Assessment of the Microbial Spoilage and Quality of Marinated Chicken Souvlaki through Spectroscopic and Biomimetic Sensors and Data Fusion
Abstract
1. Introduction
2. Materials and Methods
2.1. Experimental Design
2.2. Microbiological Analysis
2.3. Sensors
2.3.1. Spectral Acquisition
2.3.2. Electronic Nose (E-Nose)
2.4. Data Processing
2.5. Model Development and Performance Assessment
3. Results
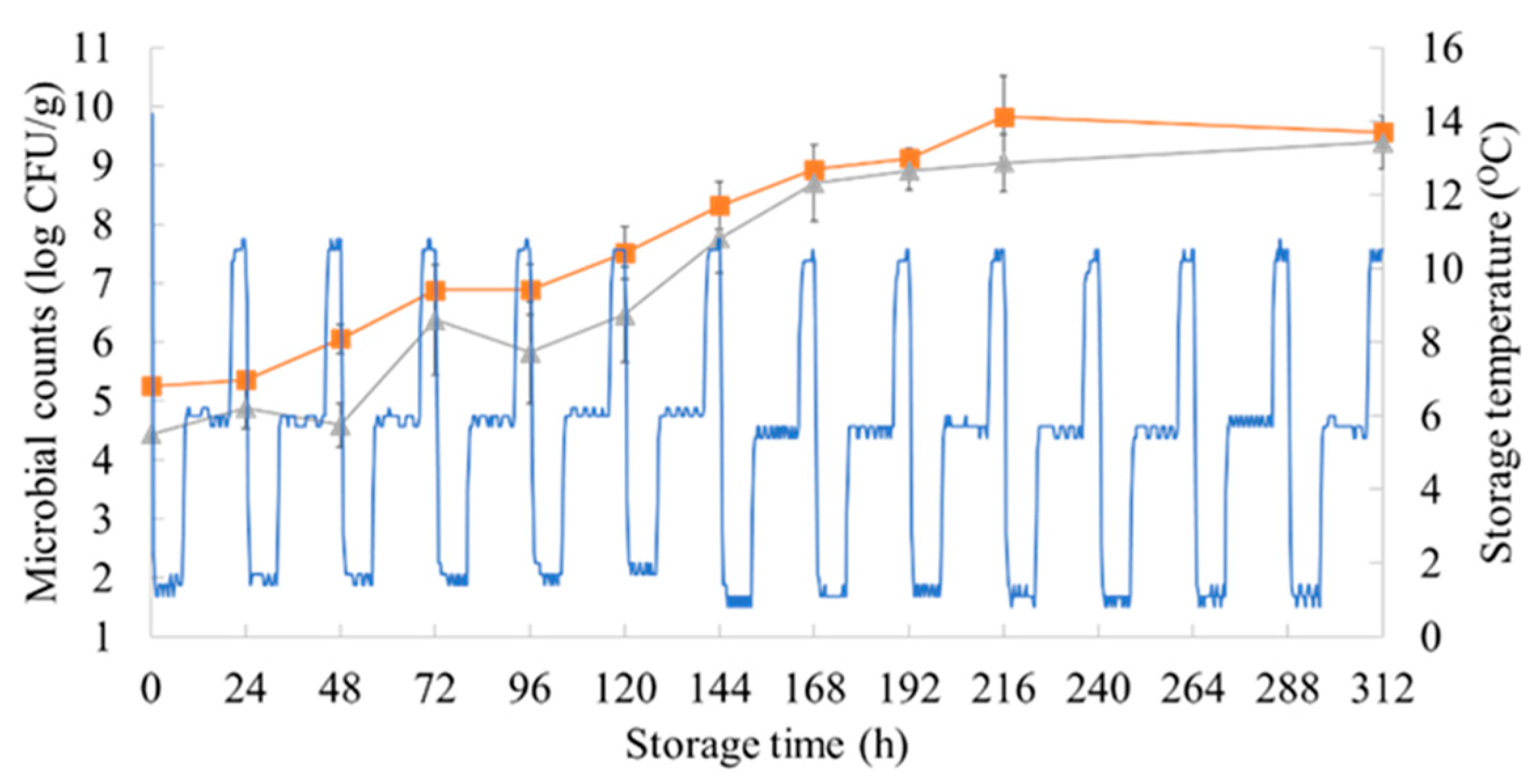
3.1. Microbiological Results
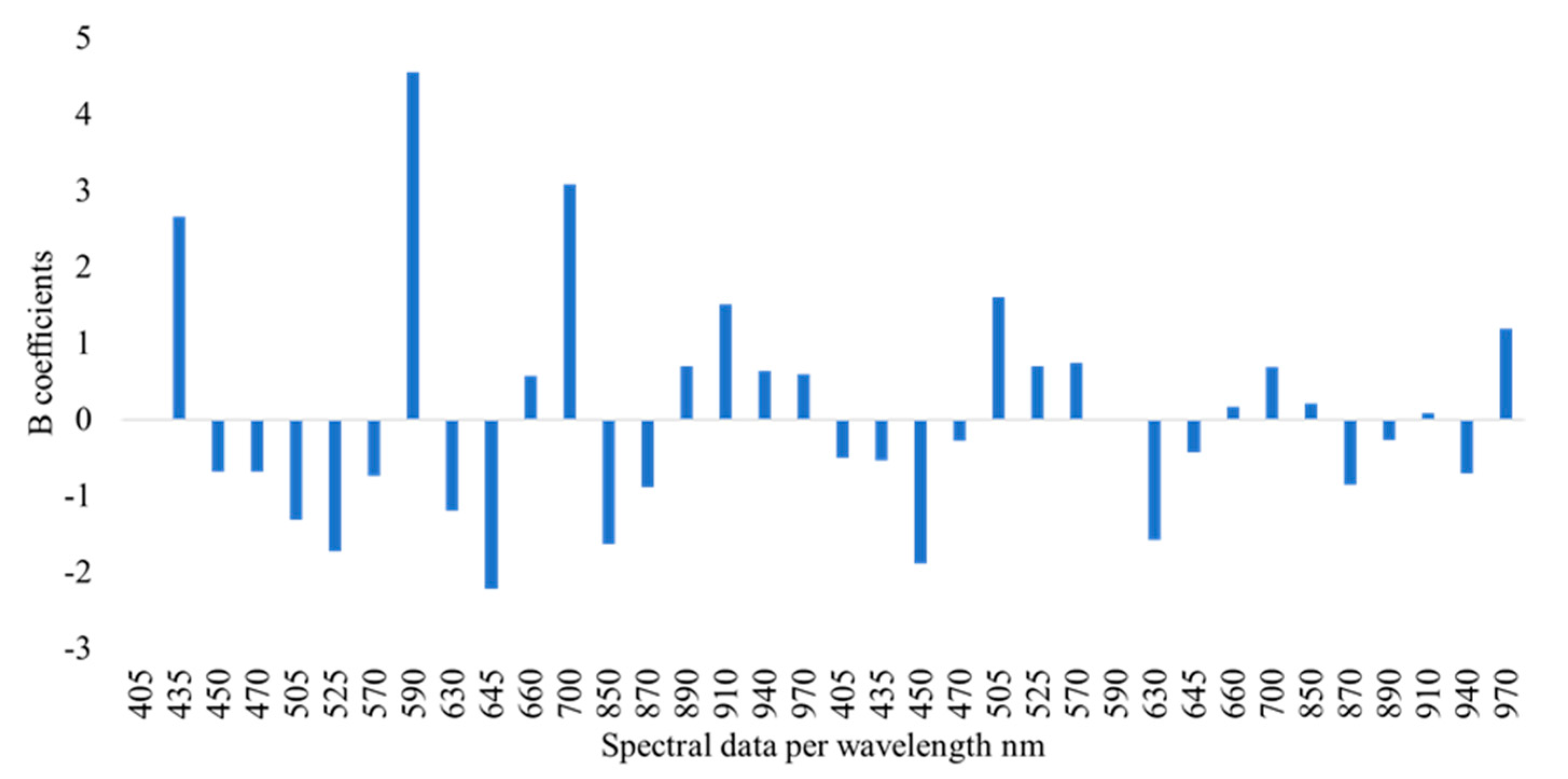
3.2. Spectra and E-Nose Signals
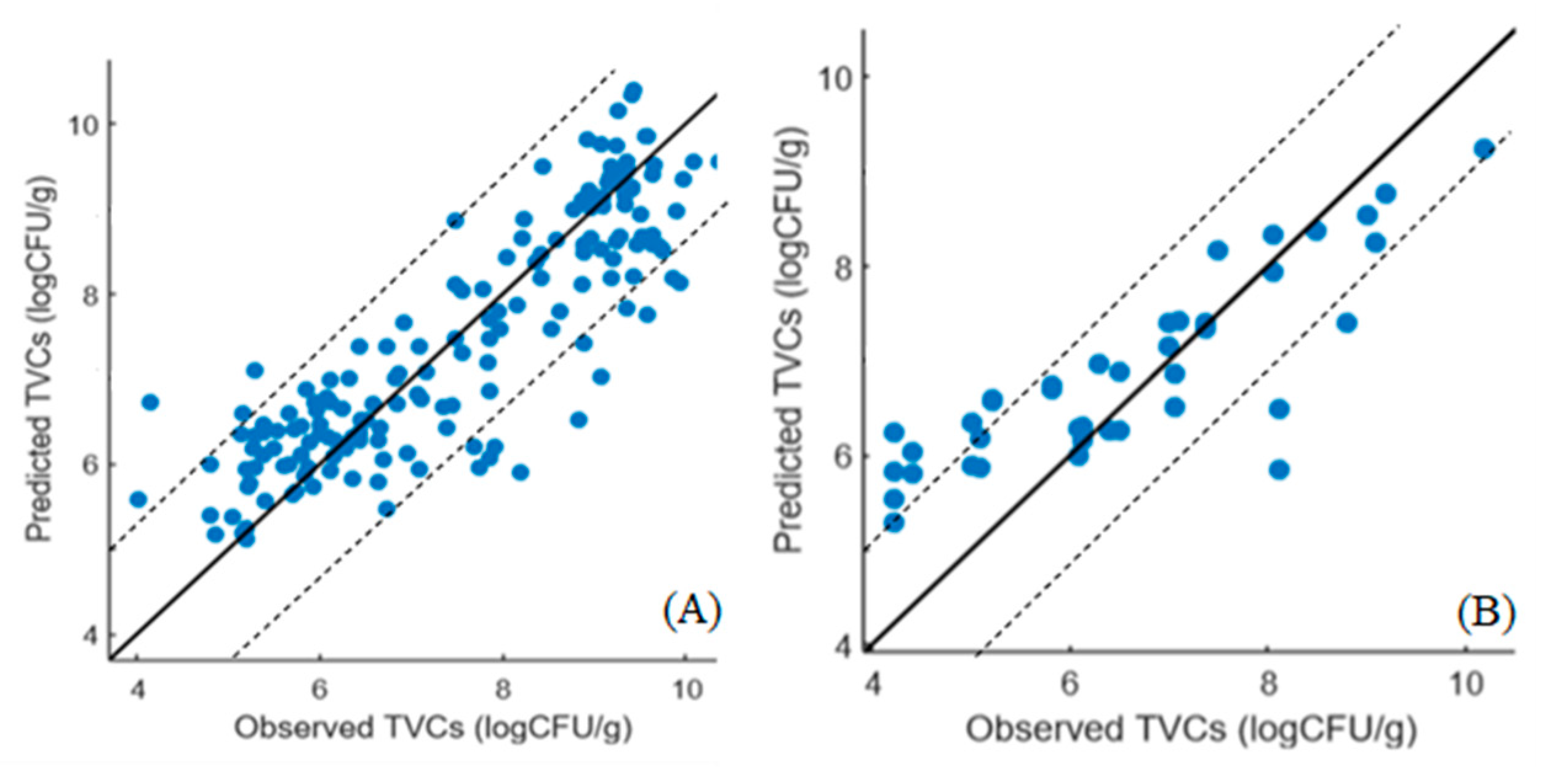
3.3. PLS-R Models for Assessing TVC Loads in Marinated Chicken Souvlaki
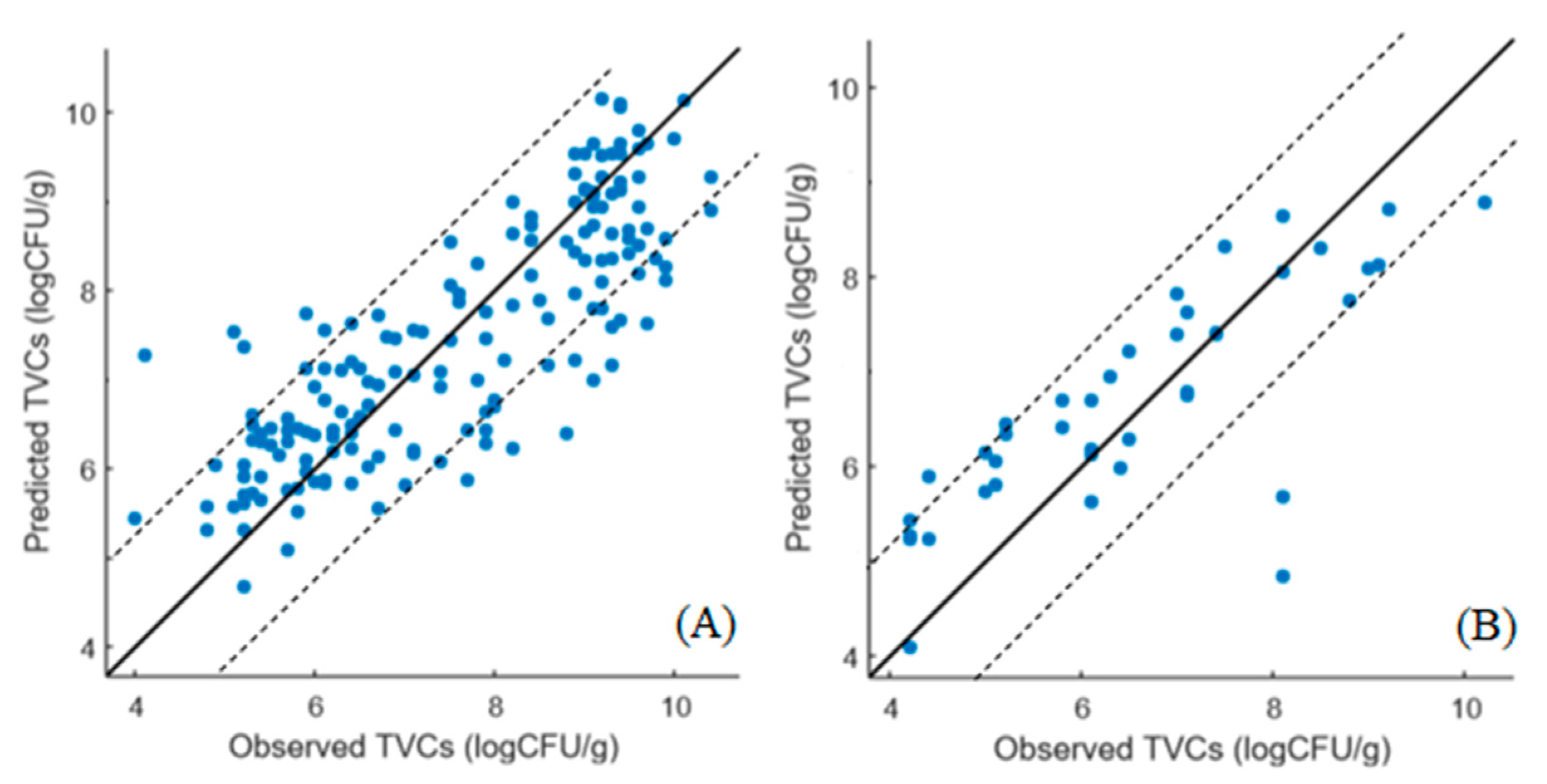
3.4. SVM-R Models for Assessing TVC Loads in Marinated Chicken Souvlaki
3.5. Classification Models for Assessing Spoilage in Marinated Chicken Souvlaki
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Berrueta, L.A.; Alonso-Salces, R.M.; Héberger, K. Supervised pattern recognition in food analysis. J. Chromatogr. A 2007, 1158, 196–214. [Google Scholar] [CrossRef] [PubMed]
- Candoğan, K.; Altuntas, E.G.; İğci, N. Authentication and quality assessment of meat products by fourier-transform infrared (FTIR) spectroscopy. Food Eng. Rev. 2021, 13, 66–91. [Google Scholar] [CrossRef]
- Kamruzzaman, M.; Makino, Y.; Oshita, S. Rapid and non-destructive detection of chicken adulteration in minced beef using visible near-infrared hyperspectral imaging and machine learning. J. Food Eng. 2016, 170, 8–15. [Google Scholar] [CrossRef]
- Alexandrakis, D.; Downey, G.; Scannell, A.G. Rapid non-destructive detection of spoilage of intact chicken breast muscle using near-infrared and Fourier transform mid-infrared spectroscopy and multivariate statistics. Food Bioproc. Technol. 2012, 5, 33–65. [Google Scholar] [CrossRef]
