Degradation of Cellulose and Hemicellulose by Ruminal Microorganisms
Abstract
1. Introduction
2. The Ruminant Digestive System
3. Ruminal Cellulolytic Microbes
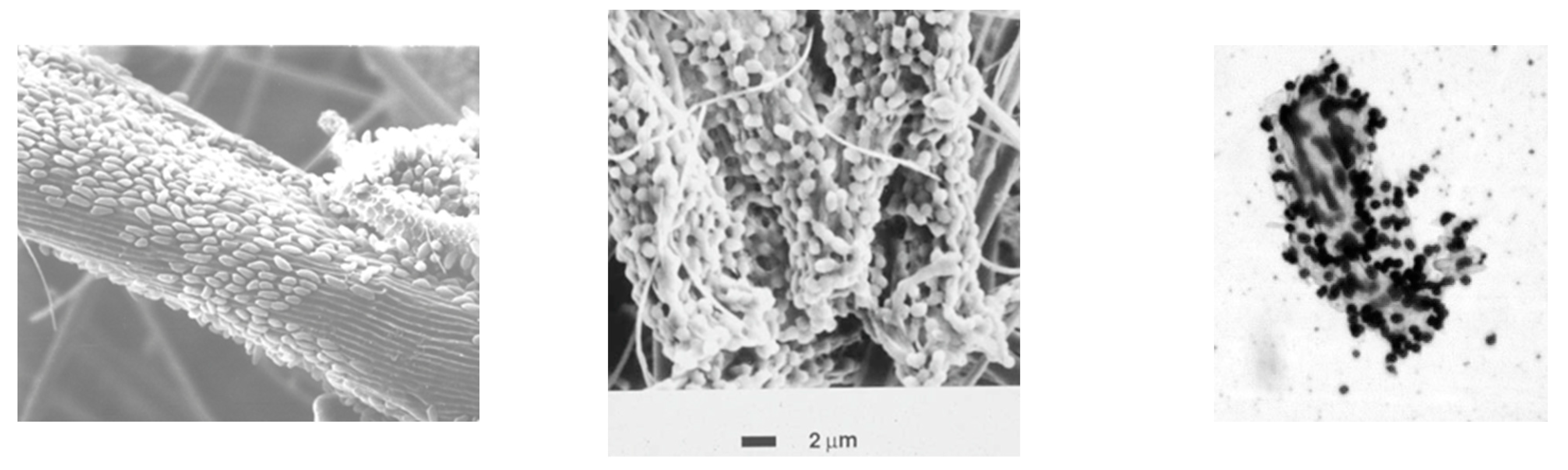
3.1. Bacteria
3.1.1. Fibrobacter Succinogenes
3.1.2. Ruminococcus
3.1.3. Other Cellulolytic Bacteria
3.1.4. Bacterial Utilization of Hydrolytic Products
3.2. Protists
3.3. Fungi
3.4. Kinetics of Ruminal Cellulose Degradation
| Inoculum | Substrate | Initial Concentration (g L−1) | k (d−1) c | References |
|---|---|---|---|---|
| Mixed ruminal | Alfalfa cellulose | 1.46–2.45 | [74] | |
| Mixed ruminal | Whatman filter paper | 1.68 | [79] | |
| Mixed ruminal | Cotton | 0.96 | [79] | |
| Mixed ruminal | Sigmacell 50 MCC a | 10 | 1.85 | [75] |
| Mixed ruminal | Sigmacell 50 MCC | 10 | 2.40 | [77] |
| Sludge digester | ND b | 0.04–0.13 | [84] | |
| Sludge digester | “Cellulose powder” | 5 | 0.94 | [85] |
| Sludge digester | Filter paper | 2 | 0.247 | [86] |
| Sludge digester | ND b | 0.1 | [87] | |
| Sludge digester | ND b | 0.066 | [88] | |
| Sludge digester | Sigmacell 20 MCC | 20 | 0.252 | [89] |
3.5. Quantitative Comparison of Ruminal versus Non-Ruminal Cellulose Degradation
3.6. Hemicellulose Degradation
3.6.1. Bacterial Hemicellulose Degradation
3.6.2. Hemicellulose Degradation by Eucaryotes
3.7. Kinetics of Hemicellulose Degradation
3.8. Microbial Interactions
3.8.1. Interactions among Cellulolytic Bacteria
3.8.2. Interactions between Cellulolytic and Non-Cellulolytic Bacteria
3.8.3. Interactions among Cellulolytic Eucaryotes
3.8.4. Inter-Kingdom Interactions
3.8.5. Ecological Implications
3.9. Potential for Industrial Exploitation of Ruminal Fiber Fermentation
3.9.1. Carboxylate Production
3.9.2. Ethanol Production
3.9.3. Microbial Fuel Cells
3.9.4. Bioaugmentation of Anaerobic Digesters
3.10. Knowledge Gaps and Research Opportunities
3.10.1. Undiscovered Cellulolytic and Hemicellulolytic Species
3.10.2. Quantitative Aspects of Cellulosics Degradation by Ruminal Eucaryotes
3.10.3. Overcoming the Recalcitrance of Cellulosic Substrates
4. Concluding Remarks
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hungate, R.E. The Rumen and Its Microbes; Academic Press: New York, NY, USA, 1966. [Google Scholar]
- Russell, J.B. Rumen Microbiology and Its Role in Ruminant Nutrition; James B. Russell: Ithaca, NY, USA, 2002. [Google Scholar]
- Lynd, L.R.; Weimer, P.J.; Van Zyl, W.H.; Pretorius, I.S. Microbial cellulose utilization: Fundamentals and biotechnology. Microbiol. Mol. Biol. Rev. 2002, 66, 506–577. [Google Scholar] [CrossRef] [PubMed]
- Wilson, J.R. Organization of forage plant tissues. In Forage Cell Wall Structure and Digestibility; Jung, H.G., Buxton, D.R., Hatfield, R.D., Ralph, J., Eds.; American Society of Agronomy; Crop Science Society of America; Soil Science Society of America: Madison, WI, USA, 1993; pp. 1–32. [Google Scholar] [CrossRef]
- Van Soest, P.J. Feeding strategies, taxonomy, and evolution. In Nutritional Ecology of the Ruminant, 2nd ed.; Cornell University Press: Ithaca, NY, USA, 1994; pp. 22–39. [Google Scholar]
- Hartnell, G.F.; Satter, L.D. Determination of rumen fill, retention tme and ruminal turnover rates of ingesta at different stages of lactation in dairy cows. J. Anim. Sci. 1979, 48, 381–392. [Google Scholar] [CrossRef] [PubMed]
- Udén, P.; Rounsaville, T.; Wiggans, G.; Van Soest, P. The measurement of liquid and solid digesta retention in ruminants, equines and rabbits given timothy (Phleum pratense) hay. Br. J. Nutr. 1982, 48, 329–339. [Google Scholar] [CrossRef]
- Mambrini, M.; Peyraud, J.L. Retention time of feed particles and liquids in the stomachs and intestines of dairy cows. Direct measurement and calculations based on faecal collection. Reprod. Nutr. Dev. 1997, 37, 427–442. [Google Scholar] [CrossRef] [PubMed]
- Jouany, J.P.; Demeyer, D.I.; Grain, J. Effect of defaunating the rumen. Anim. Feed Sci. Technol. 1988, 21, 229–265. [Google Scholar] [CrossRef]
- Weimer, P.J.; Hall, M.B. The potential for biomimetic application of rumination to bioreactor design. Biomass Bioenergy 2020, 143, 105882. [Google Scholar] [CrossRef]
- Weimer, P.J.; Russell, J.B.; Muck, R.E. Lessons from the cow: What the rumen fermentation can teach us about consolidated bioprocessing of cellulosic biomass. Bioresour. Technol. 2009, 100, 5323–5331. [Google Scholar] [CrossRef]
- Beauchemin, K.A. Invited review: Current perspectives on eating and rumination activity in dairy cows. J. Dairy Sci. 2018, 101, 4762–4784. [Google Scholar] [CrossRef]
- Jami, E.; Israel, A.; Kotser, A.; Mizrahi, I. Exploring the bovine rumen bacterial community from birth to adulthood. ISME J. 2013, 7, 1069–1079. [Google Scholar] [CrossRef]
- Moraïs, S.; Mizrahi, I. Islands in the stream: From individual to communal fiber degradation in the rumen ecosystem. FEMS Microbiol. Rev. 2019, 43, 362–379. [Google Scholar] [CrossRef]
- Delfosse-Debusscher, J.; Thines-Sempoux, D.; Vanbelle, M.; Latteur, B. Contribution of protozoa to the rumen cellulolytic activity. Ann. Res. Vet. 1979, 10, 255–257. [Google Scholar]
- Lee, S.S.; Ha, J.K.; Cheng, K. Relative contributions of bacteria, protozoa, and fungi to in vitro degradation of orchard grass cell walls and their interactions. Appl. Environ. Microbiol. 2000, 66, 3807–3813. [Google Scholar] [CrossRef]
