Alhagi sparsifolia Harbors a Different Root-Associated Mycobiome during Different Development Stages
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site and Experimental Design
2.2. Sample Collection
2.3. Soil Physical and Chemical Property Measurements
2.4. Molecular Methods
2.5. Bioinformatics Analyses
2.6. Data Analysis
3. Results
3.1. Soil Properties and Plant Traits
3.2. Community Structure of Root-Associated Fungi among Compartments and Host Growth Times
3.3. The Relationships between the Root-Associated Mycobiome and Environmental Factors
3.4. Co-Occurrence Networks
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tedersoo, L.; Bahram, M.; Zobel, M. How mycorrhizal associations drive plant population and community biology. Science 2020, 367, eaba1223. [Google Scholar] [CrossRef]
- Bever, J.D.; Mangan, S.A.; Alexander, H.M. Maintenance of Plant Species Diversity by Pathogens. Annu. Rev. Ecol. Evol. Syst. 2015, 46, 305–325. [Google Scholar] [CrossRef]
- Weyens, N.; van der Lelie, D.; Taghavi, S.; Newman, L.; Vangronsveld, J. Exploiting plant-microbe partnerships to improve biomass production and remediation. Trends Biotechnol. 2009, 27, 591–598. [Google Scholar] [CrossRef] [PubMed]
- Edwards, J.; Johnson, C.; Santos-Medellin, C.; Lurie, E.; Podishetty, N.K.; Bhatnagar, S.; Eisen, J.A.; Sundaresan, V. Structure, variation, and assembly of the root-associated microbiomes of rice. Proc. Natl. Acad. Sci. USA 2015, 112, E911–E920. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef]
- Xiong, C.; He, J.-Z.; Singh, B.K.; Zhu, Y.-G.; Wang, J.-T.; Li, P.-P.; Zhang, Q.-B.; Han, L.-L.; Shen, J.-P.; Ge, A.-H.; et al. Rare taxa maintain the stability of crop mycobiomes and ecosystem functions. Environ. Microbiol. 2021, 23, 1907–1924. [Google Scholar] [CrossRef]
- Alsharif, W.; Saad, M.M.; Hirt, H. Desert Microbes for Boosting Sustainable Agriculture in Extreme Environments. Front. Microbiol. 2020, 11, 1666. [Google Scholar] [CrossRef]
- Makhalanyane, T.P.; Valverde, A.; Gunnigle, E.; Frossard, A.; Ramond, J.-B.; Cowan, D.A. Microbial ecology of hot desert edaphic systems. FEMS Microbiol. Rev. 2015, 39, 203–221. [Google Scholar] [CrossRef]
- Zhang, Z.; Tariq, A.; Zeng, F.; Graciano, C.; Sun, F.; Chai, X.; Ahmed, Z. Nitrogen and water addition regulate fungal community and microbial co-occurrence network complexity in the rhizosphere of Alhagi sparsifolia seedlings. Appl. Soil Ecol. 2021, 164, 103940. [Google Scholar] [CrossRef]
- Rosling, A.; Midgley, M.G.; Cheeke, T.; Urbina, H.; Fransson, P.; Phillips, R.P. Phosphorus cycling in deciduous forest soil differs between stands dominated by ecto- and arbuscular mycorrhizal trees. New Phytol. 2016, 209, 1184–1195. [Google Scholar] [CrossRef]
- Coleman-Derr, D.; Desgarennes, D.; Fonseca-Garcia, C.; Gross, S.; Clingenpeel, S.; Woyke, T.; North, G.; Visel, A.; Partida-Martinez, L.P.; Tringe, S.G. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol. 2016, 209, 798–811. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lennon, J.T.; Jones, S.E. Microbial seed banks: The ecological and evolutionary implications of dormancy. Nat. Rev. Microbiol. 2011, 9, 119–130. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Robert, C.A.M.; Cadot, S.; Zhang, X.; Ye, M.; Li, B.; Manzo, D.; Chervet, N.; Steinger, T.; van der Heijden, M.G.A.; et al. Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 2018, 9, 2738. [Google Scholar] [CrossRef] [Green Version]