- Ellis, D.I.; Broadhurst, D.; Kell, D.B.; Rowland, J.J.; Goodacre, R. Rapid and quantitative detection of the microbial spoilage of meat by Fourier transform infrared spectroscopy and machine learning. Appl. Environ. Microbiol. 2002, 68, 2822–2828. [Google Scholar] [CrossRef] [PubMed]
- Vasconcelos, H.; Saraiva, C.; de Almeida, J.M. Evaluation of the spoilage of raw chicken breast fillets using Fourier transform infrared spectroscopy in tandem with chemometrics. Food Bioproc. Tech. 2014, 7, 2330–2341. [Google Scholar] [CrossRef]
- Tsakanikas, P.; Karnavas, A.; Panagou, E.Z.; Nychas, G.J. A machine learning workflow for raw food spectroscopic classification in a future industry. Sci. Rep. 2020, 10, 11212. [Google Scholar] [CrossRef]
- Feng, Y.Z.; Sun, D.W. Determination of total viable count (TVC) in chicken breast fillets by near-infrared hyperspectral imaging and spectroscopic transforms. Talanta 2013, 105, 244–249. [Google Scholar] [CrossRef]
- Feng, Y.Z.; Sun, D.W. Near-infrared hyperspectral imaging in tandem with partial least squares regression and genetic algorithm for non-destructive determination and visualization of Pseudomonas loads in chicken fillets. Talanta 2013, 109, 74–83. [Google Scholar] [CrossRef]
- Ye, X.; Iino, K.; Zhang, S. Monitoring of bacterial contamination on chicken meat surface using a novel narrowband spectral index derived from hyperspectral imagery data. Meat Sci. 2016, 122, 25–31. [Google Scholar] [CrossRef]
- Spyrelli, E.D.; Doulgeraki, A.I.; Argyri, A.A.; Tassou, C.C.; Panagou, E.Z.; Nychas, G.J.E. Implementation of Multispectral Imaging (MSI) for Microbiological Quality Assessment of Poultry Products. Microorganisms 2020, 8, 552. [Google Scholar] [CrossRef] [PubMed]
