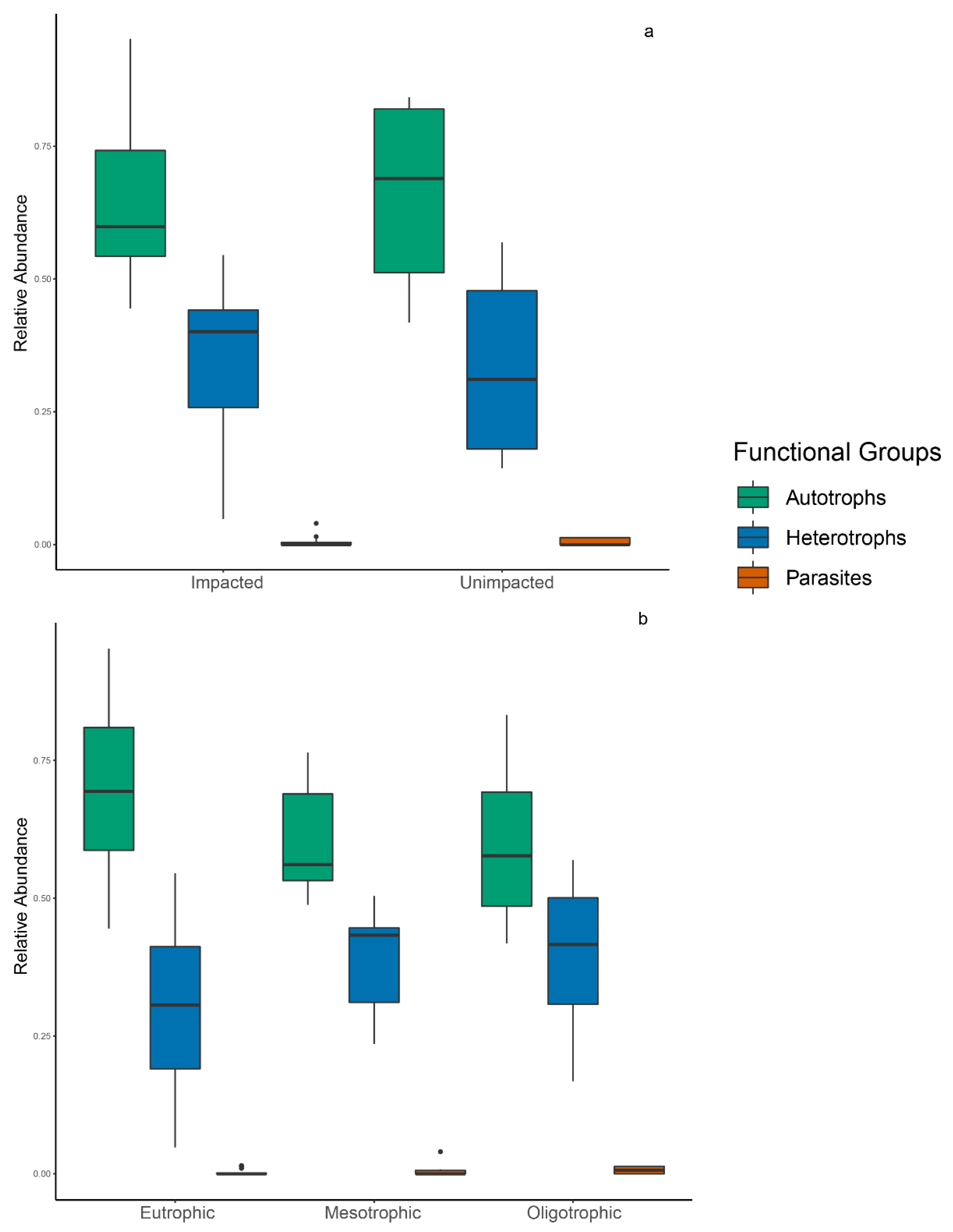
3.4. Spatial Distribution of Functional Groups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dudgeon, D.; Arthington, A.H.; Gessner, M.O.; Kawabata, Z.-I.; Knowler, D.J.; Lévêque, C.; Naiman, R.J.; Prieur-Richard, A.-H.; Soto, D.; Stiassny, M.L.J.; et al. Freshwater biodiversity: Importance, threats, status and conservation challenges. Biol. Rev. 2006, 81, 163–182. [Google Scholar] [CrossRef] [PubMed]
- Gleick, P.H. Basic Water Requirements for Human Activities: Meeting Basic Needs. Water Int. 2009, 21, 83–92. [Google Scholar] [CrossRef]
- Covich, A.P.; Ewel, K.C.; Hall, R.O.; Giller, P.E.; Goedkoop, W.; Merritt, D.M. Ecosystem services provided by freshwater benthos. In Sustaining Biodiversity and Ecosystem Services in Soils and Sediments; Wall, D.H., Ed.; Island Press: Washington, DC, USA, 2004; Volume 64, pp. 45–72. [Google Scholar]
- Jackson, R.B.; Carpenter, S.R.; Dahm, C.N.; McKnight, D.M.; Naiman, R.J.; Postel, S.L.; Running, S.W. Water in a changing world. Ecol. Appl. 2001, 11, 1027–1045. [Google Scholar] [CrossRef]
- Vorosmarty, C.J.; Sahagian, D. Anthropogenic Disturbance of the Terrestrial Water Cycle. Bioscience 2000, 50, 753–765. [Google Scholar] [CrossRef]
- Strayer, D.L.; Dudgeon, D. Freshwater biodiversity conservation: Recent progress and future challenges. J. N. Am. Benthol. Soc. 2010, 29, 344–358. [Google Scholar] [CrossRef]
- Joshi, P.; Pande, V.; Joshi, P. Microbial diversity of aquatic ecosystem and its industrial potential. J. Bacteriol. Mycol. Open Access 2016, 3, 177–179. [Google Scholar]
- Falkowski, P.G.; Fenchel, T.; Delong, E.F. The microbial engines that drive earth’s biogeochemical cycles. Science 2008, 320, 1034–1039. [Google Scholar] [CrossRef]
- Findlay, S. Stream microbial ecology. J. N. Am. Benthol. Soc. 2010, 29, 170–181. [Google Scholar] [CrossRef]
- Keeling, P.J.; Burki, F. Progress towards the Tree of Eukaryotes. Curr. Biol. 2019, 29, R808–R817. [Google Scholar] [CrossRef]