- Stevenson, D.M.; Weimer, P.J. Dominance of Prevotella and low abundance of classical ruminal bacterial species in the bovine rumen revealed by relative quantification real-time PCR. Appl. Microbiol. Biotechnol. 2007, 75, 165–174. [Google Scholar] [CrossRef]
- Weimer, P.J.; Stevenson, D.M.; Mertens, D.R.; Thomas, E.E. Effect of monensin feeding and withdrawal on populations of individual bacterial species in the rumen of lactating dairy cows fed high-starch rations. Appl. Microbiol. Biotechnol. 2008, 80, 135–145. [Google Scholar] [CrossRef]
- Weimer, P.J.; Stevenson, D.M.; Mertens, D.R.; Hall, M.B. Fiber digestion, VFA production, and microbial population changes during in vitro ruminal fermentations of mixed rations in the absence or presence of monensin. Anim. Feed Sci. Technol. 2011, 169, 68–78. [Google Scholar] [CrossRef]
- Rico, D.E.; Preston, S.H.; Risser, J.M.; Harvatine, K.J. Rapid changes in key ruminal microbial populations during the induction of and recovery from diet-induced milk fat depression in dairy cows. Br. J. Nutr. 2015, 114, 358–367. [Google Scholar] [CrossRef]
- Liu, Y.R.; Du, H.S.; Wu, Z.Z.; Wang, C.; Liu, Q.; Guo, G.; Huo, W.J.; Zhang, Y.I.; Pei, C.X.; Zhang, S.L. Branched-chain volatile fatty acids and folic acid accelerated the growth of Holstein dairy calves by stimulating nutrient digestion and rumen metabolism. Animal 2020, 14, 1176–1183. [Google Scholar] [CrossRef]
- Wells, J.E.; Russell, J.B.; Shi, Y.; Weimer, P.J. Cellodextrin efflux by the ruminal cellulolytic bacterium Fibrobacter succinogenes and its potential role in the growth of nonadherent bacteria. Appl. Environ. Microbiol. 1995, 61, 1757–1762. [Google Scholar] [CrossRef]
- Gong, J.; Forsberg, C.W. Factors affecting adhesion of Fibrobacter succinogenes subsp. succinogenes S85 and adherence-defective mutants to cellulose. Appl. Environ. Microbiol. 1989, 55, 3039–3044. [Google Scholar] [CrossRef]
- Rasmussen, M.A.; Hespell, R.B.; White, B.A.; Bothast, R.J. Inhibitory effects of methylcellulose on cellulose degradation by Ruminococcus flavefaciens. Appl. Environ. Microbiol. 1988, 54, 890–897. [Google Scholar] [CrossRef]
- Stack., R.J.; Cotta, M.A. Effect of 3-phenylpropanoic acid on growth of and cellulose utilization by cellulolytic ruminal bacteria. Appl. Environ. Microbiol. 1986, 52, 209–210. [Google Scholar] [CrossRef]
- Morrison, M.; Mackie, R.I.; Kistner, A. 3-Phenylpropanoic acid improves the affinity of Ruminococcus albus for cellulose in continuous culture. Appl. Environ. Microbiol. 1990, 56, 3220–3222. [Google Scholar] [CrossRef] [PubMed]
- Weimer, P.J. Effect of dilution rate and pH on the ruminal cellulolytic bacterium Fibrobacter succinogenes S85 in cellulose-fed continuous culture. Arch. Microbiol. 1993, 160, 88–294. [Google Scholar] [CrossRef] [PubMed]
- Pavlostathis, S.G.; Miller, Y.L.; Wolin, M.J. Kinetics of insoluble cellulose fermentation by continuous cultures of Ruminococcus albus. Appl. Environ. Microbiol. 1988, 54, 2660–2663. [Google Scholar] [CrossRef] [PubMed]
- Weimer, P.J.; Shi, Y.; Odt, C.L. A segmented gas/liquid delivery system for continuous culture of microorganisms on solid substrates, and its use for growth of Ruminococcus flavefaciens on cellulose. Appl. Microbiol. Biotechnol. 1991, 36, 178–183. [Google Scholar] [CrossRef]
- Weimer, P.J.; French, A.D.; Calamari, T.A., Jr. Differential fermentation of cellulose allomorphs by ruminal cellulolytic bacteria. Appl. Environ. Microbiol. 1991, 57, 3101–3106. [Google Scholar] [CrossRef]
- Montgomery, L.; Flesher, B.; Stahl, D. Transfer of Bacteriodes succinogenes (Hungate) to Fibrobacter gen. nov. as Fibrobacter succinogenes comb. nov. and description of Fibrobacter intestinalis sp. nov. Int. J. Syst. Bacteriol. 1988, 38, 430–435. [Google Scholar] [CrossRef]
- Garrity, G.M.; Holt, J.G. The road map to the manual. In Bergey’s Manual of Systematic Bacteriology, 2nd ed.; Boone, D.R., Castenholz, R.W., Garrity, G.M., Eds.; Springer: New York, NY, USA, 2001; Volume 1, pp. 119–166. [Google Scholar]
- Ransom-Jones, E.; Jones, D.L.; McCarthy, A.J.; McDonald, J.E. The Fibrobacteres: An important phylum of cellulose-degrading bacteria. Microb. Ecol. 2012, 63, 267–281. [Google Scholar] [CrossRef]
- Suen, G.; Weimer, P.J.; Stevenson, D.M.; Aylward, F.O.; Boyum, J.; Deneke, J.; Drinkwater, C.; Mikhailova, N.; Ivanova, N.; Chertkov, I.; et al. The complete genome sequence of Fibrobacter succinogenes S85 reveals a cellulolytic and metabolic specialist. PLoS ONE 2011, 6, e0018814. [Google Scholar] [CrossRef]
- Burnet, M.C.; Dohnalkova, A.C.; Neumann, A.P.; Lipton, M.S.; Smith, R.D.; Suen, G.; Callister, S.J. Evaluating models of cellulose degradation by Fibrobacter succinogenes S85. PLoS ONE 2015, 10, e0143809. [Google Scholar] [CrossRef]
- Raut, M.P.; Couto, N.; Karunakaran, E.; Biggs, C.A.; Wright, P.C. Deciphering the unique cellulose degradation mechanism of the ruminal bacterium Fibrobacter succinogenes S85. Sci. Rep. 2019, 9, 16542. [Google Scholar] [CrossRef]
- Neumann, A.P.; Weimer, P.J.; Suen, G. A global analysis of gene expression in Fibrobacter succinogenes S85 grown on cellulose and soluble sugars at different growth rates. Biotechnol. Biofuels 2018, 11, 295. [Google Scholar] [CrossRef]