- Fengou, L.C.; Tsakanikas, P.; Nychas, G.J.E. Rapid detection of minced pork and chicken adulteration in fresh, stored and cooked ground meat. Food Control 2021, 125, 108002. [Google Scholar] [CrossRef]
- Spyrelli, E.D.; Papachristou, C.; Nychas, G.J.E.; Panagou, E.Z. Microbiological Quality Assessment of Chicken Thigh Fillets Using Spectroscopic Sensors and Multivariate Data Analysis. Foods 2021, 10, 2723. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi-Varnamkhasti, M.; Mohtasebi, S.S.; Siadat, M.; Balasubramanian, S. Meat quality assessment by electronic nose (machine olfaction technology). Sensors 2009, 9, 6058. [Google Scholar] [CrossRef]
- Shi, H.; Zhang, M.; Adhikari, B. Advances of electronic nose and its application in fresh foods: A review. Crit. Rev. Food Sci. Nutr. 2018, 58, 2700–2710. [Google Scholar] [CrossRef]
- Song, S.; Yuan, L.; Zhang, X.; Hayat, K.; Chen, H.; Liu, F.; Xiao, Z.; Niu, Y. Rapid measuring and modelling flavour quality changes of oxidised chicken fat by electronic nose profiles through the partial least squares regression analysis. Food Chem. 2013, 141, 4278–4288. [Google Scholar] [CrossRef]
- Di Rosa, A.R.; Leone, F.; Cheli, F.; Chiofalo, V. Fusion of electronic nose, electronic tongue and computer vision for animal source food authentication and quality assessment—A review. J. Food Eng. 2017, 210, 62–75. [Google Scholar] [CrossRef]
- Loutfi, A.; Coradeschi, S.; Mani, G.K.; Shankar, P.; Rayappan, J.B.B. Electronic noses for food quality: A review. J. Food Eng. 2015, 144, 103–111. [Google Scholar] [CrossRef]