- Schlemper, T.R.; Leite, M.F.A.; Lucheta, A.R.; Shimels, M.; Bouwmeester, H.J.; van Veen, J.A.; Kuramae, E.E. Rhizobacterial community structure differences among sorghum cultivars in different growth stages and soils. FEMS Microbiol. Ecol. 2017, 93, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Zhalnina, K.; Louie, K.B.; Hao, Z.; Mansoori, N.; da Rocha, U.N.; Shi, S.; Cho, H.; Karaoz, U.; Loque, D.; Bowen, B.P.; et al. Dynamic root exudate chemistry and microbial substrate preferences drive patterns in rhizosphere microbial community assembly. Nat. Microbiol. 2018, 3, 470–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delgado-Baquerizo, M.; Fry, E.L.; Eldridge, D.J.; de Vries, F.T.; Manning, P.; Hamonts, K.; Kattge, J.; Boenisch, G.; Singh, B.K.; Bardgett, R.D. Plant attributes explain the distribution of soil microbial communities in two contrasting regions of the globe. New Phytol. 2018, 219, 574–587. [Google Scholar] [CrossRef] [Green Version]
- Thomas, F.M.; Arndt, S.K.; Bruelheide, H.; Foetzki, A.; Gries, D.; Huang, J.; Popp, M.; Wang, G.; Zhang, X.M.; Runge, M. Ecological basis for a sustainable management of the indigenous vegetation in a Central-Asian desert: Presentation and first results. J. Appl. Bot.-Angew. Bot. 2000, 74, 212–219. [Google Scholar]
- Liu, B.; Zeng, F.J.; Arndt, S.K.; He, J.X.; Luo, W.C.; Song, C. Patterns of root architecture adaptation of a phreatophytic perennial desert plant in a hyperarid desert. S. Afr. J. Bot. 2013, 86, 56–62. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Tariq, A.; Zeng, F.; Graciano, C.; Zhang, B. Nitrogen application mitigates drought-induced metabolic changes in Alhagi sparsifolia seedlings by regulating nutrient and biomass allocation patterns. Plant Physiol. Biochem. 2020, 155, 828–841. [Google Scholar] [CrossRef]
- Zhang, Z.; Chai, X.; Gao, Y.; Zhang, B.; Lu, Y.; Huang, C.; Li, L.; Tariq, A.; Li, X.; Zeng, F. Dynamics in diversity, co-occurrence pattern, and community assembly of a perennial desert plant root-associated bacteria. Rhizosphere 2022, 22, 100526. [Google Scholar] [CrossRef]
- Gui, D.; Zeng, F.; Liu, Z.; Zhang, B. Characteristics of the clonal propagation of Alhagi sparsifolia Shap. (Fabaceae) under different groundwater depths in Xinjiang, China. Rangel. J. 2013, 35, 355–362. [Google Scholar] [CrossRef]
- Chen, S.; Waghmode, T.R.; Sun, R.; Kuramae, E.E.; Hu, C.; Liu, B. Root-associated microbiomes of wheat under the combined effect of plant development and nitrogen fertilization. Microbiome 2019, 7, 136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, G.; Wang, L.; Jiang, L.; Pan, X.; Huang, Z.; Dong, M.; Cornelissen, J.H.C. Specific leaf area predicts dryland litter decomposition via two mechanisms. J. Ecol. 2018, 106, 218–229. [Google Scholar] [CrossRef] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bokulich, N.A.; Subramanian, S.; Faith, J.J.; Gevers, D.; Gordon, J.I.; Knight, R.; Mills, D.A.; Caporaso, J.G. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 2013, 10, 57–59. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; Publisher Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 20 August 2020).
- Mendiburu, F. Agricolae: Statistical Procedures for Agricultural Research. R Package Version 1.3-3. 2020. Available online: https://cran.r-project.org/package=agricolae (accessed on 20 August 2020).
- Taiyun, W.; Viliam, S. R Package “Corrplot”: Visualization of a Correlation Matrix. R Package Version 084. 2017. Available online: https://cran.r-project.org/package=corrplot (accessed on 20 August 2020).
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; O’Hara, R.B.; Simpson, G.; Solymos, P.; Stevens, H.; Wagner, H. Vegan: Community Ecology Package. Ordination Methods, Diversity Analysis and Other Functions for Community and Vegetation Ecologists. Version 2.3-1. 2012. Available online: https://cran.r-project.org/package=vegan (accessed on 20 August 2020).