- Riederer, A.; Takasuka, T.E.; Makino, S.; Stevenson, D.M.; Bukhman, Y.; Elsen, N.L.; Fox, B.G. Global gene expression patterns in Clostridium thermocellum as determined by microarray analysis of chemostat cultures on cellulose or cellobiose. Appl. Environ. Microbiol. 2011, 77, 1243–1253. [Google Scholar] [CrossRef]
- Kudo, H.; Cheng, K.-J.; Costerton, J.W. Electron microscopic study of the methylcellulose-mediated detachment of cellulolytic rumen bacteria. Can. J. Microbiol. 1987, 33, 287–292. [Google Scholar] [CrossRef]
- Weimer, P.J.; Hatfield, R.D.; Buxton, D.R. Inhibition of ruminal cellulose fermentation by extracts of the perennial legume cicer milkvetch (Astragalus cicer). Appl. Environ. Microbiol. 1993, 59, 405–409. [Google Scholar] [CrossRef]
- Weimer, P.J.; Odt, C.L. Cellulose degradation by ruminal microbes: Physiological and hydrolytic diversity among ruminal cellulolytic bacteria. Am. Chem. Soc. Symp. Ser. 1995, 618, 291–304. [Google Scholar]
- Christopherson, M.R.; Dawson, J.A.; Stevenson, D.M.; Cunningham, A.C.; Bramhacharya, S.; Weimer, P.J.; Kendziorski, C.; Suen, G. Unique aspects of fiber degradation by the ruminal ethanologen Ruminococcus albus 7 revealed by physiological and transcriptomic analysis. BMC Genom. 2014, 15, 1066. [Google Scholar] [CrossRef]
- Latham, M.J.; Brooker, B.E.; Pettipher, G.L.; Harris, P.J. Ruminococcus flavefaciens cell coat and adhesion to cotton cellulose and to cell walls in leaves of perennial ryegrass (Lolium perenne). Appl. Environ. Microbiol. 1978, 35, 156–165. [Google Scholar] [CrossRef]
- Suen, G.; Stevenson, D.M.; Bruce, D.C.; Chertkov, O.; Copeland, A.; Cheng, J.F.; Detter, C.; Detter, J.C.; Goodwin, L.A.; Han, C.S.; et al. Complete genome of the cellulolytic ruminal bacterium Ruminococcus albus 7. J. Bacteriol. 2011, 193, 5574–5575. [Google Scholar] [CrossRef]
- Morrison, M.; Pope, P.B.; Denman, S.E.; McSweeney, C.S. Plant biomass degradation by gut microbiomes: More of the same or something new? Curr. Opin. Biotechnol. 2009, 20, 358–363. [Google Scholar] [CrossRef]
- Israeli-Ruimy, V.; Bule, P.; Jindou, S.; Dassa, B.; Moraïs, S.; Borovok, I.; Barak, Y.; Slutzki, M.; Hamberg, Y.; Cardoso, V.; et al. Complexity of the Ruminococcus flavefaciens FD-1 cellulosome reflects an expansion of family-related protein-protein interactions. Sci. Rep. 2017, 7, 42355. [Google Scholar] [CrossRef] [PubMed]
- Bule, P.; Alves, V.D.; Israeli-Ruimy, V.; Carvalho, A.L.; Ferreira, L.M.A.; Smith, S.P.; Gilbert, H.J.; Najmudin, S.; Bayer, E.A.; Fontes, C.M.G.A. Assembly of Ruminococcus flavefaciens cellulosome revealed by structures of two cohesion-dockerin complexes. Sci. Rep. 2017, 7, 759. [Google Scholar] [CrossRef] [PubMed]
- Hungate, R.E. Microorganisms in the rumen of cattle fed a constant ration. Can. J. Microbiol. 1957, 3, 289–311. [Google Scholar] [CrossRef] [PubMed]
- Varel, V.H. Reisolation and characterization of Clostridium longisporum, a ruminal sporeforming cellulolytic anaerobe. Arch. Microbiol. 1989, 152, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Varel, V.H.; Yen, J.T.; Kreikemeier, K.K. Addition of cellulolytic clostridia to the bovine rumen and pig intestinal tract. Appl. Environ. Microbiol. 1995, 61, 1116–1119. [Google Scholar] [CrossRef]
- Van Gyslwyk, N.O.; Van Der Toorn, J.J.T.K. Description and designation of a neotype strain of Eubacterium cellulosolvens (Cillobacterium cellulosolvens Bryant, Small, Bouma and Robinson) Holdeman and Moore. Int. J. Syst. Microbiol. 1986, 36, 275–277. [Google Scholar] [CrossRef][Green Version]
- Ludwig, W.; Viver, T.; Westram, R.; Francisco Gago, J.; Bustos-Caparros, E.; Knittel, K.; Amann, R.; Rossello-Mora, R. Release LTP_12_2020, featuring a new ARB alignment and improved 16S rRNA tree for prokaryotic type strains. Syst. Appl. Microbiol. 2021, 44, 126218. [Google Scholar] [CrossRef]
- Shi, Y.; Weimer, P.J. Utilization of individual cellodextrins by three predominant ruminal cellulolytic bacteria. Appl. Environ. Microbiol. 1996, 62, 1084–1088. [Google Scholar] [CrossRef]
- Gaudet., G.; Forano, E.; Dauphin, G.; Delort, A.M. Futile cycling of glycogen in Fibrobacter succinogenes as shown by in situ 1H-NMR and 13C-NMR investigation. Eur. J. Biochem. 1992, 207, 155–162. [Google Scholar] [CrossRef]
- Bibollet, X.; Bosc, N.; Matulova, M.; Delort, A.M.; Gaudet, G.; Forano, E. 13C and 1H NMR study of cellulose metabolism by Fibrobacter succinogenes S85. J. Biotechnol. 2000, 77, 37–47. [Google Scholar] [CrossRef]
- Weimer, P.J.; Price, N.; Kroukamp, O.; Joubert, L.M.; Wolfaardt, G.M.; Van Zyl, W.H. Studies of the extracellular glycocalyx of the anaerobic cellulolytic bacterium Ruminococcus albus. Appl. Environ. Microbiol. 2006, 72, 7559–7566. [Google Scholar] [CrossRef][Green Version]
- Williams, A.G.; Coleman, G.S. The Rumen Protozoa; Springer: New York, NY, USA, 1992. [Google Scholar]
- Dehority, B.A. Improved in vitro procedure for maintaining stock cultures of three genera of rumen protozoa. J. Anim. Sci. 2008, 86, 1395–1401. [Google Scholar] [CrossRef]
