Micro”bee”ota: Honey Bee Normal Microbiota as a Part of Superorganism
Abstract
1. Introduction
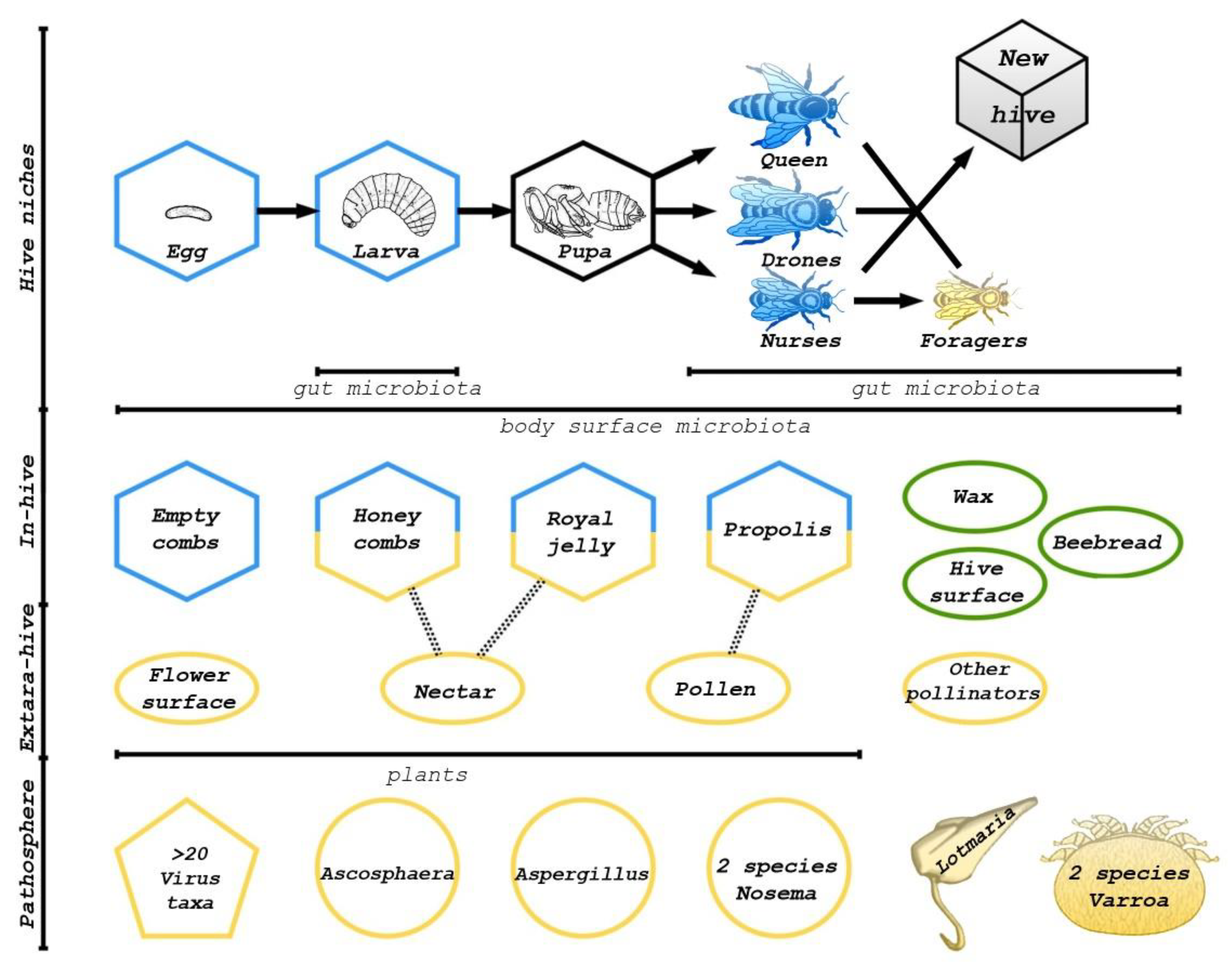
2. Types of Honey Bee Microbiota
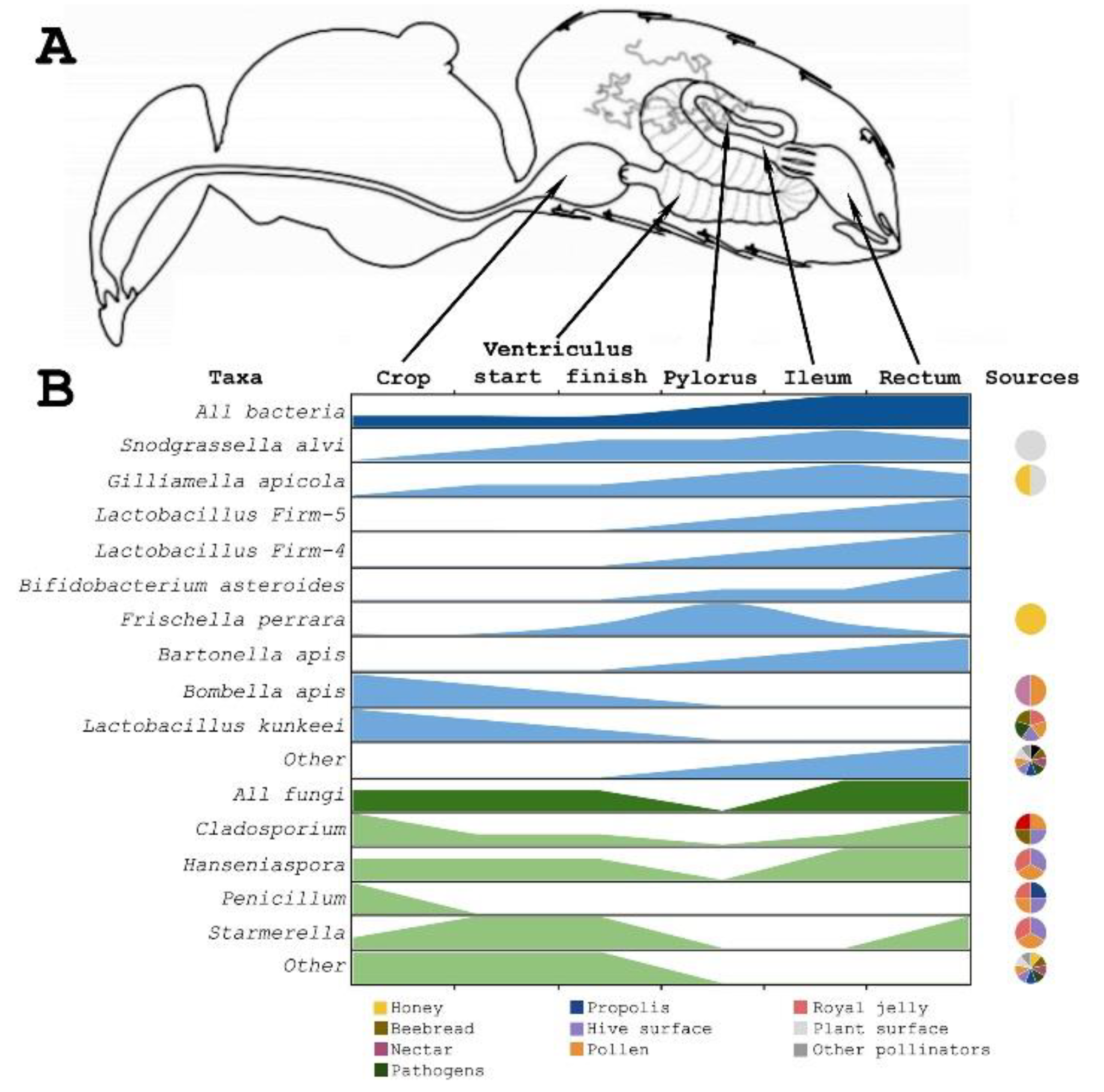
3. Gut Microbiota
4. Body Surface Microbiota
5. In-Hive Environments Microbiota
6. Microbiota from the Environment, including Pathogens
7. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dale, C.; Moran, N.A. Molecular Interactions between Bacterial Symbionts and Their Hosts. Cell 2006, 126, 453–465. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.A.; Hoffmann, A.R.; Neufeld, J.D. The skin microbiome of vertebrates. Microbiome 2019, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.; Lozupone, C.A.; Hamady, M.; Knight, R.; Gordon, J.I. Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat. Rev. Genet. 2008, 6, 776–788. [Google Scholar] [CrossRef] [PubMed]
- Papp, M.; Békési, L.; Farkas, R.; Makrai, L.; Maróti, G.; Tőzsér, D.; Solymosi, N. Natural diversity of honey bee (Apis mellifera) gut bacteriome in various climatic and seasonal states. bioRxiv 2021, 27, 428438. [Google Scholar] [CrossRef]
- Schmidt, K.; Engel, P. Mechanisms underlying gut microbiota–host interactions in insects. J. Exp. Biol. 2021, 224 Pt 2, jeb207696. [Google Scholar] [CrossRef]
- Liberti, J.; Engel, P. The gut microbiota—Brain axis of insects. Curr. Opin. Insect Sci. 2020, 39, 6–13. [Google Scholar] [CrossRef]
- Engel, P.; James, R.R.; Koga, R.; Kwong, W.K.; McFrederick, Q.S.; Moran, N.A. Standard methods for research on Apis mellifera gut symbionts. J. Apic. Res. 2013, 52, 1–24. [Google Scholar] [CrossRef]
- Rangberg, A.; Diep, D.B.; Rudi, K.; Amdam, G.V. Paratransgenesis: An Approach to Improve Colony Health and Molecular Insight in Honey Bees (Apis mellifera)? Integr. Comp. Biol. 2012, 52, 89–99. [Google Scholar] [CrossRef]
- Miller, D.L.; Parish, A.J.; Newton, I.L. Transitions and transmission: Behavior and physiology as drivers of honey bee-associated microbial communities. Curr. Opin. Microbiol. 2019, 50, 1–7. [Google Scholar] [CrossRef]
- Schwarz, R.S.; Huang, Q.; Evans, J.D. Hologenome theory and the honey bee pathosphere. Curr. Opin. Insect Sci. 2015, 10, 1–7. [Google Scholar] [CrossRef]
- Moran, N.A.; Hansen, A.; Powell, J.; Sabree, Z.L. Distinctive Gut Microbiota of Honey Bees Assessed Using Deep Sampling from Individual Worker Bees. PLoS ONE 2012, 7, e36393. [Google Scholar] [CrossRef] [PubMed]
- Kwong, W.K.; Moran, N.A. Gut microbial communities of social bees. Nat. Rev. Genet. 2016, 14, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Ribière, C.; Hegarty, C.; Stephenson, H.; Whelan, P.; O’Toole, P.W. Gut and Whole-Body Microbiota of the Honey Bee Separate Thriving and Non-thriving Hives. Microb. Ecol. 2018, 78, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Subotic, S.; Boddicker, A.M.; Nguyen, V.M.; Rivers, J.; Briles, C.E.; Mosier, A.C. Honey bee microbiome associated with different hive and sample types over a honey production season. PLoS ONE 2019, 14, e0223834. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Wang, L.; Jin, Y.; Zhang, J.; Su, L.; Zhang, X.; Zhou, J.; Li, Y. The Microbial Community Dynamics during the Vitex Honey Ripening Process in the Honeycomb. Front. Microbiol. 2017, 8, 1649. [Google Scholar] [CrossRef] [PubMed]
- Anderson, K.E.; Sheehan, T.H.; Mott, B.M.; Maes, P.; Snyder, L.; Schwan, M.R.; Walton, A.; Jones, B.M.; Corby-Harris, V. Microbial Ecology of the Hive and Pollination Landscape: Bacterial Associates from Floral Nectar, the Alimentary Tract and Stored Food of Honey Bees (Apis mellifera). PLoS ONE 2013, 8, e83125. [Google Scholar] [CrossRef]
