Patterns of Genomic Variations in the Plant Pathogen Dickeya solani
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains
2.2. Genome DNA Extraction, Sequencing and Assembly
2.3. Phylogenomics
2.4. Variant Calling and Analysis
2.5. Phylogenetic Analysis and Annotation of Variants’ Hotspot Regions within D. solani Strains
3. Results
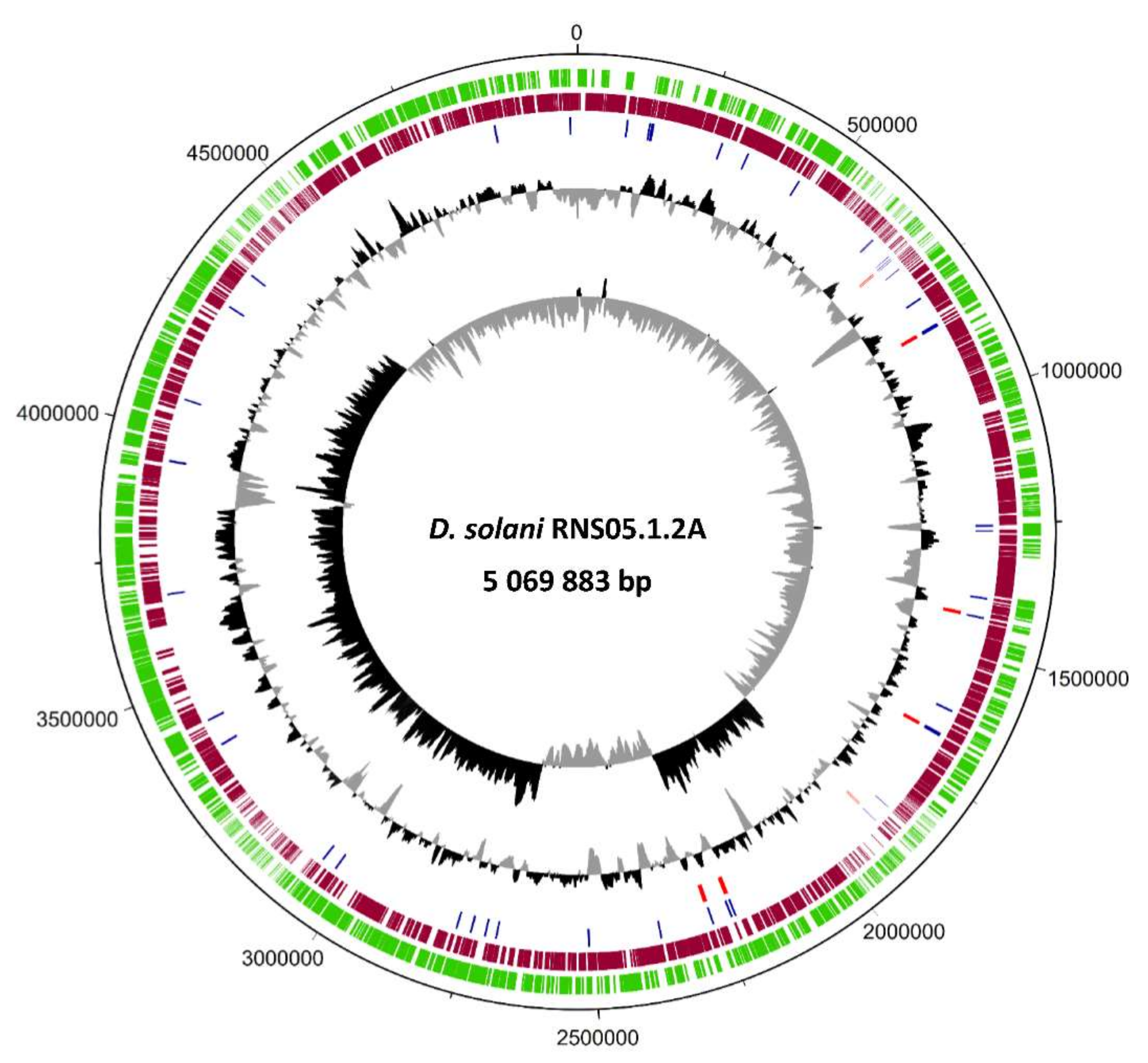
3.1. Complete Genome Sequence of Atypical Strain D. solani RNS05.1.2A
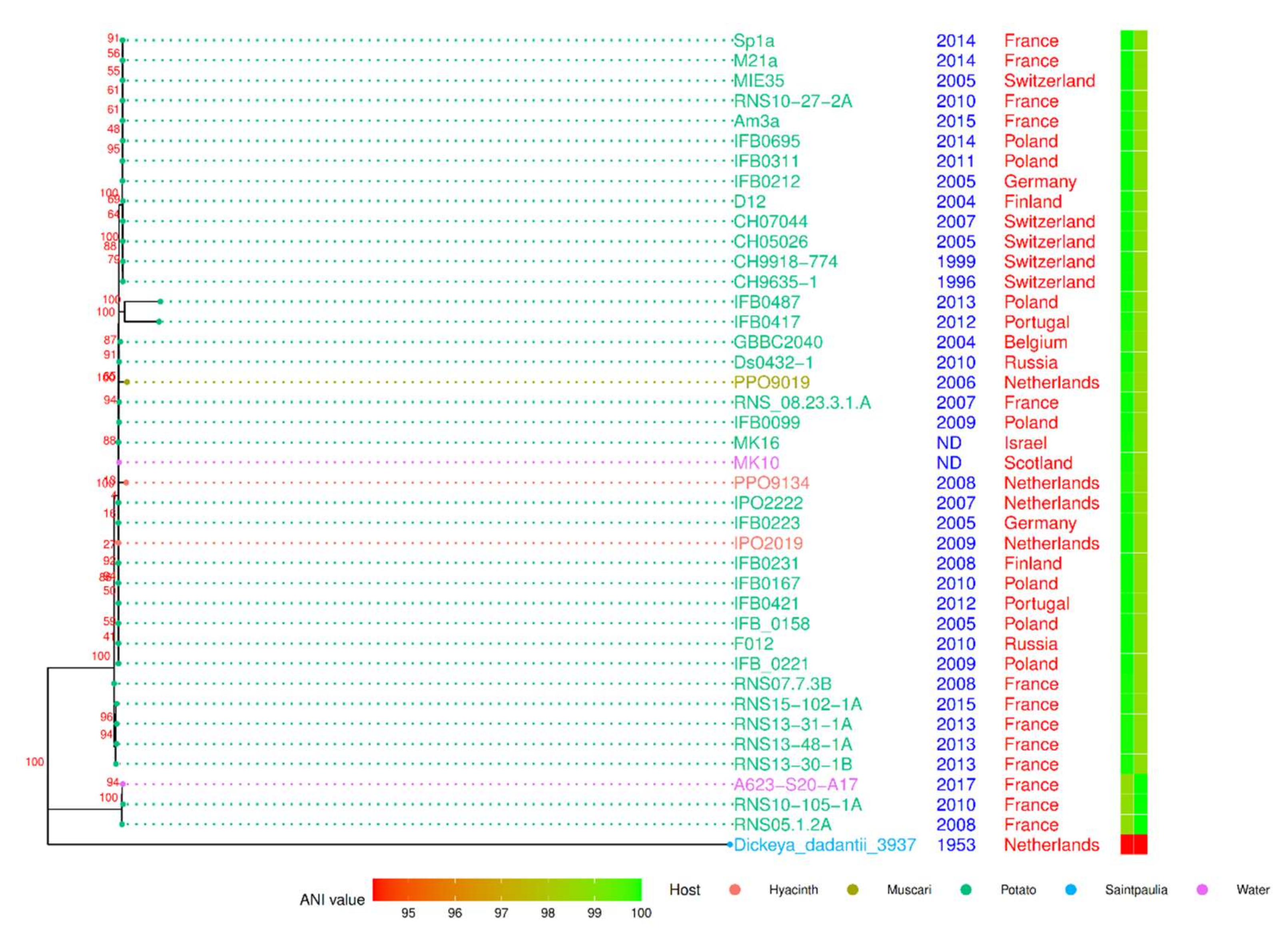
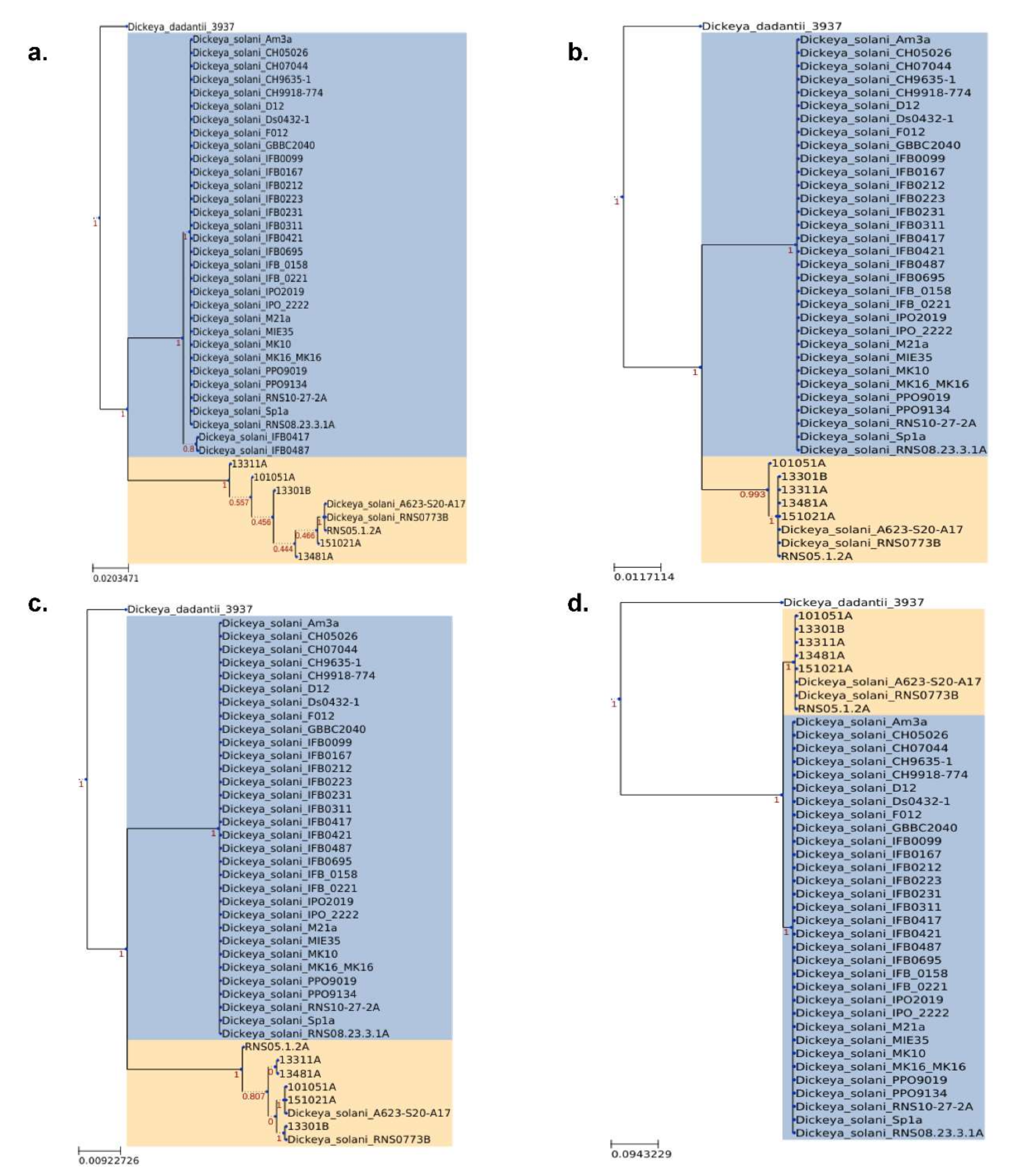
3.2. Phylogenomics of D. solani Strains RNS10-105-1A, RNS13-30-1B, RNS13-31-1A, RNS13-48-1A and RNS15-102-1A
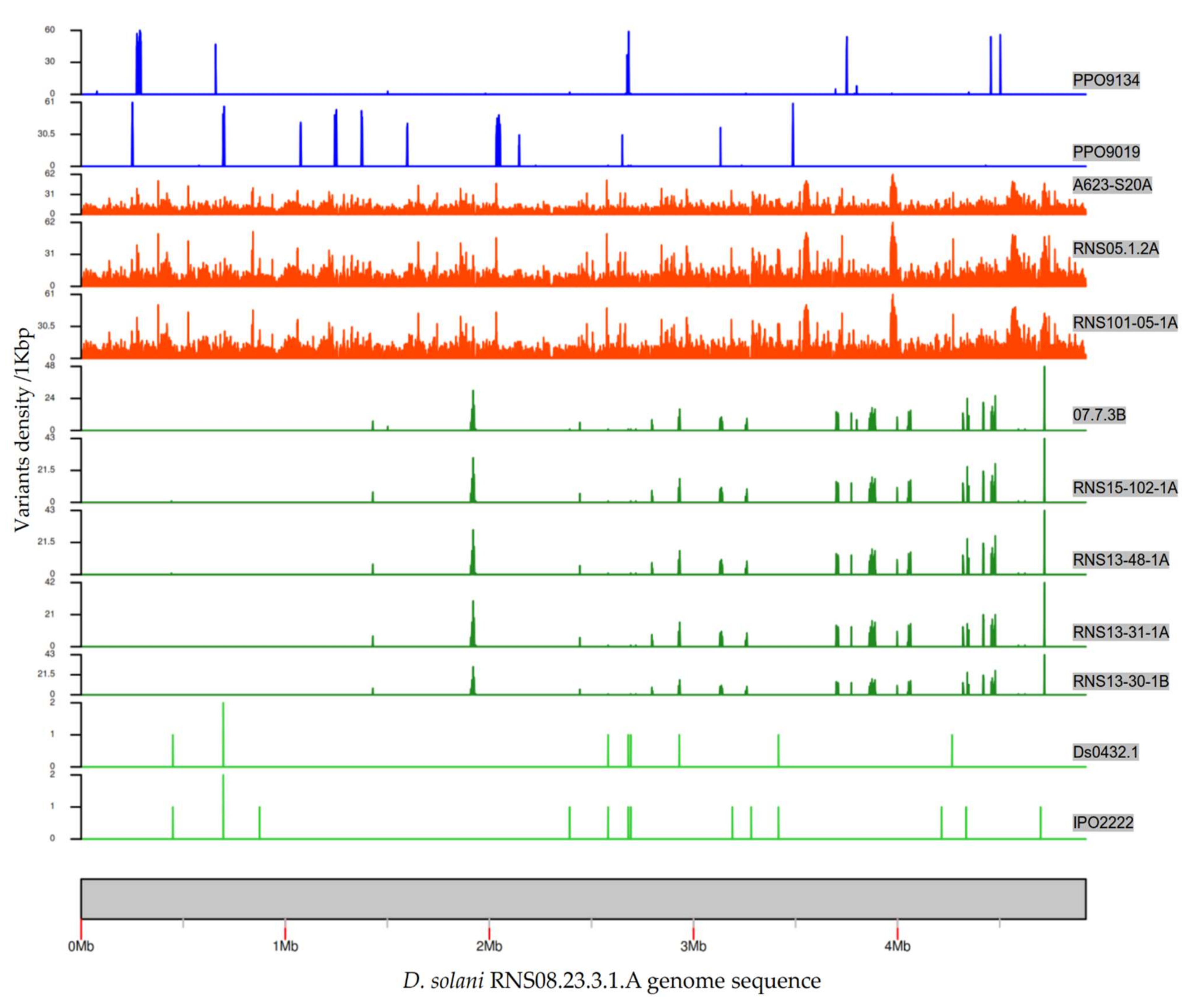
3.3. SNP and Indel Variations in D. solani Strains