- Patel, S.; Ambalam, P. Role of rumen protozoa: Metabolic and fibrolytic. Adv. Biotechnol. Microbiol. 2018, 10, AIMB.MS.ID.555793. [Google Scholar] [CrossRef]
- Newbold, C.J.; de la Fuente, G.; Belanche, A.; Ramos-Morales, E.; McEwan, N.R. The role of ciliate protozoa in the rumen. Front. Microbiol. 2015, 6, 1313. [Google Scholar] [CrossRef]
- Ankrah, P.; Loerch, S.C.; Dehority, B.A. Sequestration, migration and lysis of protozoa in the rumen. J. Gen. Microbiol. 1990, 136, 1869–1875. [Google Scholar] [CrossRef]
- Dehority, B.A. Physiological characteristics of several rumen protozoa grown in vitro with observations on within and among species variation. Eur. J. Protistol. 2010, 46, 271–279. [Google Scholar] [CrossRef]
- Akin, D.E.; Amos, H.E. Mode of attack on orchardgrass leaf blades by rumen protozoa. Appl. Environ. Microbiol. 1979, 37, 332–338. [Google Scholar] [CrossRef]
- Morgavi, D.P.; Sakurada, M.; Mizokami, M.; Tomita, Y.; Onodera, R. Effects of ruminal protozoa on cellulose degradation and the growth of an anaerobic ruminal fungus, Piromyces sp. strain OTS1, in vitro. Appl. Environ. Microbiol. 1994, 60, 3718–3723. [Google Scholar] [CrossRef]
- Michalowski, T.; Rybicka, K.; Wereszka, K.; Kaperowicz, A. Ability of the rumen ciliate Epidinium ecaudatum to digest and use crystalline cellulose and xylan for in vitro growth. Acta Protozool. 2001, 40, 203–210. [Google Scholar]
- Czauderna, M.; Wereszka, K.; Michałowski, T. The utilization and digestion of cellulose by the rumen ciliate Diploplastron affine. Eur. J. Protistol. 2019, 68, 17–24. [Google Scholar] [CrossRef]
- Coleman, G.S. The rate of uptake and metabolism of starch grains and cellulose particles by Entodinium species, Eudiplodinium maggii, some other entodiniomorphid protozoa and natural protozoal populations taken from the ovine rumen. J. Appl Bacteriol. 1992, 73, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Orpin, C.G. The occurrence of chitin in the cell walls of the rumen organisms Neocallimastix frontalis, Piromonas communis and Sphaeromonas communis. J. Gen. Microbiol. 1977, 99, 215–218. [Google Scholar] [CrossRef] [PubMed]
- Hess, M.; Paul, S.S.; Puniya, A.K.; van der Giezen, M.; Shaw, C.; Edwards, J.E.; Fliegerová, K. Anaerobic fungi: Past, present, and future. Front. Microbiol. 2020, 11, 584893. [Google Scholar] [CrossRef] [PubMed]
- Cheng, K.J.; Kudo, H.; Duncan, S.H.; Mesbah, A.; Stewart, C.S.; Bernalier, A.; Fonty, G.; Costerton, J.W. Prevention of fungal colonization and digestion of cellulose by the addition of methylcellulose. Can. J. Microbiol. 1991, 37, 484–487. [Google Scholar] [CrossRef] [PubMed]
- Vinzelj, J.; Joshi, A.; Young, D.; Begovic, L.; Peer, N.; Mosberger, L.; Luedi, K.C.S.; Insam, H.; Flad, V.; Nagler, M.; et al. No time to die: Comparative study on preservation protocols for anaerobic fungi. Front. Microbiol. 2022, 13, 978028. [Google Scholar] [CrossRef]
- Borneman, W.S.; Akin, D.E.; Ljungdahl, L.G. Fermentation products and plant cell wall-degrading enzymes produced by monocentric and polycentric anaerobic ruminal fungi. Appl. Environ. Microbiol. 1989, 55, 1066–1073. [Google Scholar] [CrossRef]
- Rezaeian, M.; Beakes, G.W.; Parker, D.S. Distribution and estimation of anaerobic zoosporic fungi along the digestive tracts of sheep. Mycol. Res. 2004, 108, 1227–1233. [Google Scholar] [CrossRef]
- Waldo, D.R.; Smith, L.W.; Cox, E.L. Models of cellulose disappearance from the rumen. J. Dairy Sci. 1972, 55, 125–128. [Google Scholar] [CrossRef]
- Weimer, P.J.; Lopez-Guisa, J.M.; French, A.D. Effect of cellulose fine structure on kinetics of its digestion by mixed ruminal microorganisms in vitro. Appl. Environ. Microbiol. 1990, 56, 2421–2429. [Google Scholar] [CrossRef]
- Van Soest, P.J. The uniformity and nutritive availability of cellulose. Fed. Proc. 1973, 32, 1804–1808. [Google Scholar]
- Mouriño, F.; Akkarawongsa, R.; Weimer, P.J. pH at the initiation of cellulose digestion determines cellulose digestion rate in vitro. J. Dairy Sci. 2001, 48, 848–859. [Google Scholar] [CrossRef]
- Jensen, P.D.; Hardin, M.T.; Clarke, W.P. Effect of biomass concentration and inoculum source on the rate of anaerobic cellulose solubilization. Bioresour. Technol. 2009, 100, 5219–5225. [Google Scholar] [CrossRef]
- Van Soest, P.J. Plant fiber and its role in herbivore nutrition. Cornell Vet. 1977, 67, 307–326. [Google Scholar]
- Russell, J.B.; Dombrowski, D.N. Effect of pH on the efficiency of growth by pure cultures of rumen bacteria in continuous culture. Appl. Environ. Microbiol. 1980, 39, 604–610. [Google Scholar] [CrossRef]
- Slyter, L.L. Ability of pH-selected mixed ruminal microbial populations to digest fiber at various pHs. Appl. Environ. Microbiol. 1986, 52, 390–391. [Google Scholar] [CrossRef]
- Hu, Z.-H.; Wang, G.; Yu, H.-Q. Anaerobic degradation of cellulose by rumen microorganisms at various pH values. Biochem. Eng. J. 2004, 21, 59–62. [Google Scholar] [CrossRef]
- Hatfield, R.D. Cell wall polysaccharide interactions and degradability. In Forage Cell Wall Structure and Digestibility; Jung, H.G., Buxton, D.R., Hatfield, R.D., Ralph, J., Eds.; American Society of Agronomy; Crop Science Society of America; Soil Science Society of America: Madison, WI, USA, 1993; pp. 285–313. [Google Scholar] [CrossRef]