- Ghasemi-Varnamkhasti, M.; Mohtasebi, S.S.; Siadat, M. Biomimetic-based odor and taste sensing systems to food quality and safety characterization: An overview on basic principles and recent achievements. J. Food Eng. 2010, 100, 377–387. [Google Scholar] [CrossRef]
- Kutsanedzie, F.Y.; Guo, Z.; Chen, Q. Advances in nondestructive methods for meat quality and safety monitoring. Food Rev. Int. 2019, 35, 536–562. [Google Scholar] [CrossRef]
- Wojnowski, W.; Kalinowska, K.; Majchrzak, T.; Płotka-Wasylka, J.; Namieśnik, J. Prediction of the biogenic amines index of poultry meat using an electronic nose. Sensors 2019, 19, 1580. [Google Scholar] [CrossRef]
- Rajamäki, T.; Alakomi, H.L.; Ritvanen, T.; Skyttä, E.; Smolander, M.; Ahvenainen, R. Application of an electronic nose for quality assessment of modified atmosphere packaged poultry meat. Food Control 2016, 17, 5–13. [Google Scholar] [CrossRef]
- Wang, D.; Wang, X.; Liu, T.; Liu, Y. Prediction of total viable counts on chilled pork using an electronic nose combined with support vector machine. Meat Sci. 2012, 90, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulou, O.S.; Panagou, E.Z.; Mohareb, F.R.; Nychas, G.J.E. Sensory and microbiological quality assessment of beef fillets using a portable electronic nose in tandem with support vector machine analysis. Food Res. Int. 2013, 50, 241–249. [Google Scholar] [CrossRef]
- Timsorn, K.; Thoopboochagorn, T.; Lertwattanasakul, N.; Wongchoosuk, C. Evaluation of bacterial population on chicken meats using a briefcase electronic nose. Biosyst. Eng. 2016, 151, 116–125. [Google Scholar] [CrossRef]
- Estelles-Lopez, L.; Ropodi, A.; Pavlidis, D.; Fotopoulou, J.; Gkousari, C.; Peyrodie, A.; Panagou, E.Z.; Nychas, G.-J.E.; Mohareb, F. An automated ranking platform for machine learning regression models for meat spoilage prediction using multi-spectral imaging and metabolic profiling. Food Res. Int. 2017, 99, 206–215. [Google Scholar] [CrossRef]