- McMenamin, A.J.; Flenniken, M.L. Recently identified bee viruses and their impact on bee pollinators. Curr. Opin. Insect Sci. 2018, 26, 120–129. [Google Scholar] [CrossRef]
- Ullah, A.; Gajger, I.T.; Majoros, A.; Dar, S.A.; Khan, S.; Shah, A.H.; Khabir, M.N.; Hussain, R.; Khan, H.U.; Hameed, M.; et al. Viral impacts on honey bee populations: A review. Saudi J. Biol. Sci. 2020, 28, 523–530. [Google Scholar] [CrossRef]
- Fünfhaus, A.; Ebeling, J.; Genersch, E. Bacterial pathogens of bees. Curr. Opin. Insect Sci. 2018, 26, 89–96. [Google Scholar] [CrossRef]
- Leonard, S.P.; Powell, J.E.; Perutka, J.; Geng, P.; Heckmann, L.C.; Horak, R.D.; Davies, B.W.; Ellington, A.D.; Barrick, J.E.; Moran, N.A. Engineered symbionts activate honey bee immunity and limit pathogens. Science 2020, 367, 573–576. [Google Scholar] [CrossRef]
- Mondet, F.; Beaurepaire, A.; McAfee, A.; Locke, B.; Alaux, C.; Blanchard, S.; Danka, B.; Le Conte, Y. Honey bee survival mechanisms against the parasite Varroa destructor: A systematic review of phenotypic and genomic research efforts. Int. J. Parasitol. 2020, 50, 433–447. [Google Scholar] [CrossRef] [PubMed]
- Diaz, T.; Del-Val, E.; Ayala, R.; Larsen, J. Alterations in honey bee gut microorganisms caused by Nosema spp. and pest control methods. Pest Manag. Sci. 2018, 75, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Moran, N.A. The gut microbiota of insects—Diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735. [Google Scholar] [CrossRef] [PubMed]
- Kwong, W.K.; Engel, P.; Koch, H.; Moran, N.A. Genomics and host specialization of honey bee and bumble bee gut symbionts. Proc. Natl. Acad. Sci. USA 2014, 111, 11509–11514. [Google Scholar] [CrossRef] [PubMed]
- Donkersley, P.; Rhodes, G.; Pickup, R.W.; Jones, K.C.; Wilson, K. Bacterial communities associated with honeybee food stores are correlated with land use. Ecol. Evol. 2018, 8, 4743–4756. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.A.; Newton, I.L.G. Genomic Signatures of Honey Bee Association in an Acetic Acid Symbiont. Genome Biol. Evol. 2020, 12, 1882–1894. [Google Scholar] [CrossRef] [PubMed]
- Hilgarth, M.; Redwitz, J.; Ehrmann, M.A.; Vogel, R.F.; Jakob, F. Bombella favorum sp. nov. and Bombella mellum sp. nov., two novel species isolated from the honeycombs of Apis mellifera. Int. J. Syst. Evol. Microbiol. 2021, 71, 004633. [Google Scholar] [CrossRef]
- Miller, D.L.; Smith, E.A.; Newton, I.L.G. A Bacterial Symbiont Protects Honey Bees from Fungal Disease. mBio 2021, 12, e0050321. [Google Scholar] [CrossRef]
- Anderson, K.E.; Carroll, M.J.; Sheehan, T.; Mott, B.M.; Maes, P.; Corby-Harris, V. Hive-stored pollen of honey bees: Many lines of evidence are consistent with pollen preservation, not nutrient conversion. Mol. Ecol. 2014, 23, 5904–5917. [Google Scholar] [CrossRef]
- Anderson, K.E.; Sheehan, T.H.; Eckholm, B.J.; Mott, B.M.; Degrandihoffman, G. An emerging paradigm of colony health: Microbial balance of the honey bee and hive (Apis mellifera). Insectes Sociaux 2011, 58, 431–444. [Google Scholar] [CrossRef]
- Neveling, D.P.; Endo, A.; Dicks, L.M.T. Fructophilic Lactobacillus kunkeei and Lactobacillus brevis Isolated from Fresh Flowers, Bees and Bee-hives. Curr. Microbiol. 2012, 65, 507–515. [Google Scholar] [CrossRef] [PubMed]
- Moran, N.A. Genomics of the honey bee microbiome. Curr. Opin. Insect Sci. 2015, 10, 22–28. [Google Scholar] [CrossRef] [PubMed]
- Casalone, E.; Cavalieri, D.; Daly, G.; Vitali, F.; Perito, B. Propolis hosts a diversemicrobial community. World J. Microbiol. Biotechnol. 2020, 36, 50. [Google Scholar] [CrossRef] [PubMed]