- Gujer, W.; Zehnder, A.J.B. Conversion processes in anaerobic digestion. Water Sci. Technol. 1983, 15, 127–167. [Google Scholar] [CrossRef]
- Noike, T.; Endo, G.; Chang, J.-E.; Matsumoto, J.I. Characteristics of carbohydrate degradation and the rate-limiting step in anaerobic digestion. Biotechnol. Bioeng. 1985, 27, 1482–1489. [Google Scholar] [CrossRef]
- Tong, X.; Smith, L.H.; McCarty, P.L. Methane fermentation of selected lignocellulosic materials. Biomass 1990, 21, 239–255. [Google Scholar] [CrossRef]
- Vavilin, V.A.; Rytov, S.V.; Lokshina, L.Y. A description of hydrolysis kinetics in anaerobic degradation of particulate organic matter. Bioresour. Technol. 1996, 56, 229–237. [Google Scholar] [CrossRef]
- Liebetrau, J.; Kraft, E.; Bidlingmaier, W. The influence of the hydrolysis rate of co-substrates on process behaviour. In Proceedings of the Tenth World Congress on Anaerobic Digestion; Guiot, S.G., Ed.; Canadian Association on Water Quality: Montreal, QC, Canada, 2004; pp. 1296–1300. [Google Scholar]
- Bolaji, I.O.; Dionisi, D. Experimental investigation and mathematical modelling of batch and semi-continuous anaerobic digestion of cellulose at high concentrations and long residence times. SN Appl. Sci. 2021, 3, 778. [Google Scholar] [CrossRef]
- Gonzalez-Estrella, J.; Asato, C.M.; Stone, J.J.; Gilcrease, P.C. A review of anaerobic digestion of paper and paper board waste. Rev. Environ. Sci. Bio Technol. 2017, 16, 569–590. [Google Scholar] [CrossRef]
- Bargo, F.; Muller, L.D.; Delahoy, J.E.; Cassidy, T.W. Milk response to concentrate supplementation of high producing dairy cows grazing at two pasture allowances. J. Dairy Sci. 2002, 85, 1777–1792. [Google Scholar] [CrossRef] [PubMed]
- Hristov, A.N.; Broderick, G.A. Synthesis of microbial protein in ruminally cannulated cows fed alfalfa silage, alfalfa hay, or corn silage. J. Dairy Sci. 1996, 79, 1627–1637. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, T.L.; Lee, D.D. Anaerobic Digestion of Cellulosic Wastes. In Proceedings of the Sixth Annual DOE LLWMP Participants’ Information Meeting; Denver, CO, USA, 11–13 September 1984. Available online: https://www.osti.gov/servlets/purl/6605155 (accessed on 28 October 2022).
- Russell, J.B.; Muck, R.E.; Weimer, P.J. Quantitative analysis of cellulose degradation and growth of cellulolytic bacteria in the rumen. FEMS Microbiol. Ecol. 2009, 67, 183–197. [Google Scholar] [CrossRef]
- O’Sullivan, C.A.; Burrell, P.C.; Clarke, W.P.; Blackall, L.L. Structure of a cellulose degrading bacterial community during anaerobic digestion. Biotechnol. Bioeng. 2005, 92, 871–878. [Google Scholar] [CrossRef]
- Jeihanipour, A.; Niklasson, C.; Taherzadeh, M.J. Enhancement of solubilization rate of cellulose in anaerobic digestion and its drawbacks. Process Biochem. 2011, 46, 1509–1514. [Google Scholar] [CrossRef]
- Stephen, A.M. Other plant polysaccharides. In The Polysaccharides; Aspinall, G.O., Ed.; Academic Press: New York, NY, USA, 1983; Volume 2, pp. 97–193. [Google Scholar]
- Scheller, H.V.; Ulvskov, P. Hemicelluloses. Ann. Rev. Plant Biol. 2010, 61, 263–289. [Google Scholar] [CrossRef]
- Goering, H.K.; Van Soest, P.J. Forage Fiber Analysis: Apparatus, Reagents, Procedures and Some Applications. In Handbook 379; Agricultural Research Service, U.S. Department of Agriculture: Washington, DC, USA, 1970. [Google Scholar]
- Eda, S.; Ohnishi, A.; Kato, K. Xylan isolated from the stalk of Nicotiana tabacum. Agric. Biol Chem. 1976, 40, 359–364. [Google Scholar] [CrossRef]
- Kozioł, A.; Cybulska, J.; Pieczywek, P.M.; Zdunek, A. Evaluation of structure and assembly of xyloglucan from tamarind seed (Tamarindus indica L.) with atomic force microscopy. Food Biophys. 2015, 10, 396–402. [Google Scholar] [CrossRef]
- Dehority, B.A. Rate of isolated hemicellulose degradation and utilization by pure cultures of rumen bacteria. Appl. Microbiol. 1967, 15, 987–993. [Google Scholar] [CrossRef]
- Coen, J.A.; Dehority, B.A. Degradation and utilization of hemicellulose from intact forages by pure cultures of rumen bacteria. Appl. Microbiol. 1970, 20, 362–368. [Google Scholar] [CrossRef]
- Venditto, I.; Luis, A.S.; Rydahl, M.; Schückel, J.; Fernandes, V.O.; Vidal-Melgosa, S.; Bule, P.; Goyal, A.; Pires, V.M.R.; Dourado, C.G.; et al. Complexity of the Ruminococcus flavefaciens cellulosome reflects an expansion in glycan recognition. Proc. Natl. Acad. Sci. USA 2016, 113, 7136–7141. [Google Scholar] [CrossRef]
- Dehority, B.A. Characterization of several bovine rumen bacteria isolated with a xylan medium. J. Bacteriol. 1966, 91, 1724–1729. [Google Scholar] [CrossRef]
- Emerson, E.L.; Weimer, P.J. Fermentation of model hemicelluloses by Prevotella strains and Butyrivibrio fibrisolvens in pure culture and in ruminal enrichment cultures. Appl. Microbiol. Biotechnol. 2017, 101, 4269–4278. [Google Scholar] [CrossRef]
- Takizawa, S.; Asani, R.; Fukuda, Y.; Baba, Y.; Tada, C.; Nakai, Y. Shifts in xylanases and the microbial community associated with xylan biodegradation during treatment with rumen fluid. Microb. Biotechnol. 2022, 15, 1729–1743. [Google Scholar] [CrossRef]
- Hespell, R.B.; Cotta, M.A. Degradation and utilization by Butyrivibrio fibrisolvens H17c of xylans with different chemical and physical properties. Appl. Environ. Microbiol. 1995, 61, 3042–3050. [Google Scholar] [CrossRef]