- Li, H.; Chen, Q.; Zhao, J.; Ouyang, Q. Non-destructive evaluation of pork freshness using a portable electronic nose (E-nose) based on a colorimetric sensor array. Anal. Methods 2014, 6, 6271–6277. [Google Scholar] [CrossRef]
- Huang, L.; Zhao, J.; Chen, Q.; Zhang, Y. Nondestructive measurement of total volatile basic nitrogen (TVB-N) in pork meat by integrating near infrared spectroscopy, computer vision and electronic nose techniques. Food Chem. 2014, 145, 228–236. [Google Scholar] [CrossRef]
- Borràs, E.; Ferré, J.; Boqué, R.; Mestres, M.; Aceña, L.; Busto, O. Data fusion methodologies for food and beverage authentication and quality assessment—A review. Anal. Chim. Acta 2015, 891, 1–14. [Google Scholar] [CrossRef]
- Liu, D.; Sun, D.W.; Zeng, X.A. Recent advances in wavelength selection techniques for hyperspectral image processing in the food industry. Food Bioprocess Technol. 2014, 7, 307–323. [Google Scholar] [CrossRef]
- Alamprese, C.; Casale, M.; Sinelli, N.; Lanteri, S.; Casiraghi, E. Detection of minced beef adulteration with turkey meat by UV–vis, NIR and MIR spectroscopy. LWT 2013, 53, 225–232. [Google Scholar] [CrossRef]
- Weng, X.; Luan, X.; Kong, C.; Chang, Z.; Li, Y.; Zhang, S.; Al-Majeed, S.; Xiao, Y. A comprehensive method for assessing meat freshness using fusing electronic nose, computer vision, and artificial tactile technologies. J. Sens. 2020, 2020, 8838535. [Google Scholar] [CrossRef]