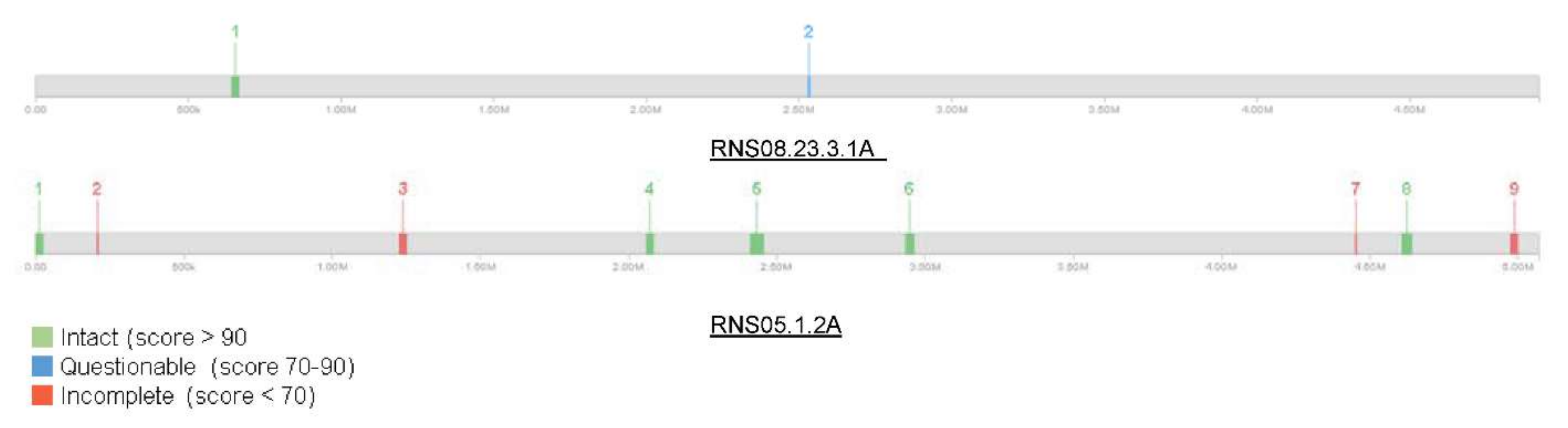
3.4. Characteristics of HGT Events in D. solani Strains RNS13-30-1B, RNS13-31-1A, RNS13-48-1A and RNS15-102-1A
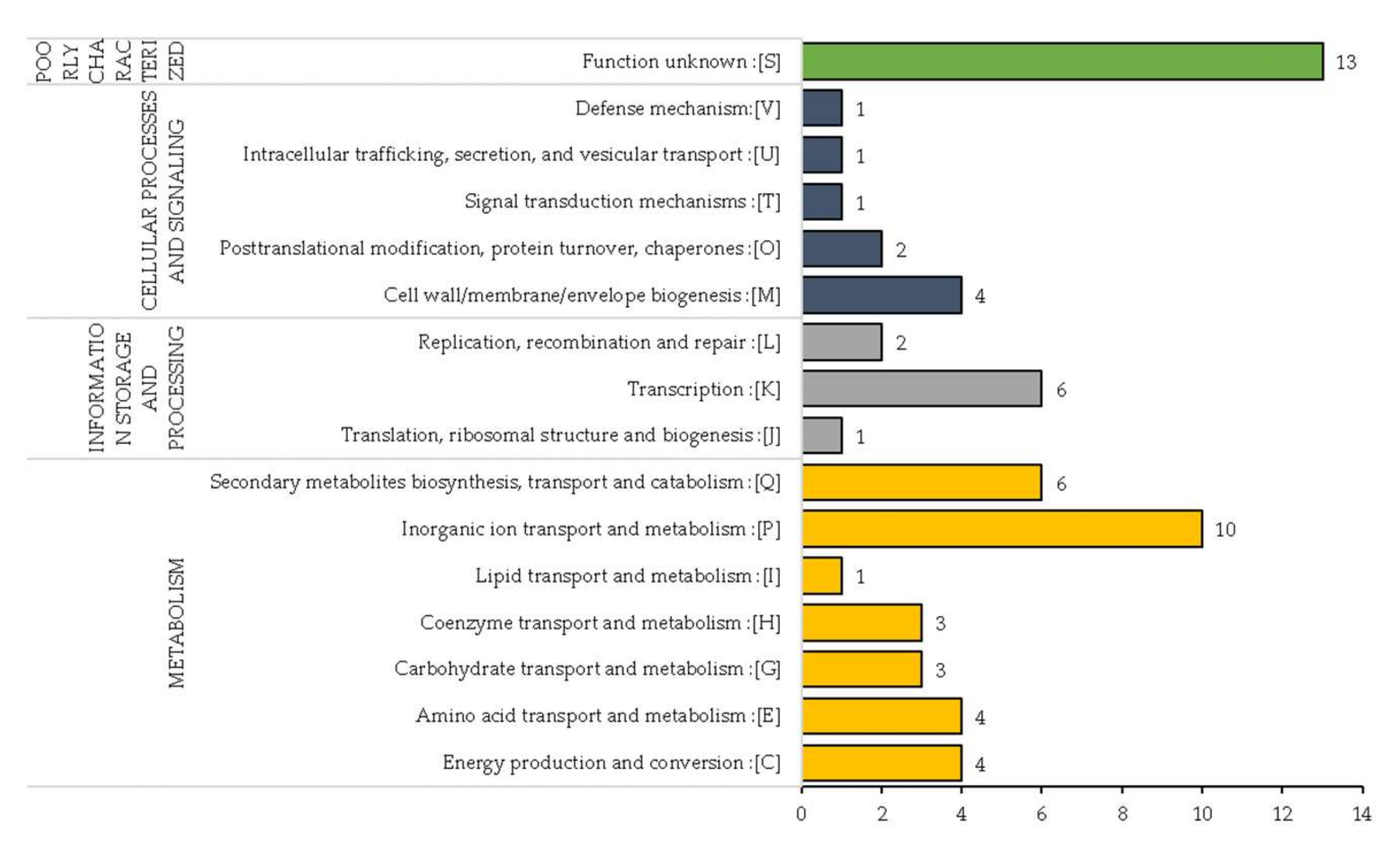
3.5. Functional Annotation of the Variable Regions in D. solain Strains
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Van der Wolf, J.M.; Acuña, I.; De Boer, S.H.; Brurberg, M.B.; Cahill, G.; Charkowski, A.O.; Coutinho, T.; Davey, T.; Dees, M.W.; Degefu, Y.; et al. Diseases Caused by Pectobacterium and Dickeya Species Around the World. In Plant Diseases Caused by Dickeya and Pectobacterium Species; Van Gijsegem, F., van der Wolf, J.M., Toth, I.K., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 215–261. ISBN 978-3-030-61458-4. [Google Scholar]