- Hespell, R.B.; Wolf, R.; Bothast, R.J. Fermentation of xylans by Butyrivibrio fibrisolvens and other ruminal bacteria. Appl. Environ. Microbiol. 1987, 53, 2849–2853. [Google Scholar] [CrossRef]
- Cotta, M.A. Utilization of xylooligosaccharides by selected ruminal bacteria. Appl. Environ. Microbiol. 1993, 59, 3557–3663. [Google Scholar] [CrossRef]
- Weimer, P.J.; Hackney, J.M.; Jung, H.J.G.; Hatfield, R.D. Fermentation of a bacterial cellulose/xylan composite by mixed ruminal microflora: Implications for the role of polysaccharide matrix interactions in plant cell wall biodegradability. J. Agric. Food Chem. 2000, 48, 1727–1733. [Google Scholar] [CrossRef]
- Odenyo, A.A.; Mackie, R.I.; Stahl, D.A.; White, B.A. The use of 16S rRNA-targeted oligonucleotide probes to study competition between ruminal fibrolytic bacteria: Development of probes for Ruminococcus species and evidence for bacteriocin production. Appl. Environ. Microbiol. 1994, 60, 3688–3696. [Google Scholar] [CrossRef] [PubMed]
- Odenyo, A.A.; Mackie, R.I.; Stahl, D.A.; White, B.A. The use of 16S rRNA-targeted oligonucleotide probes to study competition between ruminal fibrolytic bacteria: Pure-culture studies with cellulose and alkaline peroxide-treated wheat straw. Appl. Environ. Microbiol. 1994, 60, 3697–3703. [Google Scholar] [CrossRef] [PubMed]
- Chan, W.W.; Dehority, B.A. Production of Ruminococcus flavefaciens growth inhibitor(s) by Ruminococcus albus. Anim. Feed Sci. Technol. 1999, 77, 61–71. [Google Scholar] [CrossRef]
- Chen, J.; Stevenson, D.M.; Weimer, P.J. Albusin B, a bacteriocin from Ruminococcus albus 7 that inhibits the growth of Ruminococcus flavefaciens. Appl. Environ. Microbiol. 2004, 70, 3167–3170. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Odt, C.L.; Weimer, P.J. Competition for cellulose among three predominant ruminal cellulolytic bacteria under substrate-excess and substrate-limited conditions. Appl. Environ. Microbiol. 1997, 63, 734–742. [Google Scholar] [CrossRef]
- Shi, Y.; Weimer, P.J. Competition for cellobiose among three predominant ruminal cellulolytic bacteria under substrate-excess and substrate-limited conditions. Appl. Environ. Microbiol. 1997, 63, 743–748. [Google Scholar] [CrossRef]
- Chen, J.; Weimer, P.J. Competition among three ruminal cellulolytic bacteria in the absence or presence of noncellulolytic bacteria. Microbiology 2001, 147, 21–30. [Google Scholar] [CrossRef][Green Version]
- Kalmokoff, M.L.; Teather, R.M. Isolation and characterization of a bacteriocin (Butyrivibriocin AR10) from the ruminal anaerobe Butyrivibrio fibrisolvens AR10: Evidence in support of the widespread occurrence of bacteriocin-like activity among ruminal isolates of B. fibrisolvens. Appl. Environ. Microbiol. 1997, 63, 394–402. [Google Scholar] [CrossRef]
- Scheifinger, C.C.; Wolin, M.J. Propionate formation from cellulose and soluble sugars by combined cultures of Bacteroides succinogenes and Selenomonas ruminantium. Appl. Microbiol. 1973, 26, 789–795. [Google Scholar] [CrossRef]
- Russell, J.B. Fermentation of cellodextrins by cellulolytic and noncellulolytic rumen bacteria. Appl. Environ. Microbiol. 1985, 49, 572–576. [Google Scholar] [CrossRef]
- Cotta, M.A.; Zeltwanger, R.L. Degradation and utilization of xylan by the ruminal bacteria Butyrivibrio fibrisolvens and Selenomons ruminantium. Appl. Environ. Microbiol. 1995, 61, 4396–4402. [Google Scholar] [CrossRef]
- Stewart, C.S.; Duncan, S.H.; Richardson, A.J.; Backwell, C.; Begbie, R. The inhibition of fungal cellulolysis by cell-free preparations from ruminococci. FEMS Microbiol. Lett. 1992, 97, 83–88. [Google Scholar] [CrossRef]
- Bernalier, A.; Fonty, G.; Bonnemoy, F.; Gouet, P. Inhibition of the cellulolytic activity of Neocallimastix frontalis by Ruminococcus flavefaciens. J. Gen. Microbiol. 1993, 139, 873–880. [Google Scholar] [CrossRef]
- Dehority, B.A.; Tirabasso, P.A. Antibiosis between ruminal bacteria and ruminal fungi. Appl. Environ. Microbiol. 2000, 66, 2921–2927. [Google Scholar] [CrossRef]
- Yarlett, N.; Hann, A.C.; Lloyd, D.; Williams, A. Hydrogenosomes in the rumen protozoan Dasytricha ruminantium Schulberg. Biochem. J. 1981, 200, 365–372. [Google Scholar] [CrossRef]
- Yarlett, N.; Orpin, C.G.; Munn, E.A.; Yarlett, N.C.; Greenwood, C.A. Hydrogenosomes in the rumen fungus Neocallimastix patriciarum. Biochem. J. 1986, 236, 729–739. [Google Scholar] [CrossRef]
- Paul, R.G.; Williams, A.G.; Butler, R.D. Hydrogenosomes in the rumen entodiniomorph ciliate Polyplastron multivesiculatum. J. Gen. Microbiol. 1990, 136, 1981–1989. [Google Scholar] [CrossRef]
- Levy, B.; Jami, E. Exploring the prokaryotic community associated with the rumen ciliate protozoa population. Front. Microbiol. 2018, 9, 2526. [Google Scholar] [CrossRef]
- Coytke, K.Z.; Schluter, J.; Foster, K.R. The ecology of the microbiome: Networks, competition, and stability. Science 2015, 350, 663–666. [Google Scholar] [CrossRef]
- Palmer, J.D.; Foster, K.R. Bacterial species rarely work together. Science 2022, 376, 581–582. [Google Scholar] [CrossRef]