- Bar-On, Y.M.; Phillips, R.; Milo, R. The biomass distribution on Earth. Proc. Natl. Acad. Sci. USA 2018, 115, 6506–6511. [Google Scholar] [CrossRef]
- Borics, G.; Abonyi, A.; Salmaso, N.; Ptacnik, R. Freshwater phytoplankton diversity: Models, drivers and implications for ecosystem properties. Hydrobiologia 2021, 848, 53–75. [Google Scholar] [CrossRef] [PubMed]
- Debroas, D.; Domaizon, I.; Humbert, J.-F.; Jardillier, L.; Lepère, C.; Oudart, A.; Taïb, N. Overview of freshwater microbial eukaryotes diversity: A first analysis of publicly available metabarcoding data. FEMS Microbiol. Ecol. 2017, 93, 23. [Google Scholar] [CrossRef] [PubMed]
- Andersson, A.F.; Riemann, L.; Bertilsson, S. Pyrosequencing reveals contrasting seasonal dynamics of taxa within Baltic Sea bacterioplankton communities. ISME J. 2010, 4, 171–181. [Google Scholar] [CrossRef]
- Ptacnik, R.; Lepistö, L.; Willén, E.; Brettum, P.; Andersen, T.; Rekolainen, S.; Solheim, A.L.; Carvalho, L. Quantitative responses of lake phytoplankton to eutrophication in Northern Europe. Aquat. Ecol. 2008, 42, 227–236. [Google Scholar] [CrossRef]
- Grossart, H.-P.; Wyngaert, S.V.D.; Kagami, M.; Wurzbacher, C.; Cunliffe, M.; Rojas-Jimenez, K. Fungi in aquatic ecosystems. Nat. Rev. Microbiol. 2019, 17, 339–354. [Google Scholar] [CrossRef] [PubMed]
- Shalchian-Tabrizi, K.; Eikrem, W.; Klaveness, D.; Vaulot, D.; Minge, M.; Le Gall, F.; Romari, K.; Throndsen, J.; Botnen, A.; Massana, R.; et al. Telonemia, a new protist phylum with affinity to chromist lineages. Proc. R. Soc. B Biol. Sci. 2006, 273, 1833–1842. [Google Scholar] [CrossRef]
- López-García, P.; Moreira, D. Tracking microbial biodiversity through molecular and genomic ecology. Res. Microbiol. 2008, 159, 67–73. [Google Scholar] [CrossRef]
- Massana, R. Eukaryotic picoplankton in surface oceans. Annu. Rev. Microbiol. 2011, 65, 91–110. [Google Scholar] [CrossRef]