- Deng, Y.; Jiang, Y.-H.; Yang, Y.; He, Z.; Luo, F.; Zhou, J. Molecular ecological network analyses. BMC Bioinform. 2012, 13, 113. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, N.H.; Song, Z.; Bates, S.T.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.S.; Kennedy, P.G. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 2016, 20, 241–248. [Google Scholar] [CrossRef]
- Liu, C.; Cui, Y.; Li, X.; Yao, M. microeco: An R package for data mining in microbial community ecology. FEMS Microbiol. Ecol. 2021, 97, fiaa255. [Google Scholar] [CrossRef]
- Csardi, G.; Nepusz, T. The Igraph Software Package for Complex Network Research. R Package Version 084 1695. 2006. Available online: https://cran.r-project.org/package=igraph (accessed on 20 August 2020).
- Duran, P.; Thiergart, T.; Garrido-Oter, R.; Agler, M.; Kemen, E.; Schulze-Lefert, P.; Hacquard, S. Microbial Interkingdom Interactions in Roots Promote Arabidopsis Survival. Cell 2018, 175, 973–983.e914. [Google Scholar] [CrossRef]
- Xiong, C.; Zhu, Y.-G.; Wang, J.-T.; Singh, B.; Han, L.-L.; Shen, J.-P.; Li, P.-P.; Wang, G.-B.; Wu, C.-F.; Ge, A.-H.; et al. Host selection shapes crop microbiome assembly and network complexity. New Phytol. 2021, 229, 1091–1104. [Google Scholar] [CrossRef]
- Guttman, D.S.; McHardy, A.C.; Schulze-Lefert, P. Microbial genome-enabled insights into plant-microorganism interactions. Nat. Rev. Genet. 2014, 15, 797–813. [Google Scholar] [CrossRef] [PubMed]