- Weimer, P.J. Redundancy, resiliency and host specificity of the ruminal microbiome and their implications for engineering improved ruminal fermentations. Front. Microbiol. 2015, 6, 296. [Google Scholar] [CrossRef] [PubMed]
- Taxis, T.M.; Wolff, S.; Gregg, S.J.; Minton, N.O.; Zhang, C.; Dai, J.; Schnabel, R.D.; Taylor, J.F.; Kerley, M.S.; Pires, J.C.; et al. The players may change but the game remains: Network analyses of ruminal microbiomes suggest taxonomic differences mask functional similarity. Nucleic Acids Res. 2015, 43, 9600–9620. [Google Scholar] [CrossRef] [PubMed]
- Hungate, R.E. The anaerobic mesophilic cellulolytic bacteria. Bacteriol. Rev. 1950, 14, 1–49. [Google Scholar] [CrossRef] [PubMed]
- Njokweni, S.G.; Weimer, P.J.; Botes, M.; Van Zyl, W.H. Effects of preservation of rumen inoculum on volatile fatty acids production and the community dynamics during batch fermentation of fruit pomace. Bioresour. Technol. 2021, 321, 124518. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Dai, X.; Weimer, P.J. Shifts in fermentation end products and bacterial community composition in long-term, sequentially transferred in vitro ruminal enrichment cultures fed switchgrass with and without ethanol as a co-substrate. Bioresour. Technol. 2019, 285, 121324. [Google Scholar] [CrossRef]
- Holtzapple, M.T.; Wu, H.; Weimer, P.J.; Dalke, R.; Granda, C.B.; Mai, J.; Urgun-Demirtas, M. Microbial communities for valorizing biomass using the carboxylate platform to produce volatile fatty acids: A review. Bioresour. Technol. 2022, 344, 126253. [Google Scholar] [CrossRef]
- Levy, P.F.; Sanderson, J.E.; Ashare, F.; de Riel, S.R. Alkane liquid fuel production from biomass. In Liquid Fuel Developments; Wise, D.L., Ed.; CRC Press: Boca Raton, FL, USA, 1983; pp. 159–188. [Google Scholar]
- Holtzapple, M.T.; Granda, C.B. Carboxylate platform: The MixAlco process part 1: Comparison of three biomass conversion platforms. Appl. Biochem. Biotechnol. 2009, 156, 95–106. [Google Scholar] [CrossRef]
- Xu, J.; Bian, B.; Angenent, L.T.; Saikaly, P.E. Long-term continuous extraction of medium-chain carboxylates by pertraction with submerged hollow-fiber membranes. Front. Bioeng. Biotechnol. 2021, 9, 726946. [Google Scholar] [CrossRef]
- Allison, M.J.; Dougherty, R.W.; Bucklin, J.A.; Snyder, E.E. Ethanol accumulation in the rumen after overfeeding with readily fermentable carbohydrate. Science 1964, 144, 54–55. [Google Scholar] [CrossRef]
- Czerkawski, J.W.; Breckenridge, G. Fermentation of various glycolytic intermediates and other compounds by rumen micro-organisms, with particular reference to methane production. Br. J. Nutr. 1972, 27, 131–146. [Google Scholar] [CrossRef]
- Rabaey, K.; Verstraete, W. Microbial fuel cells: Novel biotechnology for energy generation. Trends Biotechnol. 2005, 23, 291–298. [Google Scholar] [CrossRef]
- Rismani-Yazdi, H.; Christy, A.D.; Dehority, B.A.; Morrison, M.; Yu, Z.; Tuovinen, O.H. Electricity generation from cellulose by rumen microorganisms in microbial fuel cells. Biotechnol. Bioeng. 2007, 97, 1398–1407. [Google Scholar] [CrossRef]
- Wang, C.-T.; Yang, C.-M.J.; Chen, Z.-S. Rumen microbial volatile fatty acids in relation to oxidation reduction potential and electricity generation from straw in microbial fuel cells. Biomass Bioenergy 2012, 37, 318–329. [Google Scholar] [CrossRef]
- Basak, B.; Ahn, Y.; Kumar, R.; Hwang, J.-H.; Kim, K.-H.; Jeon, B.-H. Lignocellulolytic microbiomes for augmenting lignocellulose degradation in anaerobic digestion. Trends Biotechnol. 2022, 30, 6–9. [Google Scholar] [CrossRef]
- Gijzen, H.J.; Zwart, K.; Teunissen, M.J.; Vogels, G.D. Anaerobic digestion of cellulose fraction of domestic refuse by means of rumen microorganisms. Biotechnol. Bioeng. 1988, 32, 749–755. [Google Scholar] [CrossRef]
- Barnes, S.P.; Keller, J. Cellulosic waste degradation by rumen-enhanced anaerobic digestion. Water Sci Technol. 2003, 48, 155–162. [Google Scholar] [CrossRef]
- Ozbayram, E.G.; Ince, O.; Ince, B.; Harms, H.; Kleisteuber, S. Comparison of rumen and manure microbiomes and implications for the inoculation of anaerobic digesters. Microorganisms 2018, 6, 15. [Google Scholar] [CrossRef]
- Ozbayram, E.G.; Akyol, C.; Ince, B.; Karakoc, C.; Ince, O. Rumen bacteria at work: Bioaugmentation strategies to enhance biogas production from cow manure. J. Appl. Microbiol. 2017, 124, 491–502. [Google Scholar] [CrossRef]
- Akyol, C.; Ince, O.; Bozan, M.; Ozbayram, E.G.; Ince, B. Fungal bioaugmentation of anaerobic digesters fed with lignocellulosic biomass: What to expect from anaerobic fungus Orpinomyces sp. Bioresour. Technol. 2019, 277, 1–10. [Google Scholar] [CrossRef]
- Zehavi, T.; Probst, M.; Mizrahi, I. Insights into culturomics of the rumen microbiome. Front. Microbiol. 2018, 9, 1999. [Google Scholar] [CrossRef]
- Opdahl, L.J.; Gonda, M.G.; St.-Pierre, B. Identification of uncultured bacterial species from Firmicutes, Bacteroidetes and Candiidatus Saccharibacteria as candidate cellulose utilizers from the rumen of beef cows. Microorganisms 2018, 6, 17. [Google Scholar] [CrossRef] [PubMed]
- Hess, M.; Sczyrba, A.; Egan, R.; Kim, T.W.; Chokhawala, H.; Schroth, G.; Luo, S.; Clark, D.S.; Chen, F.; Zhang, T.; et al. Metagenomic discovery of biomass-degrading genes and genomes from cow rumen. Science 2011, 331, 463–467. [Google Scholar] [CrossRef] [PubMed]