- Callegari, M.; Crotti, E.; Fusi, M.; Marasco, R.; Gonella, E.; De Noni, I.; Romano, D.; Borin, S.; Tsiamis, G.; Cherif, A.; et al. Compartmentalization of bacterial and fungal microbiomes in the gut of adult honeybees. NPJ Biofilms Microbiomes 2021, 7, 42. [Google Scholar] [CrossRef]
- Cui, P.; Kong, K.; Yao, Y.; Huang, Z.; Shi, S.; Liu, P.; Huang, Y.; Abbas, N.; Yu, L.; Zhang, Y. Community composition, bacterial symbionts, antibacterial and antioxidant activities of honeybee-associated fungi. BMC Microbiol. 2022, 22, 168. [Google Scholar] [CrossRef] [PubMed]
- Decker, L.E.; Juan, P.A.S.; Warren, M.L.; Duckworth, C.E.; Gao, C.; Fukami, T. Higher Variability in Fungi Compared to Bacteria in the Foraging Honey Bee Gut. Microb. Ecol. 2022, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Khan, K.A.; Al-Ghamdi, A.A.; Ghramh, H.A.; Ansari, M.J.; Ali, H.; Alamri, S.A.; Kahtani, S.N.A.; Adgaba, N.; Qasim, M.; Hafeez, M. Structural diversity and functional variability of gut microbial communities associated with honey bees. Microb. Pathog. 2019, 138, 103793. [Google Scholar] [CrossRef] [PubMed]
- Ludvigsen, J.; Andersen, Å.; Hjeljord, L.; Rudi, K. The Honeybee Gut Mycobiota Cluster by Season Versus the Microbiota Which Cluster by Gut Segment. Vet.-Sci. 2020, 8, 4. [Google Scholar] [CrossRef]
- Rosso, G.B.; Engel, P. Functional roles and metabolic niches in the honey bee gut microbiota. Curr. Opin. Microbiol. 2018, 43, 69–76. [Google Scholar] [CrossRef]
- Martinson, V.G.; Moy, J.; Moran, N.A. Establishment of Characteristic Gut Bacteria during Development of the Honeybee Worker. Appl. Environ. Microbiol. 2012, 78, 2830–2840. [Google Scholar] [CrossRef]
- Olofsson, T.C.; Vásquez, A. Detection and Identification of a Novel Lactic Acid Bacterial Flora Within the Honey Stomach of the Honeybee Apis mellifera. Curr. Microbiol. 2008, 57, 356–363. [Google Scholar] [CrossRef] [PubMed]
- Corby-Harris, V.; Maes, P.; Anderson, K.E. The Bacterial Communities Associated with Honey Bee (Apis mellifera) Foragers. PLoS ONE 2014, 9, e95056. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.E.; Martinson, V.G.; Urban-Mead, K.; Moran, N.A. Routes of Acquisition of the Gut Microbiota of the Honey Bee Apis mellifera. Appl. Environ. Microbiol. 2014, 80, 7378–7387. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Bartlett, K.D.; Moran, N.A. The Bacterium Frischella perrara Causes Scab Formation in the Gut of its Honeybee Host. mBio 2015, 6, e00193-15. [Google Scholar] [CrossRef]
- Emery, O.; Schmidt, K.; Engel, P. Immune system stimulation by the gut symbiont Frischella perrara in the honey bee (Apis mellifera). Mol. Ecol. 2017, 26, 2576–2590. [Google Scholar] [CrossRef] [PubMed]
- Engel, P.; Martinson, V.G.; Moran, N.A. Functional diversity within the simple gut microbiota of the honey bee. Proc. Natl. Acad. Sci. USA 2012, 109, 11002–11007. [Google Scholar] [CrossRef]
- Steele, M.I.; Kwong, W.K.; Whiteley, M.; Moran, N.A. Diversification of Type VI Secretion System Toxins Reveals Ancient Antagonism among Bee Gut Microbes. mBio 2017, 8, e01630-17. [Google Scholar] [CrossRef]
- Steele, M.I.; Moran, N.A. Evolution of Interbacterial Antagonism in Bee Gut Microbiota Reflects Host and Symbiont Diversification. mSystems 2021, 6, e00063-21. [Google Scholar] [CrossRef]
- Lugli, G.A.; Fontana, F.; Tarracchini, C.; Mancabelli, L.; Milani, C.; Turroni, F.; Ventura, M. Exploring the biodiversity of Bifidobacterium asteroides among honey bee microbiomes. Environ. Microbiol. 2022. [Google Scholar] [CrossRef]
- Chen, J.; Wang, J.; Zheng, H. Characterization of Bifidobacterium apousia sp. nov., Bifidobacterium choladohabitans sp. nov., and Bifidobacterium polysaccharolyticum sp. nov., three novel species of the genus Bifidobacterium from honey bee gut. Syst. Appl. Microbiol. 2021, 44, 126247. [Google Scholar] [CrossRef]