- Van der Wolf, J.M.; Nijhuis, E.H.; Kowalewska, M.J.; Saddler, G.S.; Parkinson, N.; Elphinstone, J.G.; Pritchard, L.; Toth, I.K.; Lojkowska, E.; Potrykus, M.; et al. Dickeya Solani Sp. Nov., a Pectinolytic Plant-Pathogenic Bacterium Isolated from Potato (Solanum tuberosum). Int. J. Syst. Evol. Microbiol. 2014, 64, 768–774. [Google Scholar] [CrossRef] [PubMed]
- Van Gijsegem, F.; Hugouvieux-Cotte-Pattat, N.; Kraepiel, Y.; Lojkowska, E.; Moleleki, L.N.; Gorshkov, V.; Yedidia, I. Molecular Interactions of Pectobacterium and Dickeya with Plants. In Plant Diseases Caused by Dickeya and Pectobacterium Species; Van Gijsegem, F., van der Wolf, J.M., Toth, I.K., Eds.; Springer International Publishing: Cham, Switzerland, 2021; pp. 85–147. ISBN 978-3-030-61458-4. [Google Scholar]
- Blin, P.; Robic, K.; Khayi, S.; Cigna, J.; Munier, E.; Dewaegeneire, P.; Laurent, A.; Jaszczyszyn, Y.; Hong, K.W.; Chan, K.G.; et al. Pattern and Causes of the Establishment of the Invasive Bacterial Potato Pathogen Dickeya solani and of the Maintenance of the Resident Pathogen D. dianthicola. Mol. Ecol. 2021, 30, 608–624. [Google Scholar] [CrossRef] [PubMed]
- Khayi, S.; Blin, P.; Pédron, J.; Chong, T.-M.; Chan, K.-G.; Moumni, M.; Hélias, V.; Gijsegem, F.V.; Faure, D. Population Genomics Reveals Additive and Replacing Horizontal Gene Transfers in the Emerging Pathogen Dickeya Solani. BMC Genom. 2015, 16, 788. [Google Scholar] [CrossRef]