- Seenivasan, R.; Sausen, N.; Medlin, L.K.; Melkonian, M. Picomonas judraskeda Gen. Et Sp. Nov.: The First Identified Member of the Picozoa Phylum Nov., a Widespread Group of Picoeukaryotes, Formerly Known as ‘Picobiliphytes’. PLoS ONE 2013, 8, e59565. [Google Scholar] [CrossRef]
- Moreira, D.; López-García, P. The rise and fall of Picobiliphytes: How assumed autotrophs turned out to be heterotrophs. BioEssays 2014, 36, 468–474. [Google Scholar] [CrossRef]
- Mäki, A.; Salmi, P.; Mikkonen, A.; Kremp, A.; Tiirola, M. Sample Preservation, DNA or RNA extraction and data analysis for high-throughput phytoplankton community sequencing. Front. Microbiol. 2017, 8, 1848. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Morgan-Kiss, R.M. Influence of environmental drivers and potential interactions on the distribution of microbial communities from three permanently stratified Antarctic lakes. Front. Microbiol. 2019, 10, 1067. [Google Scholar] [CrossRef]
- Salmaso, N.; Boscaini, A.; Pindo, M. Unraveling the Diversity of Eukaryotic Microplankton in a Large and Deep Perialpine Lake Using a High Throughput Sequencing Approach. Front. Microbiol. 2020, 11, 789. [Google Scholar] [CrossRef] [PubMed]
- Fermani, P.; Metz, S.; Balagué, V.; Descy, J.P.; Morana, C.; Logares, R.; Massana, R.; Sarmento, H. Microbial eukaryote assemblages and potential novel diversity in four tropical East African Great Lakes. FEMS Microbiol. Ecol. 2021, 97, 114. [Google Scholar] [CrossRef] [PubMed]
- Santoferrara, L.; Burki, F.; Filker, S.; Logares, R.; Dunthorn, M.; McManus, G.B. Perspectives from Ten Years of Protist Studies by High-Throughput Metabarcoding. J. Eukaryot Microbiol. 2020, 67, 612–622. [Google Scholar] [CrossRef]
- Van der Hammen, T. The Pleistocene changes of vegetation and climate in tropical South America. J. Biogegr. 1974, 1, 3–26. [Google Scholar] [CrossRef]
- Ameel, J.J.; Axler, R.P.; Owen, C.J. Persulfate digestion for determination of total nitrogen and phosphorus in low-nutrient waters. Am. Environ. Lab. 1993, 10, 1–11. [Google Scholar]