- Hayashi, K.; Maekawa, I.; Tanaka, K.; Ijyuin, S.; Shiwa, Y.; Suzuki, I.; Niimura, Y.; Kawasaki, S. Purification and characterization of oxygen-inducible haem catalase from oxygen-tolerant Bifidobacterium asteroides. Microbiology 2013, 159, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Ellegaard, K.M.; Brochet, S.; Bonilla-Rosso, G.; Emery, O.; Glover, N.; Hadadi, N.; Jaron, K.S.; van der Meer, J.R.; Robinson-Rechavi, M.; Sentchilo, V.; et al. Genomic changes underlying host specialization in the bee gut symbiont Lactobacillus Firm5. Mol. Ecol. 2019, 28, 2224–2237. [Google Scholar] [CrossRef] [PubMed]
- Jones, J.C.; Fruciano, C.; Hildebrand, F.; Al Toufalilia, H.; Balfour, N.J.; Bork, P.; Engel, P.; Ratnieks, F.L.; Hughes, W.O. Gut microbiota composition is associated with environmental landscape in honey bees. Ecol. Evol. 2017, 8, 441–451. [Google Scholar] [CrossRef]
- Kešnerová, L.; Emery, O.; Troilo, M.; Liberti, J.; Erkosar, B.; Engel, P. Gut microbiota structure differs between honeybees in winter and summer. ISME J. 2019, 14, 801–814. [Google Scholar] [CrossRef] [PubMed]
- Ricigliano, V.A.; Anderson, K.E. Probing the Honey Bee Diet-Microbiota-Host Axis Using Pollen Restriction and Organic Acid Feeding. Insects 2020, 11, 291. [Google Scholar] [CrossRef] [PubMed]
- Kwong, W.K.; Mancenido, A.L.; Moran, N.A. Immune system stimulation by the native gut microbiota of honey bees. R. Soc. Open Sci. 2017, 4, 170003. [Google Scholar] [CrossRef] [PubMed]
- Raymann, K.; Moran, N.A. The role of the gut microbiome in health and disease of adult honey bee workers. Curr. Opin. Insect. Sci. 2018, 26, 97–104. [Google Scholar] [CrossRef]
- Forsgren, E.; Olofsson, T.C.; Váasquez, A.; Fries, I. Novel lactic acid bacteria inhibiting Paenibacillus larvae in honey bee larvae. Apidologie 2009, 41, 99–108. [Google Scholar] [CrossRef]
- Killer, J.; Dubná, S.; Sedláček, I.; Švec, P. Lactobacillus apis sp. nov., from the stomach of honeybees (Apis mellifera), having an in vitro inhibitory effect on the causative agents of American and European foulbrood. Int. J. Syst. Evol. Microbiol. 2014, 64, 152–157. [Google Scholar] [CrossRef]
- Vásquez, A.; Forsgren, E.; Fries, I.; Paxton, R.; Flaberg, E.; Szekely, L.; Olofsson, T.C. Symbionts as Major Modulators of Insect Health: Lactic Acid Bacteria and Honeybees. PLoS ONE 2012, 7, e33188. [Google Scholar] [CrossRef]
- Desai, S.D.; Currie, R.W. Effects of Wintering Environment and Parasite–Pathogen Interactions on Honey Bee Colony Loss in North Temperate Regions. PLoS ONE 2016, 11, e0159615. [Google Scholar] [CrossRef] [PubMed]
- Maes, P.W.; Rodrigues, P.A.P.; Oliver, R.; Mott, B.M.; Anderson, K.E. Diet-related gut bacterial dysbiosis correlates with impaired development, increased mortality and Nosema disease in the honeybee (Apis mellifera). Mol. Ecol. 2016, 25, 5439–5450. [Google Scholar] [CrossRef] [PubMed]
- Hroncova, Z.; Havlik, J.; Killer, J.; Doskocil, I.; Tyl, J.; Kamler, M.; Titera, D.; Hakl, J.; Mrazek, J.; Bunesova, V.; et al. Variation in Honey Bee Gut Microbial Diversity Affected by Ontogenetic Stage, Age and Geographic Location. PLoS ONE 2015, 10, e0118707. [Google Scholar] [CrossRef] [PubMed]
- Rokop, Z.P.; Horton, M.A.; Newton, I.L.G. Interactions between Cooccurring Lactic Acid Bacteria in Honey Bee Hives. Appl. Environ. Microbiol. 2015, 81, 7261–7270. [Google Scholar] [CrossRef]
- Vojvodic, S.; Rehan, S.M.; Anderson, K.E. Microbial Gut Diversity of Africanized and European Honey Bee Larval Instars. PLoS ONE 2013, 8, e72106. [Google Scholar] [CrossRef]