- Golanowska, M.; Potrykus, M.; Motyka-Pomagruk, A.; Kabza, M.; Bacci, G.; Galardini, M.; Bazzicalupo, M.; Makalowska, I.; Smalla, K.; Mengoni, A.; et al. Comparison of Highly and Weakly Virulent Dickeya Solani Strains, with a View on the Pangenome and Panregulon of This Species. Front. Microbiol. 2018, 9, 1940. [Google Scholar] [CrossRef] [PubMed]
- Hélias, V.; Hamon, P.; Huchet, E.; Wolf, J.V.D.; Andrivon, D. Two New Effective Semiselective Crystal Violet Pectate Media for Isolation of Pectobacterium and Dickeya: Isolating Pectolytic Bacteria on CVP. Plant Pathol. 2012, 61, 339–345. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A Better, Faster Version of the PHAST Phage Search Tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: A Fast Phage Search Tool. Nucleic Acids Res. 2011, 39, W347–W352. [Google Scholar] [CrossRef]
- Emms, D.M.; Kelly, S. OrthoFinder: Phylogenetic Orthology Inference for Comparative Genomics. Genome Biol. 2019, 20, 238. [Google Scholar] [CrossRef]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Jain, C.; Rodriguez-R, L.M.; Phillippy, A.M.; Konstantinidis, K.T.; Aluru, S. High Throughput ANI Analysis of 90K Prokaryotic Genomes Reveals Clear Species Boundaries. Nat. Commun. 2018, 9, 5114. [Google Scholar] [CrossRef] [PubMed]
- Yu, G. Using Ggtree to Visualize Data on Tree-like Structures. Curr. Protoc. Bioinform. 2020, 69, e96. [Google Scholar] [CrossRef] [PubMed]
- Seemann, T. Snippy: Fast Bacterial Variant Calling from NGS Reads 2015. Available online: https://github.com/tseemann/snippy (accessed on 13 January 2022).