- Kunstler, G.; Falster, D.; Coomes, D.A.; Hui, F.; Kooyman, R.M.; Laughlin, D.C.; Poorter, L.; Vanderwel, M.; Vieilledent, G.; Wright, S.J.; et al. Plant functional traits have globally consistent effects on competition. Nature 2016, 529, 204–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, S.N.; Raghubanshi, A.S. Seedling growth of five tropical dry forest tree species in relation to light and nitrogen gradients. J. Plant Ecol. 2014, 7, 250–263. [Google Scholar] [CrossRef] [Green Version]
- Shi, S.; Nuccio, E.E.; Shi, Z.J.; He, Z.; Zhou, J.; Firestone, M.K. The interconnected rhizosphere: High network complexity dominates rhizosphere assemblages. Ecol. Lett. 2016, 19, 926–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philippot, L.; Raaijmakers, J.M.; Lemanceau, P.; van der Putten, W.H. Going back to the roots: The microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 2013, 11, 789–799. [Google Scholar] [CrossRef]
- Delpiano, C.A.; Prieto, I.; Loayza, A.P.; Carvajal, D.E.; Squeo, F.A. Different responses of leaf and root traits to changes in soil nutrient availability do not converge into a community-level plant economics spectrum. Plant Soil 2020, 450, 463–478. [Google Scholar] [CrossRef]
- Qian, X.; Li, H.; Wang, Y.; Wu, B.; Wu, M.; Chen, L.; Li, X.; Zhang, Y.; Wang, X.; Shi, M.; et al. Leaf and Root Endospheres Harbor Lower Fungal Diversity and Less Complex Fungal Co-occurrence Patterns Than Rhizosphere. Front. Microbiol. 2019, 10, 01015. [Google Scholar] [CrossRef]