- Lee, F.J.; Rusch, D.B.; Stewart, F.J.; Mattila, H.R.; Newton, I.L.G. Saccharide breakdown and fermentation by the honey bee gut microbiome. Environ. Microbiol. 2014, 17, 796–815. [Google Scholar] [CrossRef]
- Kapheim, K.M.; Rao, V.D.; Yeoman, C.J.; Wilson, B.A.; White, B.A.; Goldenfeld, N.; Robinson, G.E. Caste-Specific Differences in Hindgut Microbial Communities of Honey Bees (Apis mellifera). PLoS ONE 2015, 10, e0123911. [Google Scholar] [CrossRef]
- Tarpy, D.R.; Mattila, H.R.; Newton, I.L.G. Development of the Honey Bee Gut Microbiome throughout the Queen-Rearing Process. Appl. Environ. Microbiol. 2015, 81, 3182–3191. [Google Scholar] [CrossRef]
- Copeland, D.C.; Anderson, K.E.; Mott, B.M. Early Queen Development in Honey Bees: Social Context and Queen Breeder Source Affect Gut Microbiota and Associated Metabolism. Microbiol. Spectr. 2022, 10, e0038322. [Google Scholar] [CrossRef]
- Ding, S.-W.; Lu, R. Virus-derived siRNAs and piRNAs in immunity and pathogenesis. Curr. Opin. Virol. 2011, 1, 533–544. [Google Scholar] [CrossRef]
- Li, C.; Tang, M.; Li, X.; Zhou, X. Community Dynamics in Structure and Function of Honey Bee Gut Bacteria in Response to Winter Dietary Shift. mBio 2022, 13, e0113122. [Google Scholar] [CrossRef] [PubMed]
- Ricigliano, V.A.; Fitz, W.; Copeland, D.C.; Mott, B.M.; Maes, P.; Floyd, A.S.; Dockstader, A.; Anderson, K.E. The impact of pollen consumption on honey bee (Apis mellifera) digestive physiology and carbohydrate metabolism. Arch. Insect Biochem. Physiol. 2017, 96, e21406. [Google Scholar] [CrossRef] [PubMed]
- Castelli, L.; Branchiccela, B.; Romero, H.; Zunino, P.; Antúnez, K. Seasonal Dynamics of the Honey Bee Gut Microbiota in Colonies Under Subtropical Climate. Microb. Ecol. 2021, 83, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Ge, Y.; Jing, Z.; Diao, Q.; He, J.-Z.; Liu, Y.-J. Host Species and Geography Differentiate Honeybee Gut Bacterial Communities by Changing the Relative Contribution of Community Assembly Processes. mBio 2021, 12, e0075121. [Google Scholar] [CrossRef]
- Martinson, V.G.; Danforth, B.; Minckley, R.L.; Rueppell, O.; Tingek, S.; Moran, N.A. A simple and distinctive microbiota associated with honey bees and bumble bees. Mol. Ecol. 2010, 20, 619–628. [Google Scholar] [CrossRef]
- Kwong, W.K.; Moran, N.A. Evolution of host specialization in gut microbes: The bee gut as a model. Gut Microbes 2015, 6, 214–220. [Google Scholar] [CrossRef]
- Chen, Y.E.; Fischbach, M.A.; Belkaid, Y. Skin microbiota–host interactions. Nature 2018, 553, 427–436. [Google Scholar] [CrossRef]
- Parks, O.B.; Kothamasu, K.S.; Ziemba, M.J.; Benner, M.; Cristinziano, M.; Kantz, S.; Leger, D.; Li, J.; Patel, D.; Rabuse, W.; et al. Exposure to cuticular bacteria can alter host behavior in a funnel-weaving spider. Curr. Zool. 2017, 64, 721–726. [Google Scholar] [CrossRef]
- Keyhani, N.O. Lipid biology in fungal stress and virulence: Entomopathogenic fungi. Fungal Biol. 2018, 122, 420–429. [Google Scholar] [CrossRef]
- Birer, C.; Moreau, C.S.; Tysklind, N.; Zinger, L.; Duplais, C. Disentangling the assembly mechanisms of ant cuticular bacterial communities of two Amazonian ant species sharing a common arboreal nest. Mol. Ecol. 2020, 29, 1372–1385. [Google Scholar] [CrossRef]
- Birer, C.; Tysklind, N.; Zinger, L.; Duplais, C. Comparative analysis of DNA extraction methods to study the body surface microbiota of insects: A case study with ant cuticular bacteria. Mol. Ecol. Resour. 2017, 17, e34–e45. [Google Scholar] [CrossRef] [PubMed]
- Mokeev, V.; Flaven-Pouchon, J.; Wang, Y.; Gehring, N.; Moussian, B. Ratio between Lactobacillus plantarum and Acetobacter pomorum on the surface of Drosophila melanogaster adult flies depends on cuticle melanisation. BMC Res. Notes 2021, 14, 351. [Google Scholar] [CrossRef] [PubMed]