- Suxia, X.; Rui, W.; JiuQing, W.; PeiYuan, G. Study on chicken quality classification method based on K-means-RBF multi-source data fusion. In Proceedings of the 2018 Chinese Control and Decision Conference (CCDC), Shenyang, China, 9–11 June 2018; IEEE: Manhattan, NY, USA, 2018; pp. 405–410. [Google Scholar] [CrossRef]
- Khulal, U.; Zhao, J.; Hu, W.; Chen, Q. Intelligent evaluation of total volatile basic nitrogen (TVB-N) content in chicken meat by an improved multiple level data fusion model. Sens. Actuators B Chem. 2017, 238, 337–345. [Google Scholar] [CrossRef]
- Chung, S.; Yoon, S.C. Detection of Foreign Materials on Broiler Breast Meat Using a Fusion of Visible Near-Infrared and Short-Wave Infrared Hyperspectral Imaging. Appl. Sci. 2021, 11, 11987. [Google Scholar] [CrossRef]
- Dissing, B.S.; Papadopoulou, O.S.; Tassou, C.; Ersbøll, B.K.; Carstensen, J.M.; Panagou, E.Z.; Nychas, G.J. Using multispectral imaging for spoilage detection of pork meat. Food Bioprocess Technol. 2013, 6, 2268–2279. [Google Scholar] [CrossRef]
- Tsakanikas, P.; Pavlidis, D.; Nychas, G.J. High throughput multispectral image processing with applications in food science. PLoS ONE 2015, 10, e0140122. [Google Scholar] [CrossRef]
- Fengou, L.C.; Spyrelli, E.; Lianou, A.; Tsakanikas, P.; Panagou, E.Z.; Nychas, G.-J.E. Estimation of minced pork microbiological spoilage through fourier transform infrared and visible spectroscopy and multispectral vision technology. Foods 2019, 8, 238. [Google Scholar] [CrossRef]
- Lin, H.; Yan, Y.; Zhao, T.; Peng, L.; Zou, H.; Li, J.; Yang, X.; Xiong, Y.; Wang, M.; Wu, H. Rapid discrimination of Apiaceae plants by electronic nose coupled with multivariate statistical analyses. J. Pharm. Biomed. Anal. 2013, 84, 1–4. [Google Scholar] [CrossRef]