- Fernandez-Gonzalez, A.J.; Cardoni, M.; Gomez-Lama Cabanas, C.; Valverde-Corredor, A.; Villadas, P.J.; Fernandez-Lopez, M.; Mercado-Blanco, J. Linking belowground microbial network changes to different tolerance level towards Verticillium wilt of olive. Microbiome 2020, 8, 11. [Google Scholar] [CrossRef]
- Tao, J.; Meng, D.; Qin, C.; Liu, X.; Liang, Y.; Xiao, Y.; Liu, Z.; Gu, Y.; Li, J.; Yin, H. Integrated network analysis reveals the importance of microbial interactions for maize growth. Appl. Microbiol. Biotechnol. 2018, 102, 3805–3818. [Google Scholar] [CrossRef]
- Rybakova, D.; Mancinelli, R.; Wikstrom, M.; Birch-Jensen, A.-S.; Postma, J.; Ehlers, R.-U.; Goertz, S.; Berg, G. The structure of the Brassica napus seed microbiome is cultivar-dependent and affects the interactions of symbionts and pathogens. Microbiome 2017, 5, 104. [Google Scholar] [CrossRef]
- Li, Y.; Li, Z.; Arafat, Y.; Lin, W. Studies on fungal communities and functional guilds shift in tea continuous cropping soils by high-throughput sequencing. Ann. Microbiol. 2020, 70, 7. [Google Scholar] [CrossRef]