- Gilliam, M. Microbiology of Pollen and Bee Bread: The Yeasts. Apidologie 1979, 10, 43–53. [Google Scholar] [CrossRef]
- Regan, T.; Barnett, M.W.; Laetsch, D.R.; Bush, S.; Wragg, D.; Budge, G.E.; Highet, F.; Dainat, B.; De Miranda, J.R.; Watson, M.; et al. Characterisation of the British honey bee metagenome. Nat. Commun. 2018, 9, 4995. [Google Scholar] [CrossRef]
- Scoaris, D.D.O.; Hughes, F.M.; Silveira, M.A.; Evans, J.D.; Pettis, J.S.; Bastos, E.M.A.F.; Rosa, C.A. Microbial communities associated with honey bees in Brazil and in the United States. Braz. J. Microbiol. 2021, 52, 2097–2115. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, M.; Jützeler, M.; França, S.C.; Wäckers, F.; Andreasson, E.; Stenberg, J.A. Bee-Vectored Aureobasidium pullulans for Biological Control of Gray Mold in Strawberry. Phytopathology 2022, 112, 232–237. [Google Scholar] [CrossRef]
- Rueda-Mejia, M.P.; Nägeli, L.; Lutz, S.; Hayes, R.D.; Varadarajan, A.R.; Grigoriev, I.V.; Ahrens, C.H.; Freimoser, F.M. Genome, transcriptome and secretome analyses of the antagonistic, yeast-like fungus Aureobasidium pullulans to identify potential biocontrol genes. Microb. Cell 2021, 8, 184–202. [Google Scholar] [CrossRef]
- Saraiva, M.A.; Zemolin, A.P.P.; Franco, J.L.; Boldo, J.T.; Stefenon, V.M.; Triplett, E.W.; Camargo, F.; Roesch, L. Relationship between honeybee nutrition and their microbial communities. Antonie van Leeuwenhoek 2015, 107, 921–933. [Google Scholar] [CrossRef]
- Garcia-Mazcorro, J.F.; Kawas, J.R.; Marroquin-Cardona, A.G. Descriptive Bacterial and Fungal Characterization of Propolis Using Ultra-High-Throughput Marker Gene Sequencing. Insects 2019, 10, 402. [Google Scholar] [CrossRef] [PubMed]
- Harwood, G.; Salmela, H.; Freitak, D.; Amdam, G. Social immunity in honey bees: Royal jelly as a vehicle in transferring bacterial pathogen fragments between nestmates. J. Exp. Biol. 2021, 224, jeb231076. [Google Scholar] [CrossRef]
- Foley, K.; Fazio, G.; Jensen, A.B.; Hughes, W.O. The distribution of Aspergillus spp. opportunistic parasites in hives and their pathogenicity to honey bees. Vet.-Microbiol. 2014, 169, 203–210. [Google Scholar] [CrossRef] [PubMed]
- Bordenstein, S.R.; Reznikoff, W.S. Mobile DNA in obligate intracellular bacteria. Nat. Rev. Microbiol. 2005, 3, 688–699. [Google Scholar] [CrossRef] [PubMed]
- Bovo, S.; Ribani, A.; Utzeri, V.J.; Schiavo, G.; Bertolini, F.; Fontanesi, L. Shotgun metagenomics of honey DNA: Evaluation of a methodological approach to describe a multi-kingdom honey bee derived environmental DNA signature. PLoS ONE 2018, 13, e0205575. [Google Scholar] [CrossRef] [PubMed]
- Maes, P.; Floyd, A.; Mott, B.; Anderson, K. Overwintering Honey Bee Colonies: Effect of Worker Age and Climate on the Hindgut Microbiota. Insects 2021, 12, 224. [Google Scholar] [CrossRef]
- Corby-Harris, V.; Snyder, L.A.; Schwan, M.R.; Maes, P.; McFrederick, Q.S.; Anderson, K.E. Origin and Effect of Alpha 2.2 Acetobacteraceae in Honey Bee Larvae and Description of Parasaccharibacter apium gen. nov., sp. nov. Appl. Environ. Microbiol. 2014, 80, 7460–7472. [Google Scholar] [CrossRef]
- Ghosh, S.; Namin, S.M.; Jung, C. Differential Bacterial Community of Bee Bread and Bee Pollen Revealed by 16s rRNA High-Throughput Sequencing. Insects 2022, 13, 863. [Google Scholar] [CrossRef]
- Prado, A.; Barret, M.; Vaissière, B.E.; Torres-Cortes, G. Honey bees change the microbiota of pollen. BioRxiv 2022. [Google Scholar] [CrossRef]
- Nicolson, S.W.; Neves, S.D.S.D.; Human, H.; Pirk, C.W. Digestibility and nutritional value of fresh and stored pollen for honey bees (Apis mellifera scutellata). J. Insect Physiol. 2018, 107, 302–308. [Google Scholar] [CrossRef]