- Bi, Y.; Yuan, K.; Xiao, W.; Wu, J.; Shi, C.; Xia, J.; Chu, G.; Zhang, G.; Zhou, G. A local pre-processing method for near-infrared spectra, combined with spectral segmentation and standard normal variate transformation. Anal. Chim. Acta 2016, 909, 30–40. [Google Scholar] [CrossRef]
- Alamprese, C.; Amigo, J.M.; Casiraghi, E.; Engelsen, S.B. Identification and quantification of turkey meat adulteration in fresh, frozen-thawed and cooked minced beef by FT-NIR spectroscopy and chemometrics. Meat Sci. 2016, 121, 175–181. [Google Scholar] [CrossRef]
- Márquez, C.; López, M.I.; Ruisánchez, I.; Callao, M.P. FT-Raman and NIR spectroscopy data fusion strategy for multivariate qualitative analysis of food fraud. Talanta 2016, 161, 80–86. [Google Scholar] [CrossRef] [PubMed]
- Sokolova, M.; Lapalme, G. A systematic analysis of performance measures for classification tasks. Inf. Process. Manag. 2009, 45, 427–437. [Google Scholar] [CrossRef]
- Galarz, L.A.; Fonseca, G.G.; Prentice, C. Predicting bacterial growth in raw, salted, and cooked chicken breast fillets during storage. Food Sci. Technol. Int. 2016, 22, 461–474. [Google Scholar] [CrossRef] [PubMed]
- Gospavic, R.; Kreyenschmidt, J.; Bruckner, S.; Popov, V.; Haque, N. Mathematical modelling for predicting the growth of Pseudomonas spp. in poultry under variable temperature conditions. Food Sci. Technol. Int. 2008, 127, 290–297. [Google Scholar] [CrossRef]
- Rouger, A.; Tresse, O.; Zagorec, M. Bacterial contaminants of poultry meat: Sources, species, and dynamics. Microorganisms 2017, 5, 50. [Google Scholar] [CrossRef]
- Nychas, G.-J.E.; Skandamis, P.; Tassou, C.C.; Koutsoumanis, K. Meat spoilage during distribution. Meat Sci. 2008, 78, 77–89. [Google Scholar] [CrossRef]