- Olesen, J.M.; Bascompte, J.; Dupont, Y.L.; Jordano, P. The modularity of pollination networks. Proc. Natl. Acad. Sci. USA 2007, 104, 19891–19896. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaparro, J.M.; Badri, D.V.; Vivanco, J.M. Rhizosphere microbiome assemblage is affected by plant development. ISME J. 2014, 8, 790–803. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hinsinger, P.; Bengough, A.G.; Vetterlein, D.; Young, I.M. Rhizosphere: Biophysics, biogeochemistry and ecological relevance. Plant Soil 2009, 321, 117–152. [Google Scholar] [CrossRef]
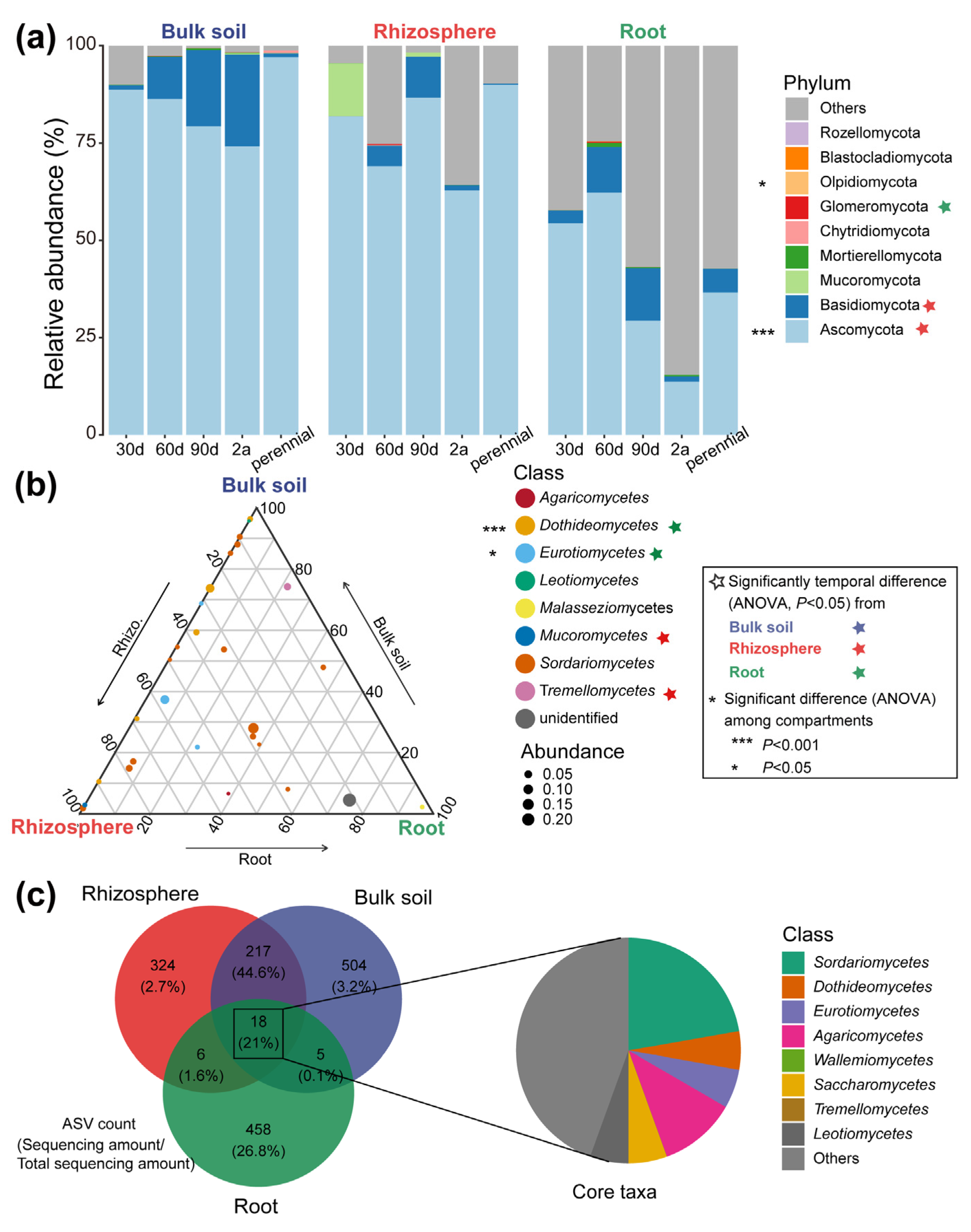

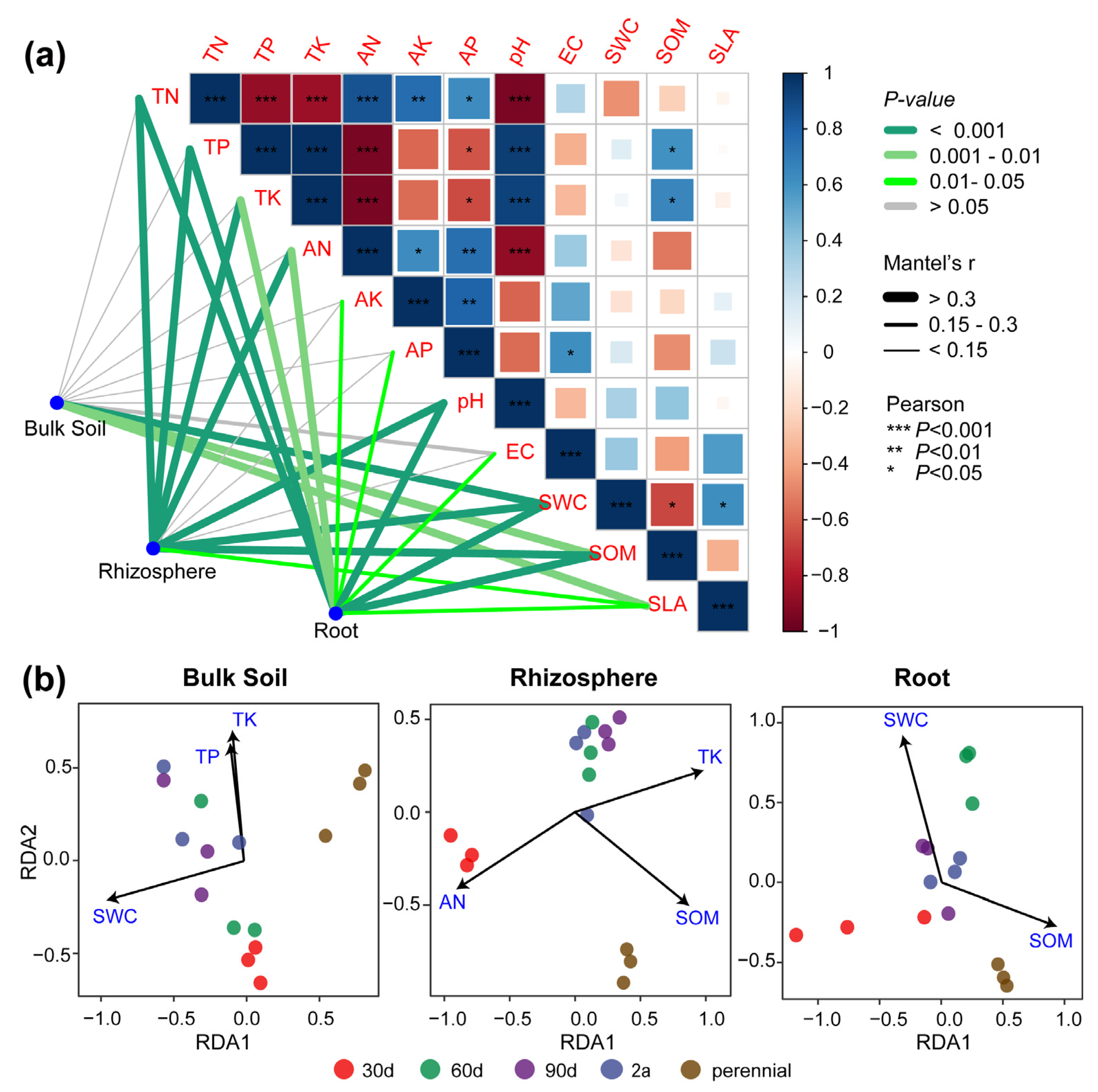
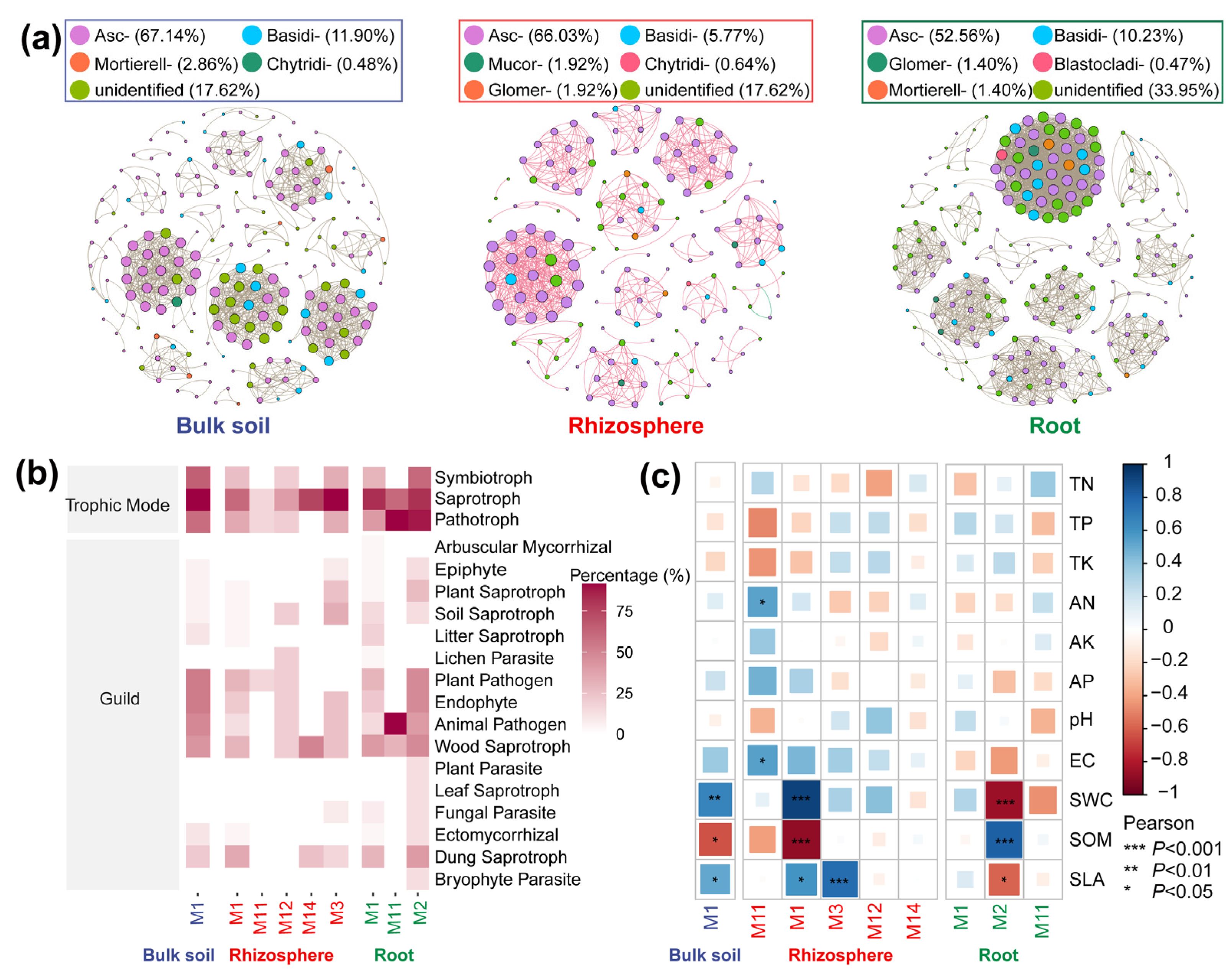
| Traits | 30 d | 60 d | 90 d | 2 a | Perennial |
|---|---|---|---|---|---|
| SOM | 1.97 ± 0.14 d | 3.03 ± 0.04 b | 3.14 ± 0.08 b | 2.73 ± 0.06 c | 4.74 ± 0.03 a |
| TN | 0.23 ± 0.02 a | 0.13 ± 0.00 c | 0.14 ± 0.00 c | 0.13 ± 0.01 c | 0.17 ± 0.00 b |
| TP | 0.31 ± 0.01 c | 0.60 ± 0.01 a | 0.58 ± 0.00 ab | 0.58 ± 0.01 b | 0.58 ± 0.00 b |
| TK | 21.24 ± 0.55 a | 17.82 ± 0.12 c | 18.11 ± 0.05 bc | 18.11 ± 0.09 bc | 18.53 ± 0.19 b |
| AN | 29.87 ± 5.52 a | 4.76 ± 0.86 b | 4.05 ± 0.24 b | 4.76 ± 0.63 b | 5.95 ± 0.24 b |
| AK | 143.17 ± 33.77 a | 103.00 ± 2.65 a | 108.33 ± 7.51 a | 90.00 ± 3.61 a | 111.00 ± 8.00 a |
| AP | 3.35 ± 0.97 a | 2.48 ± 0.14 a | 1.74 ± 0.41 a | 1.33 ± 0.07 a | 1.38 ± 0.08 a |
| pH | 8.09 ± 0.02 b | 9.16 ± 0.01 a | 9.06 ± 0.04 a | 9.23 ± 0.03 a | 8.89 ± 0.12 ab |
| EC | 0.43 ± 0.06 a | 0.36 ± 0.05 a | 0.43 ± 0.06 a | 0.33 ± 0.06 a | 0.28 ± 0.05 a |