- Dharampal, P.S.; Carlson, C.; Currie, C.R.; Steffan, S.A. Pollen-borne microbes shape bee fitness. Proc. R. Soc. B Boil. Sci. 2019, 286, 20182894. [Google Scholar] [CrossRef]
- Di Pasquale, G.; Salignon, M.; Le Conte, Y.; Belzunces, L.P.; Decourtye, A.; Kretzschmar, A.; Suchail, S.; Brunet, J.-L.; Alaux, C. Influence of Pollen Nutrition on Honey Bee Health: Do Pollen Quality and Diversity Matter? PLoS ONE 2013, 8, e72016. [Google Scholar] [CrossRef]
- Vásquez, A.; Olofsson, T.C. The lactic acid bacteria involved in the production of bee pollen and bee bread. J. Apic. Res. 2009, 48, 189–195. [Google Scholar] [CrossRef]
- Mullin, C.A.; Frazier, M.; Frazier, J.L.; Ashcraft, S.; Simonds, R.; Vanengelsdorp, D.; Pettis, J.S. High Levels of Miticides and Agrochemicals in North American Apiaries: Implications for Honey Bee Health. PLoS ONE 2010, 5, e9754. [Google Scholar] [CrossRef] [PubMed]
- Sartori, A.G.D.O.; Cesar, A.S.M.; Woitowicz, F.C.G.; Saliba, A.S.M.C.; Ikegaki, M.; Rosalen, P.L.; Coutinho, L.L.; de Alencar, S.M. Plant genetic diversity by DNA barcoding to investigate propolis origin. Phytochemistry 2022, 200, 113226. [Google Scholar] [CrossRef] [PubMed]
- Saelao, P.; Borba, R.S.; Ricigliano, V.; Spivak, M.; Simone-Finstrom, M. Honeybee microbiome is stabilized in the presence of propolis. Biol. Lett. 2020, 16, 20200003. [Google Scholar] [CrossRef] [PubMed]
- Grubbs, K.J.; Scott, J.; Budsberg, K.J.; Read, H.; Balser, T.C.; Currie, C.R. Unique Honey Bee (Apis mellifera) Hive Component-Based Communities as Detected by a Hybrid of Phospholipid Fatty-Acid and Fatty-Acid Methyl Ester Analyses. PLoS ONE 2015, 10, e0121697. [Google Scholar] [CrossRef]
- Takatani, N.; Endo, A. Viable fructophilic lactic acid bacteria present in honeybee-based food products. FEMS Microbiol. Lett. 2021, 368, fnab150. [Google Scholar] [CrossRef]
- Bovo, S.; Utzeri, V.J.; Ribani, A.; Cabbri, R.; Fontanesi, L. Shotgun sequencing of honey DNA can describe honey bee derived environmental signatures and the honey bee hologenome complexity. Sci. Rep. 2020, 10, 9279. [Google Scholar] [CrossRef]
- Yun, J.-H.; Jung, M.-J.; Kim, P.S.; Bae, J.-W. Social status shapes the bacterial and fungal gut communities of the honey bee. Sci. Rep. 2018, 8, 2019. [Google Scholar] [CrossRef]
- Wirta, H.; Abrego, N.; Miller, K.; Roslin, T.; Vesterinen, E. DNA traces the origin of honey by identifying plants, bacteria and fungi. Sci. Rep. 2021, 11, 4798. [Google Scholar] [CrossRef]
- Liu, S.; Lang, D.; Meng, G.; Hu, J.; Tang, M.; Zhou, X. Tracing the origin of honey products based on metagenomics and machine learning. Food Chem. 2021, 371, 131066. [Google Scholar] [CrossRef]
- Kačániová, M.; Kňazovická, V.; Felšöciová, S.; Rovná, K. Microscopic fungi recovered from honey and their toxinogenity. J. Environ. Sci. Health Part A 2012, 47, 1659–1664. [Google Scholar] [CrossRef] [PubMed]
- Cox-Foster, D.L.; Conlan, S.; Holmes, E.C.; Palacios, G.; Evans, J.D.; Moran, N.A.; Quan, P.-L.; Briese, T.; Hornig, M.; Geiser, D.M.; et al. A Metagenomic Survey of Microbes in Honey Bee Colony Collapse Disorder. Science 2007, 318, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Crovadore, J.; Gérard, F.; Chablais, R.; Cochard, B.; Jensen, K.K.B.; Lefort, F. Deeper Insight in Beehives: Metagenomes of Royal Jelly, Pollen, and Honey from Lavender, Chestnut, and Fir Honeydew and Epiphytic and Endophytic Microbiota of Lavender and Rose Flowers. Genome Announc. 2017, 5, e00425-17. [Google Scholar] [CrossRef] [PubMed]
- Snowdon, J.A.; Cliver, D.O. Microorganisms in honey. Int. J. Food Microbiol. 1996, 31, 1–26. [Google Scholar] [CrossRef] [PubMed]