- Baird, R.; Bridgewater, L. Standard Methods for the Examination of Water and Wastewater, 23rd ed.; American Water Works Association: Washington, DC, USA, 2017. [Google Scholar]
- Vollenweider, R.A.; Giovanardi, F.; Montanari, G.; Rinaldi, A.A. Characterization of The Trophic Conditions of Marine Coastal Waters With Special Reference To The NW Adriatic Sea: Proposal For A Trophic Scale, Turbidity And Generalized Water Quality Index. Environmetrics 1998, 9, 329–357. [Google Scholar] [CrossRef]
- Caruso, G.; Leonardi, M.; Monticelli, L.; Decembrini, F.; Azzaro, F.; Crisafi, E.; Zappalà, G.; Bergamasco, A.; Vizzini, S. Assessment of the ecological status of transitional waters in Sicily (Italy): First characterisation and classification according to a multiparametric approach. Mar. Pollut. Bull. 2010, 60, 1682–1690. [Google Scholar] [CrossRef]
- Djurhuus, A.; Port, J.; Closek, C.; Yamahara, K.M.; Romero-Maraccini, O.; Walz, K.R.; Goldsmith, D.B.; Michisaki, R.; Breitbart, M.; Boehm, A.B.; et al. Evaluation of filtration and DNA extraction methods for environmental DNA biodiversity assessments across multiple trophic levels. Front. Mar. Sci. 2017, 4, 314. [Google Scholar] [CrossRef]
- Bower, S.M.; Carnegie, R.B.; Goh, B.; Jones, S.; Lowe, G.J.; Mak, M.W.S. Preferential PCR Amplification of Parasitic Protistan Small Subunit rDNA from Metazoan Tissues. J. Eukaryot. Microbiol. 2004, 51, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Illumina. Nextera XT DNA Library Prep Reference Guide; Illumina: San Diego, CA, USA, 2019. [Google Scholar]
- Venables, W.N.; Smith, D.M. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Callahan, B.J.; Mcmurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Guillou, L.; Bachar, D.; Audic, S.; Bass, D.; Berney, C.; Bittner, L.; Boutte, C.; Burgaud, G.; de Vargas, C.; Decelle, J.; et al. The Protist Ribosomal Reference database (PR2): A catalog of unicellular eukaryote Small Sub-Unit rRNA sequences with curated taxonomy. Nucleic Acids Res. 2013, 41, D597–D604. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’hara, R.B.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H.; et al. Package ‘vegan’, Community Ecology Package. Time. Int. 2013, 2, 1–295. [Google Scholar]
- Lahti, L.; Shetty, S. Introduction to the Microbiome R Package. Bioconductor. 2018. Available online: https://www.bioconductor.org/packages/release/bioc/html/microbiome.html (accessed on 15 October 2022).
- Singer, D.; Seppey, C.V.; Lentendu, G.; Dunthorn, M.; Bass, D.; Belbahri, L.; Blandenier, Q.; Debroas, D.; de Groot, G.A.; de Vargas, C.; et al. Protist taxonomic and functional diversity in soil, freshwater and marine ecosystems. Environ. Int. 2021, 146, 106262. [Google Scholar] [CrossRef] [PubMed]
- Ford, T.E. Response of marine microbial communities to anthropogenic stress. J. Aquat. Ecosyst. Stress Recovery 2000, 7, 75–89. [Google Scholar] [CrossRef]
- Moss, B. Water pollution by agriculture. Philos. Trans. R. Soc. B Biol. Sci. 2007, 363, 659–666. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Wang, Y.; Lin, J.; Quan, C. Response of Planktonic and Benthic Microbial Community to Urban Pollution from Sewage Discharge in Jilin Reach of the Second Songhua River, China. Clean 2014, 42, 1376–1383. [Google Scholar] [CrossRef]
- Rasmussen, J.J.; Wiberg-Larsen, P.; Baattrup-Pedersen, A.; Cedergreen, N.; McKnight, U.S.; Kreuger, J.; Jacobsen, D.; Kristensen, E.A.; Friberg, N. The legacy of pesticide pollution: An overlooked factor in current risk assessments of freshwater systems. Water Res. 2015, 84, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Lai, Y.C.; Yang, C.P.; Hsieh, C.Y.; Wu, C.Y.; Kao, C.M. Evaluation of non-point source pollution and river water quality using a multimedia two-model system. J. Hydrol. 2011, 409, 583–595. [Google Scholar] [CrossRef]