- Gel, B.; Serra, E. KaryoploteR: An R/Bioconductor Package to Plot Customizable Genomes Displaying Arbitrary Data. Bioinformatics 2017, 33, 3088–3090. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing Large Minimum Evolution Trees with Profiles Instead of a Distance Matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. EggNOG-Mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Szklarczyk, D.; Heller, D.; Hernández-Plaza, A.; Forslund, S.K.; Cook, H.; Mende, D.R.; Letunic, I.; Rattei, T.; Jensen, L.J.; et al. EggNOG 5.0: A Hierarchical, Functionally and Phylogenetically Annotated Orthology Resource Based on 5090 Organisms and 2502 Viruses. Nucleic Acids Res. 2019, 47, D309–D314. [Google Scholar] [CrossRef] [PubMed]
- Buchfink, B.; Reuter, K.; Drost, H.-G. Sensitive Protein Alignments at Tree-of-Life Scale Using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef] [PubMed]
- Groth, A.C.; Calos, M.P. Phage Integrases: Biology and Applications. J. Mol. Biol. 2004, 335, 667–678. [Google Scholar] [CrossRef]
- Sahbou, A.-E.; Iraqi, D.; Mentag, R.; Khayi, S. BuscoPhylo: A Webserver for Busco-Based Phylogenomic Analysis for Non-Specialists. Sci. Rep. 2022, 12, 17352. [Google Scholar] [CrossRef]
- Motyka-Pomagruk, A.; Zoledowska, S.; Misztak, A.E.; Sledz, W.; Mengoni, A.; Lojkowska, E. Comparative Genomics and Pangenome-Oriented Studies Reveal High Homogeneity of the Agronomically Relevant Enterobacterial Plant Pathogen Dickeya Solani. BMC Genom. 2020, 21, 449. [Google Scholar] [CrossRef]
- Soucy, S.M.; Huang, J.; Gogarten, J.P. Horizontal Gene Transfer: Building the Web of Life. Nat. Rev. Genet. 2015, 16, 472–482. [Google Scholar] [CrossRef] [PubMed]
- Toth, I.K.; Pritchard, L.; Birch, P.R. Comparative Genomics Reveals What Makes an Enterobacterial Plant Pathogen. Annu. Rev. Phytopathol. 2006, 44, 305–336. [Google Scholar] [CrossRef] [PubMed]
- Barny, M.-A.; Moussa, H.B.; Bertran, C.; Rochelle-Newall, E.; Fiorini, S.; Pédron, J. The Diversity and Abundance of Soft Rot Pectobacteriaceae along the Durance River Stream in the Southeast of France Revealed by Multiple Seasonal Surveys. 2022. Available online: https://apsjournals.apsnet.org/doi/10.1094/PHYTO-12-21-0515-R (accessed on 13 January 2022).
- Pedron, J.; Gijsegem, F.V. Diversity in the Bacterial Genus Dickeya Grouping Plant Pathogens and Waterways Isolates. 2019. Available online: https://www.lidsen.com/journals/genetics/genetics-03-04-098 (accessed on 13 January 2022).