- Pope, P.B.; Smith, W.; Denman, S.E.; Tringe, S.G.; Barry, K.; Hugenholtz, P.; McSweeney, C.S.; McHardy, A.C.; Morrison, M. Isolation of Succinivibrionaceae implicated in low methane emissions from Tammar wallabies. Science 2011, 333, 646–648. [Google Scholar] [CrossRef] [PubMed]
- Bender, R.W.; Cook, D.E.; Combs, D.K. Comparison of in situ versus in vitro methods of fiber digestion at 120 and 288 hours to quantify the indigestible neutral detergent fiber fraction of corn silage samples. J. Dairy Sci. 2016, 99, 5394–5400. [Google Scholar] [CrossRef]
- Berg, B.; Ekbohm, G.; McClaugherty, C. Lignin and holocellulose relations during long-term decomposition of some forest litters: Long-term decomposition in a Scots pine forest. IV. Can. J. Bot. 1984, 62, 2540–2550. [Google Scholar] [CrossRef]
| Percentage of 16S rRNA Gene Copy Number a | Notes b | References | ||
|---|---|---|---|---|
| Fibrobacter succinogenes | Ruminococcus albus | Ruminococcus flavefaciens | ||
| 0.834 | 0.004 | 0.613 | 2 RC dairy cows, same diet | [17] |
| 0.625 | 0.034 | 1.08 | 2 RC lactating cows; MFD induction study | [18] |
| 0.92 | 0.03 | 1.47 | 2 RC lactating cows; inoculum for in vitro experiments | [19] |
| 0.01 | 0.002 | 0.1 | 5 two-year old dairy cows; time-course population assessment | [13] |
| 0.545 | 0.108 | NT c | 8 RC lactating cows, MFD induction study | [20] |
| 1.31 | 0.141 | 1.51 | 36 weaned dairy calves; effects of BCVFA and folic acid supplementation | [21] |
| Characteristic | Fibrobacter succinogenes | Ruminococcus albus | Ruminococcus flavefaciens |
|---|---|---|---|
| Phylum | Fibrobacteriota | Firmicutes | Firmicutes |
| Growth substrates | Cellulose, cellodextrins, glucose | Cellulose, cellodextrins, glucose, various hemicelluloses | Cellulose, cellodextrins, various hemicelluloses |
| Mode of adherence | Fibro-slime proteins, CBMs | CBMs, glycocalyx enriched in Glc, Xyl, Man | CBMs, glycocalyx enriched in Rha, Glc, Gal |
| Cellulase enzyme organization | Noncellulosomal; Surface bound via covalent linkages to cell wall | Cellusomal or non-cellulosomal depending on strain | True cellulosome, but employing single-binding dockerins |
| Fermentation end products | Succinate, acetate, CO2 | Acetate, ethanol, H2, CO2 | Acetate, succinate, formate, H2 |
| Rate constant for cellulose degradation (h−1) a | 0.108 | 0.049 | 0.11 |
| True growth yield (g cells [g cellulose]−1) a | 0.23–0.25 | 0.11 | 0.23–0.30 |
| Maintenance coefficient (g cellulose [g cell−1 h−1]) a | 0.04–0.06 | 0.10 | 0.05–0.07 |
| Characteristic | Cellulose | Hemicellulose |
|---|---|---|
| Substrate | Linear β-1→4 linked glucosyl units, no side chains, varying from crystalline to amorphous | Many combinations of sugars and linkages. Noncrystalline |
| Water solubility | Insoluble | Low to high |
| Localization | Mostly in secondary wall | Widely distributed in wall, sometimes abundant in non-herbaceous parts (seeds, tubers) |
| Association with lignin | Weak to none | Physical and chemical (covalent linkages) |
| Degrading microbes | Widely distributed across bacterial, fungal and protist lineages. Bacteria are nutritional specialists. | Primarily bacteria and fungi that are nutritional generalists |
| Degradation rate in pure form | ~0.1 h−1 | Up to 0.5 h−1 |
| Degradation rate in plant cell walls | ~0.1 h−1 | 0.02–0.04 h−1 |
| Substrate a | Cultivation Mode | Inoculation | Relative Abundance (%) b | Ref. | ||
|---|---|---|---|---|---|---|
| R. albus | R. flavefaciens | F. succinogenes | ||||
| ASC | Batch | Fs S85 + Ra 8 | 45 | 55 | [113] | |
| Fs S85 + Rf FD-1 | 58 | 42 | ||||
| Ra 8 + Rf FD-1 | 100 | ND | ||||
| Fs S85 + Ra 8 + Rf FD-1 | 42 | ND | 58 | |||
| MCC | Batch | Fs S85 + Ra.7 | 44 | 56 | [116] | |
| Fs S85 + Rf FD-1 | 42 | 58 | ||||
| Ra 7 + Rf FD-1 | Variable c | Variable c | ||||
| Continuous | Fs S85 + Ra 7 | 10.1–21.8 | 78.1–90.7 | |||
| Fs S85 + Rf FD-1 | >96.5 | <3.5 | ||||
| Ra 7 + Rf FD-1 | 14.9 | 85.1 | ||||
| Cellobiose | Batch | Fs S85 + Ra 8 | 50 | 50 | [112] | |
| Fs S85 + Rf FD-1 | 81 | 19 | ||||
| Ra 8 + Rf FD-1 | 100 | ND | ||||
| Fs S85 + Ra 8 + Rf FD-1 | 50 | ND | 50 | |||
| Batch | Fs S85 + Ra 7 | 28 | 72 | [117] | ||
| Fs S85 + Rf FD-1 | 49.9 | 50.1 | ||||
| Ra 7 + Rf FD-1 | Variable c | Variable c | ||||
| Continuous | Fs S85 + Ra 7 | >99.8 | <0.17 | |||
| Fs S85 + Rf FD-1 | >99.8 | <0.17 | ||||
| Ra 7 + Rf FD-1 | >99.8 | <0.14 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Weimer, P.J. Degradation of Cellulose and Hemicellulose by Ruminal Microorganisms. Microorganisms 2022, 10, 2345. https://doi.org/10.3390/microorganisms10122345
Weimer PJ. Degradation of Cellulose and Hemicellulose by Ruminal Microorganisms. Microorganisms. 2022; 10(12):2345. https://doi.org/10.3390/microorganisms10122345
Chicago/Turabian StyleWeimer, Paul J. 2022. "Degradation of Cellulose and Hemicellulose by Ruminal Microorganisms" Microorganisms 10, no. 12: 2345. https://doi.org/10.3390/microorganisms10122345
APA StyleWeimer, P. J. (2022). Degradation of Cellulose and Hemicellulose by Ruminal Microorganisms. Microorganisms, 10(12), 2345. https://doi.org/10.3390/microorganisms10122345