| SWC | 0.06 ± 0.00 c | 0.10 ± 0.00 a | 0.09 ± 0.00 b | 0.08 ± 0.00 b | 0.01 ± 0.00 d |
| SLA | 112.16 ± 0.00 b | 118.73 ± 6.83 b | 144.99 ± 10.03 a | 90.01 ± 6.86 c | 80.74 ± 5.67 c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Chai, X.; Gao, Y.; Zhang, B.; Lu, Y.; Du, Y.; Zhang, Y.; Ding, Y.; Tariq, A.; Ullah, A.; et al. Alhagi sparsifolia Harbors a Different Root-Associated Mycobiome during Different Development Stages. Microorganisms 2022, 10, 2376. https://doi.org/10.3390/microorganisms10122376
Zhang Z, Chai X, Gao Y, Zhang B, Lu Y, Du Y, Zhang Y, Ding Y, Tariq A, Ullah A, et al. Alhagi sparsifolia Harbors a Different Root-Associated Mycobiome during Different Development Stages. Microorganisms. 2022; 10(12):2376. https://doi.org/10.3390/microorganisms10122376
Chicago/Turabian StyleZhang, Zhihao, Xutian Chai, Yanju Gao, Bo Zhang, Yan Lu, Yi Du, Yulin Zhang, Ya Ding, Akash Tariq, Abd Ullah, and et al. 2022. "Alhagi sparsifolia Harbors a Different Root-Associated Mycobiome during Different Development Stages" Microorganisms 10, no. 12: 2376. https://doi.org/10.3390/microorganisms10122376
APA StyleZhang, Z., Chai, X., Gao, Y., Zhang, B., Lu, Y., Du, Y., Zhang, Y., Ding, Y., Tariq, A., Ullah, A., Li, X., & Zeng, F. (2022). Alhagi sparsifolia Harbors a Different Root-Associated Mycobiome during Different Development Stages. Microorganisms, 10(12), 2376. https://doi.org/10.3390/microorganisms10122376