- Ormerod, S.J.; Dobson, M.; Hildrew, A.G.; Townsend, C.R. Multiple stressors in freshwater ecosystems. Freshw. Biol. 2010, 55, 1–4. [Google Scholar] [CrossRef]
- Lee, T.C.; Liu, C.C.K. Assessing Eutrophication Potential of a Freshwater Lake by Relating Its Bioproductivity and Biodiversity: A Case Study of Lake Wilson on Central Oahu, Hawaii. Water 2018, 10, 296. [Google Scholar] [CrossRef]
- Lefranc, M.; Thénot, A.; Lepère, C.; Debroas, D. Genetic diversity of small eukaryotes in lakes differing by their trophic status. Appl. Environ. Microbiol. 2005, 71, 5935–5942. [Google Scholar] [CrossRef]
- Zhao, B.; Chen, M.; Sun, Y.; Yang, J.; Chen, F. Genetic diversity of picoeukaryotes in eight lakes differing in trophic status. Can. J. Microbiol. 2011, 57, 115–126. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, J.; Yang, J.; Giesy, J.P.; Yu, H.; Zhang, X. Environmental DNA metabarcoding reveals primary chemical contaminants in freshwater sediments from different land-use types. Chemosphere 2017, 172, 201–209. [Google Scholar] [CrossRef] [PubMed]
- Santos, S.S.; Schöler, A.; Nielsen, T.K.; Hansen, L.H.; Schloter, M.; Winding, A. Land use as a driver for protist community structure in soils under agricultural use across Europe. Sci. Total Environ. 2020, 717, 137228. [Google Scholar] [CrossRef] [PubMed]
- Zou, K.; Wang, R.; Xu, S.; Li, Z.; Liu, L.; Li, M.; Zhou, L. Changes in protist communities in drainages across the Pearl River Delta under anthropogenic influence. Water Res. 2021, 200, 117294. [Google Scholar] [CrossRef] [PubMed]
- Sagova-Mareckova, M.; Boenigk, J.; Bouchez, A.; Cermakova, K.; Chonova, T.; Cordier, T.; Eisendle, U.; Elersek, T.; Fazi, S.; Fleituch, T.; et al. Expanding ecological assessment by integrating microorganisms into routine freshwater biomonitoring. Water Res. 2021, 191, 116767. [Google Scholar] [CrossRef]