- Kačániová, M.; Pavličová, S.; Haščík, P.; Kociubinski, G.; Kňazovická, V.; Sudzina, M.; Sudzinová, J.; Fikselová, M. Microbial communities in bees, pollen and honey from Slovakia. Acta Microbiol. Immunol. Hung. 2009, 56, 285–295. [Google Scholar] [CrossRef]
- Shade, A.; McManus, P.S.; Handelsman, J. Unexpected Diversity during Community Succession in the Apple Flower Microbiome. mBio 2013, 4, e00602-12. [Google Scholar] [CrossRef]
- Brochet, S.; Quinn, A.; Mars, R.A.; Neuschwander, N.; Sauer, U.; Engel, P. Niche partitioning facilitates coexistence of closely related honey bee gut bacteria. eLife 2021, 10, e68583. [Google Scholar] [CrossRef]
- Fenner, E.D.; Scapini, T.; Diniz, M.D.C.; Giehl, A.; Treichel, H.; Álvarez-Pérez, S.; Alves, S.L. Nature’s Most Fruitful Threesome: The Relationship between Yeasts, Insects, and Angiosperms. J. Fungi 2022, 8, 984. [Google Scholar] [CrossRef]
- Álvarez-Pérez, S.; Lievens, B.; Fukami, T. Yeast–Bacterium Interactions: The Next Frontier in Nectar Research. Trends Plant Sci. 2019, 24, 393–401. [Google Scholar] [CrossRef]
- Rering, C.C.; Beck, J.J.; Hall, G.W.; McCartney, M.M.; Vannette, R.L. Nectar-inhabiting microorganisms influence nectar volatile composition and attractiveness to a generalist pollinator. N. Phytol. 2017, 220, 750–759. [Google Scholar] [CrossRef]
- Good, A.P.; Gauthier, M.-P.L.; Vannette, R.; Fukami, T. Honey Bees Avoid Nectar Colonized by Three Bacterial Species, But Not by a Yeast Species, Isolated from the Bee Gut. PLoS ONE 2014, 9, e86494. [Google Scholar] [CrossRef] [PubMed]
- Steele, M.I.; Motta, E.V.S.; Gattu, T.; Martinez, D.; Moran, N.A. The Gut Microbiota Protects Bees from Invasion by a Bacterial Pathogen. Microbiol. Spectr. 2021, 9, e0039421. [Google Scholar] [CrossRef] [PubMed]
- Hubert, J.; Kamler, M.; Nesvorna, M.; Ledvinka, O.; Kopecky, J.; Erban, T. Comparison of Varroa destructor and Worker Honeybee Microbiota Within Hives Indicates Shared Bacteria. Microb. Ecol. 2016, 72, 448–459. [Google Scholar] [CrossRef] [PubMed]
- Balakrishnan, B.; Wu, H.; Cao, L.; Zhang, Y.; Li, W.; Han, R. Immune Response and Hemolymph Microbiota of Apis mellifera and Apis cerana After the Challenge with Recombinant Varroa Toxic Protein. J. Econ. Entomol. 2021, 114, 1310–1320. [Google Scholar] [CrossRef]
- Huang, Q.; Lopez, D.; Evans, J.D. Shared and unique microbes between Small hive beetles (Aethina tumida) and their honey bee hosts. MicrobiologyOpen 2019, 8, e899. [Google Scholar] [CrossRef]
- De Landa, G.F.; Porrini, M.P.; Revainera, P.; Farina, J.; Correa-Benítez, A.; Maggi, M.D.; Eguaras, M.J.; Quintana, S. Pathogens Detection in the Small Hive Beetle (Aethina tumida (Coleoptera: Nitidulidae)). Neotropical Entomol. 2020, 50, 312–316. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smutin, D.; Lebedev, E.; Selitskiy, M.; Panyushev, N.; Adonin, L. Micro”bee”ota: Honey Bee Normal Microbiota as a Part of Superorganism. Microorganisms 2022, 10, 2359. https://doi.org/10.3390/microorganisms10122359
Smutin D, Lebedev E, Selitskiy M, Panyushev N, Adonin L. Micro”bee”ota: Honey Bee Normal Microbiota as a Part of Superorganism. Microorganisms. 2022; 10(12):2359. https://doi.org/10.3390/microorganisms10122359
Chicago/Turabian StyleSmutin, Daniil, Egor Lebedev, Maxim Selitskiy, Nick Panyushev, and Leonid Adonin. 2022. "Micro”bee”ota: Honey Bee Normal Microbiota as a Part of Superorganism" Microorganisms 10, no. 12: 2359. https://doi.org/10.3390/microorganisms10122359
APA StyleSmutin, D., Lebedev, E., Selitskiy, M., Panyushev, N., & Adonin, L. (2022). Micro”bee”ota: Honey Bee Normal Microbiota as a Part of Superorganism. Microorganisms, 10(12), 2359. https://doi.org/10.3390/microorganisms10122359