- Ignatov, A.N.; Lukianova, A.; Evseev, P.V.; Stakheev, A.V.; Kotova, I.B.; Zavriev, S.K.; Miroshnikov, K.A. Quantitative Real-Time PCR Assay for the Detection of Pectobacterium parmentieri, a Causal Agent of Potato Soft Rot. Plants 2021, 10, 1880. [Google Scholar]
- Pritchard, L.; Humphris, S.; Saddler, G.S.; Parkinson, N.M.; Bertrand, V.; Elphinstone, J.G.; Toth, I.K. Detection of phytopathogens of the genus Dickeya using a PCR primer prediction pipeline for draft bacterial genome sequences. Plant Pathol. 2013, 62, 587–596. [Google Scholar] [CrossRef]
- Khayi, S.; Blin, P.; Chong, T.M.; Chan, K.-G.; Faure, D. Complete genome anatomy of the emerging potato pathogen Dickeya solani type strain IPO 2222. Stand. Genom. Sci. 2016, 11, 87. [Google Scholar] [CrossRef]
- Khayi, S.; Mondy, S.; Beury-Cirou, A.; Moumni, M.; Hélias, V.; Faure, D. Genome Sequence of the Emerging Plant Pathogen Dickeya solani Strain RNS 08.23.3.1A. 2014. Available online: https://journals.asm.org/doi/10.1128/genomeA.01270-13 (accessed on 13 January 2022).
- Garlant, L.; Koskinen, P.; Rouhiainen, L.; Laine, P.; Paulin, L.; Auvinen, P.; Holm, L.; Pirhonen, M. Genome Sequence of Dickeya solani, a New soft Rot Pathogen of Potato, Suggests its Emergence May Be Related to a Novel Combination of Non-Ribosomal Peptide/Polyketide Synthetase Clusters. Diversity 2013, 5, 824–842. [Google Scholar] [CrossRef]

| Strain | Total | Genic | IG 1 | NG 2 | Syn 3 | NSy 4 | Cp 5 | SNP 6 | MNP 7 | DEL 8 | INS 9 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| IPO2222 | 14 | 11 | 3 | 10 | 6 | 3 | 0 | 10 | 0 | 3 | 1 |
| Ds0432.1 | 9 | 9 | 0 | 8 | 6 | 1 | 0 | 7 | 0 | 1 | 1 |
| RNS13-30-1B | 1268 | 1148 | 120 | 151 | 877 | 263 | 133 | 1101 | 20 | 6 | 8 |
| RNS13-31-1A | 1253 | 1144 | 109 | 151 | 877 | 261 | 128 | 1090 | 23 | 5 | 7 |
| RNS13-48-1A | 1269 | 1150 | 119 | 152 | 879 | 263 | 131 | 1102 | 23 | 6 | 7 |
| RNS15.102.1A | 1273 | 1153 | 120 | 152 | 882 | 263 | 127 | 1105 | 27 | 6 | 8 |
| RNS07.7.3B | 1278 | 1157 | 121 | 154 | 879 | 265 | 156 | 1105 | 0 | 8 | 9 |
| RNS10.105.1A | 39,235 | 34,251 | 4984 | 3877 | 26,557 | 7465 | 4367 | 33,885 | 516 | 248 | 219 |
| RNS05.1.2A | 39,743 | 34,627 | 5116 | 3883 | 26,715 | 7648 | 4445 | 34,044 | 750 | 264 | 240 |
| A623S20A17 | 38,623 | 33,803 | 4820 | 3854 | 26,126 | 7375 | 4868 | 33,163 | 1 | 301 | 290 |
| PPO9019 | 2573 | 2374 | 199 | 68 | 1906 | 449 | 589 | 1961 | 0 | 14 | 9 |
| PPO9134 | 1842 | 1751 | 91 | 47 | 1350 | 396 | 492 | 1341 | 0 | 5 | 4 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khayi, S.; Chan, K.-G.; Faure, D. Patterns of Genomic Variations in the Plant Pathogen Dickeya solani. Microorganisms 2022, 10, 2254. https://doi.org/10.3390/microorganisms10112254
Khayi S, Chan K-G, Faure D. Patterns of Genomic Variations in the Plant Pathogen Dickeya solani. Microorganisms. 2022; 10(11):2254. https://doi.org/10.3390/microorganisms10112254
Chicago/Turabian StyleKhayi, Slimane, Kok-Gan Chan, and Denis Faure. 2022. "Patterns of Genomic Variations in the Plant Pathogen Dickeya solani" Microorganisms 10, no. 11: 2254. https://doi.org/10.3390/microorganisms10112254
APA StyleKhayi, S., Chan, K.-G., & Faure, D. (2022). Patterns of Genomic Variations in the Plant Pathogen Dickeya solani. Microorganisms, 10(11), 2254. https://doi.org/10.3390/microorganisms10112254