- Stoeck, T.; Kochems, R.; Forster, D.; Lejzerowicz, F.; Pawlowski, J. Metabarcoding of benthic ciliate communities shows high potential for environmental monitoring in salmon aquaculture. Ecol. Indic. 2018, 85, 153–164. [Google Scholar] [CrossRef]
- Piwosz, K.; Shabarova, T.; Pernthaler, J.; Posch, T.; Šimek, K.; Porcal, P.; Salcher, M.M. Bacterial and Eukaryotic Small-Subunit Amplicon Data Do Not Provide a Quantitative Picture of Microbial Communities, but They Are Reliable in the Context of Ecological Interpretations. mSphere 2020, 5, e00052-20. [Google Scholar] [CrossRef]
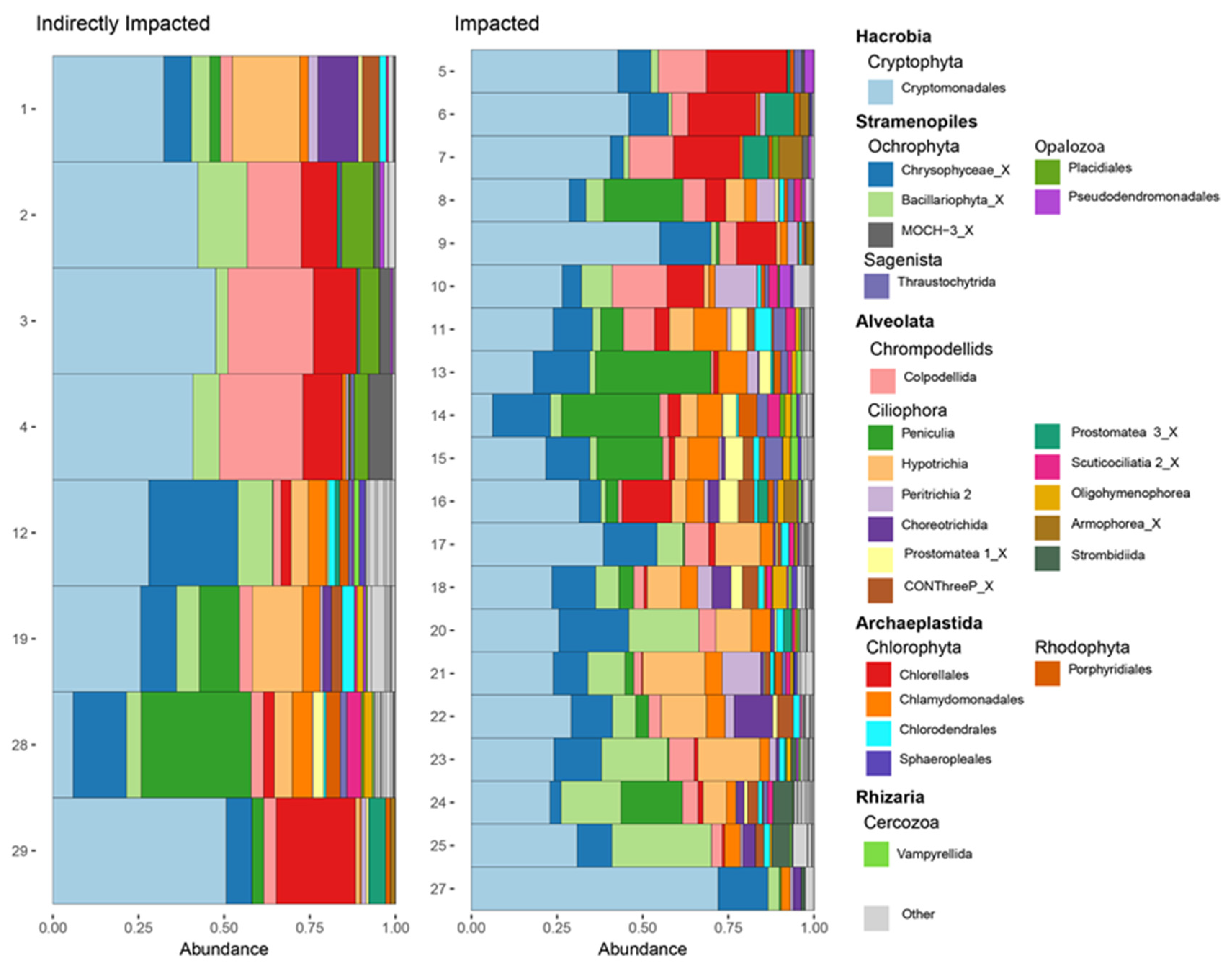
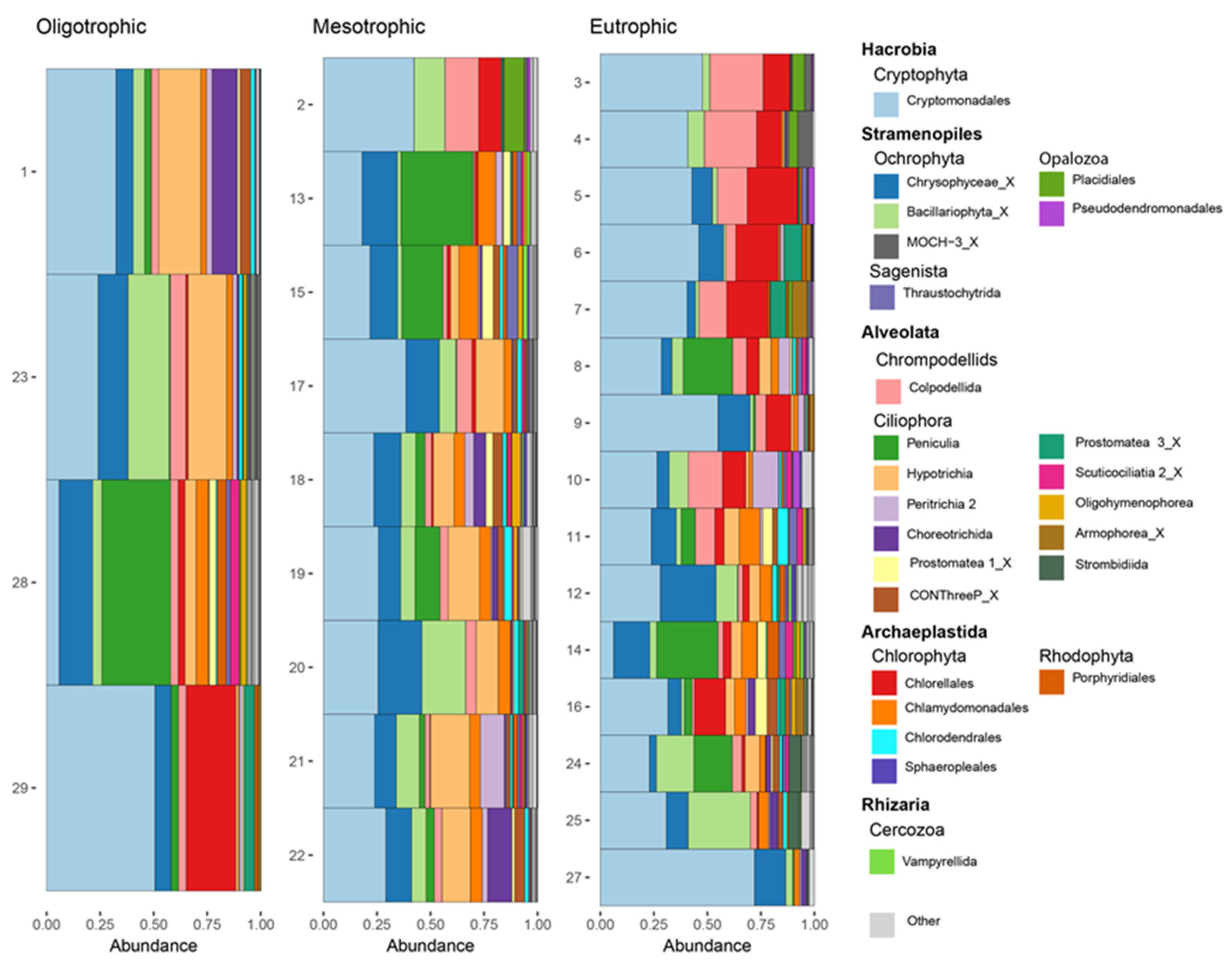

| Protist Group | Contribution% | Cumulative% | Impacted Sites Abundance | Indirectly Impacted Sites Abundance |
|---|---|---|---|---|
| Peniculia | 6.00 | 6.00 | 7.04 | 6.86 |
| Chlorellales | 6.00 | 13.00 | 5.09 | 9.94 |
| Chrysophyceae_X | 6.00 | 18.00 | 11.61 | 8.14 |
| Hypotrichia | 5.00 | 24.00 | 0.02 | 0.04 |
| Cryptomonadales | 5.00 | 28.00 | 30.68 | 35.83 |
| Colpodellida | 4.00 | 33.00 | 5.45 | 9.96 |
| Bacillariophyta_X | 4.00 | 37.00 | 8.12 | 5.76 |
| Chlamydomonadales | 4.00 | 41.00 | 4.58 | 2.12 |
| Choreotrichida | 4.00 | 44.00 | 1.88 | 1.31 |
| Placidiales | 3.00 | 48.00 | 0.24 | 2.21 |
| Prostomatea_3_X | 3.00 | 51.00 | 0.78 | 1.79 |
| Peritrichia_2 | 3.00 | 53.00 | 2.38 | 1.37 |
| CONThreeP_X | 3.00 | 56.00 | 1.64 | 0.58 |
| Prostomatea_1_X | 3.00 | 59.00 | 1.58 | 0.73 |
| Chlorodendrales | 3.00 | 61.00 | 1.45 | 0.67 |
| MOCH.3_X | 2.00 | 64.00 | 0.13 | 1.35 |
| Porphyridiales | 2.00 | 66.00 | 0.96 | 1.25 |
| Scuticociliatia_2 | 2.00 | 69.00 | 0.96 | 0.74 |
| Thraustochytrida | 2.00 | 71.00 | 1.12 | 0.78 |
| Protist Group | Contribution% | Cumulative% | Mesotrophic Sites Abundance | Eutrophic Sites Abundance |
|---|---|---|---|---|
| Peniculia | 6.00 | 6.00 | 8.37 | 5.41 |
| Chlorellales | 6.00 | 13.00 | 1.82 | 9.49 |
| Hypotrichia | 6.00 | 19.00 | 9.27 | 2.46 |
| Chrysophyceae_X | 5.00 | 24.00 | 12.27 | 9.24 |
| Colpodellida | 4.00 | 28.00 | 4.81 | 8.78 |
| Bacillariophyta_X | 4.00 | 32.00 | 8.65 | 6.62 |
| Cryptomonadales | 4.00 | 36.00 | 27.61 | 36.25 |
| Choreotrichida | 4.00 | 40.00 | 2.59 | 0.77 |
| Chlamydomonadales | 3.00 | 43.00 | 5.27 | 3.15 |
| Peritrichia_2 | 3.00 | 46.00 | 2.55 | 1.87 |
| CONThreeP_X | 3.00 | 49.00 | 2.01 | 0.88 |
| Prostomatea_1_X | 3.00 | 52.00 | 1.64 | 1.11 |
| Placidiales | 3.00 | 55.00 | 1.11 | 0.91 |
| Thraustochytrida | 2.00 | 57.00 | 0.99 | 1.17 |
| Chlorodendrales | 2.00 | 60.00 | 1.64 | 0.97 |
| Prostomatea_3_X | 2.00 | 62.00 | 0.45 | 1.43 |
| Oligohymenophorea_X | 2.00 | 64.00 | 1.28 | 0.40 |
| Scuticociliatia_2 | 2.00 | 67.00 | 0.85 | 0.83 |
| Porphyridiales | 2.00 | 69.00 | 0.71 | 1.16 |
| MOCH.3_X | 2.00 | 71.00 | 0.19 | 0.86 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barbosa, M.; Lefler, F.W.; Berthold, D.E.; Briggs-Gonzalez, V.S.; Mazzotti, F.J.; Laughinghouse, H.D., IV. Trophic State Drives the Diversity of Protists in a Tropical River (New River, Belize). Microorganisms 2022, 10, 2425. https://doi.org/10.3390/microorganisms10122425
Barbosa M, Lefler FW, Berthold DE, Briggs-Gonzalez VS, Mazzotti FJ, Laughinghouse HD IV. Trophic State Drives the Diversity of Protists in a Tropical River (New River, Belize). Microorganisms. 2022; 10(12):2425. https://doi.org/10.3390/microorganisms10122425
Chicago/Turabian StyleBarbosa, Maximiliano, Forrest W. Lefler, David E. Berthold, Venetia S. Briggs-Gonzalez, Frank J. Mazzotti, and H. Dail Laughinghouse, IV. 2022. "Trophic State Drives the Diversity of Protists in a Tropical River (New River, Belize)" Microorganisms 10, no. 12: 2425. https://doi.org/10.3390/microorganisms10122425
APA StyleBarbosa, M., Lefler, F. W., Berthold, D. E., Briggs-Gonzalez, V. S., Mazzotti, F. J., & Laughinghouse, H. D., IV. (2022). Trophic State Drives the Diversity of Protists in a Tropical River (New River, Belize). Microorganisms, 10(12), 2425. https://doi.org/10.3390/microorganisms10122425