- Xu, G.; Liao, C.; Ren, X.; Zhang, X.; Zhang, X.; Liu, S.; Fu, X.; Wu, H.; Huang, L.; Liu, C.; et al. Rapid assessment of quality of deer antler slices by using an electronic nose coupled with chemometric analysis. Rev. Bras. Farmacogn. 2014, 24, 716–721. [Google Scholar] [CrossRef]
- Wickramasinghe, N.N.; Ravensdale, J.T.; Coorey, R.; Dykes, G.A.; Scott Chandry, P. In situ characterisation of biofilms formed by psychrotrophic meat spoilage Pseudomonads. Biofouling 2019, 35, 840–855. [Google Scholar] [CrossRef]
- Böcker, U.; Ofstad, R.; Wu, Z.; Bertram, H.C.; Sockalingum, G.D.; Manfait, M.; Kohler, A. Revealing covariance structures in Fourier transform infrared and Raman microspectroscopy spectra: A study on pork muscle fiber tissue subjected to different processing parameters. Appl. Spectrosc. 2007, 61, 1032–1039. [Google Scholar] [CrossRef]
- Mohareb, F.; Iriondoa, M.; Doulgeraki, A.I.; Van Hoekc, A.; Aarts, H.; Cauchia, M.; Nychas, G.-J. Identification of meat spoilage gene biomarkers in Pseudomonas putida using gene profiling. Food Control 2015, 57, 152–160. [Google Scholar] [CrossRef]
- Nychas, G.-J.; Sims, E.; Tsakanikas, P.; Mohareb, F. Data Science in the Food Industry. Annu. Rev. Biomed. Data Sci. 2021, 4, 341–367. [Google Scholar] [CrossRef] [PubMed]
- Kamruzzaman, M.; Sun, D.W.; ElMasry, G.; Allen, P. Fast detection and visualization of minced lamb meat adulteration using NIR hyperspectral imaging and multivariate image analysis. Talanta 2013, 103, 130–136. [Google Scholar] [CrossRef] [PubMed]
- Rahman, U.; Sahar, A.; Pasha, I.; Rahman, S.; Ishaq, A. Assessing the capability of Fourier transform infrared spectroscopy in tandem with chemometric analysis for predicting poultry meat spoilage. PeerJ 2018, 6, e5376. [Google Scholar] [CrossRef] [PubMed]




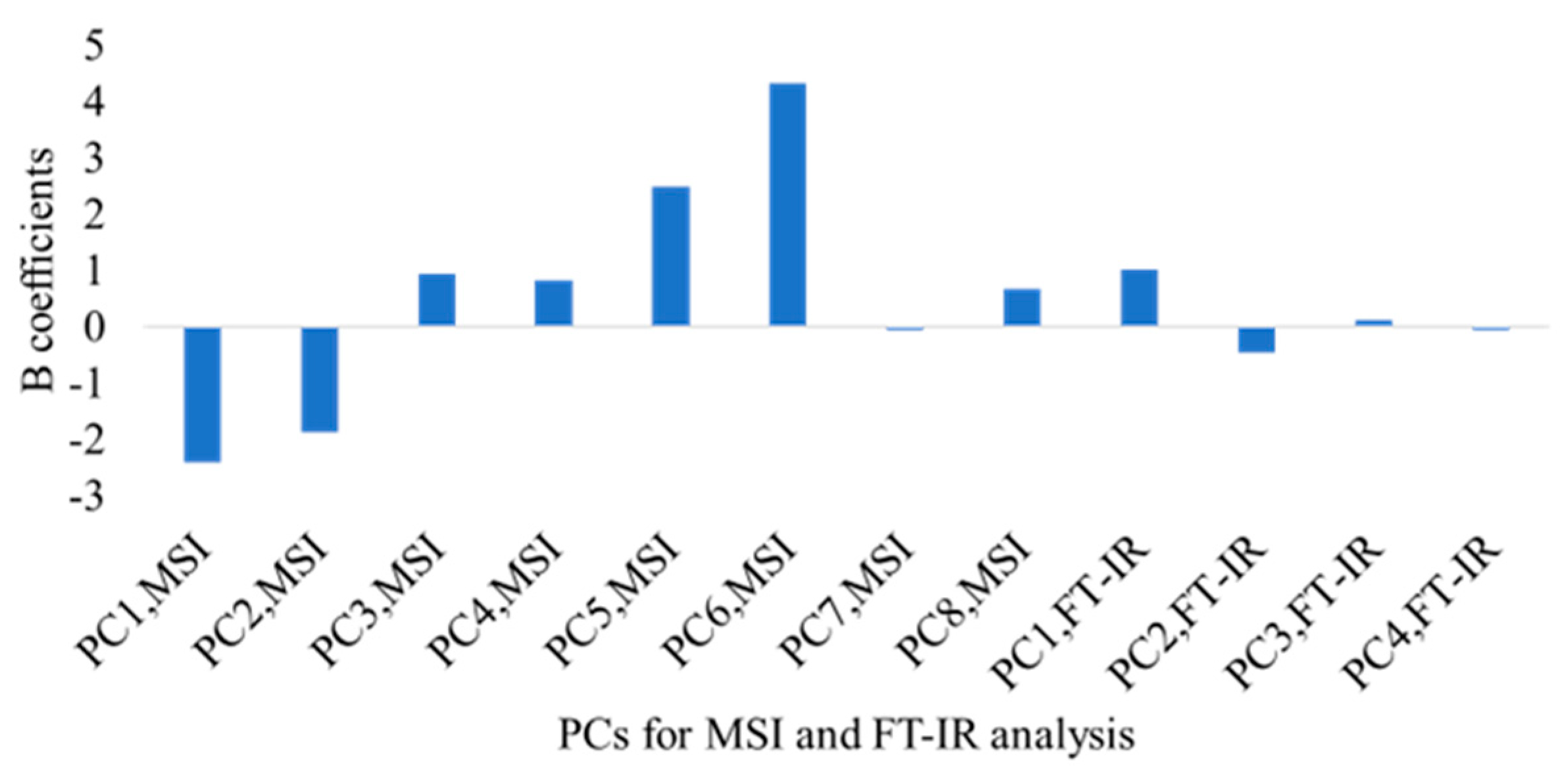
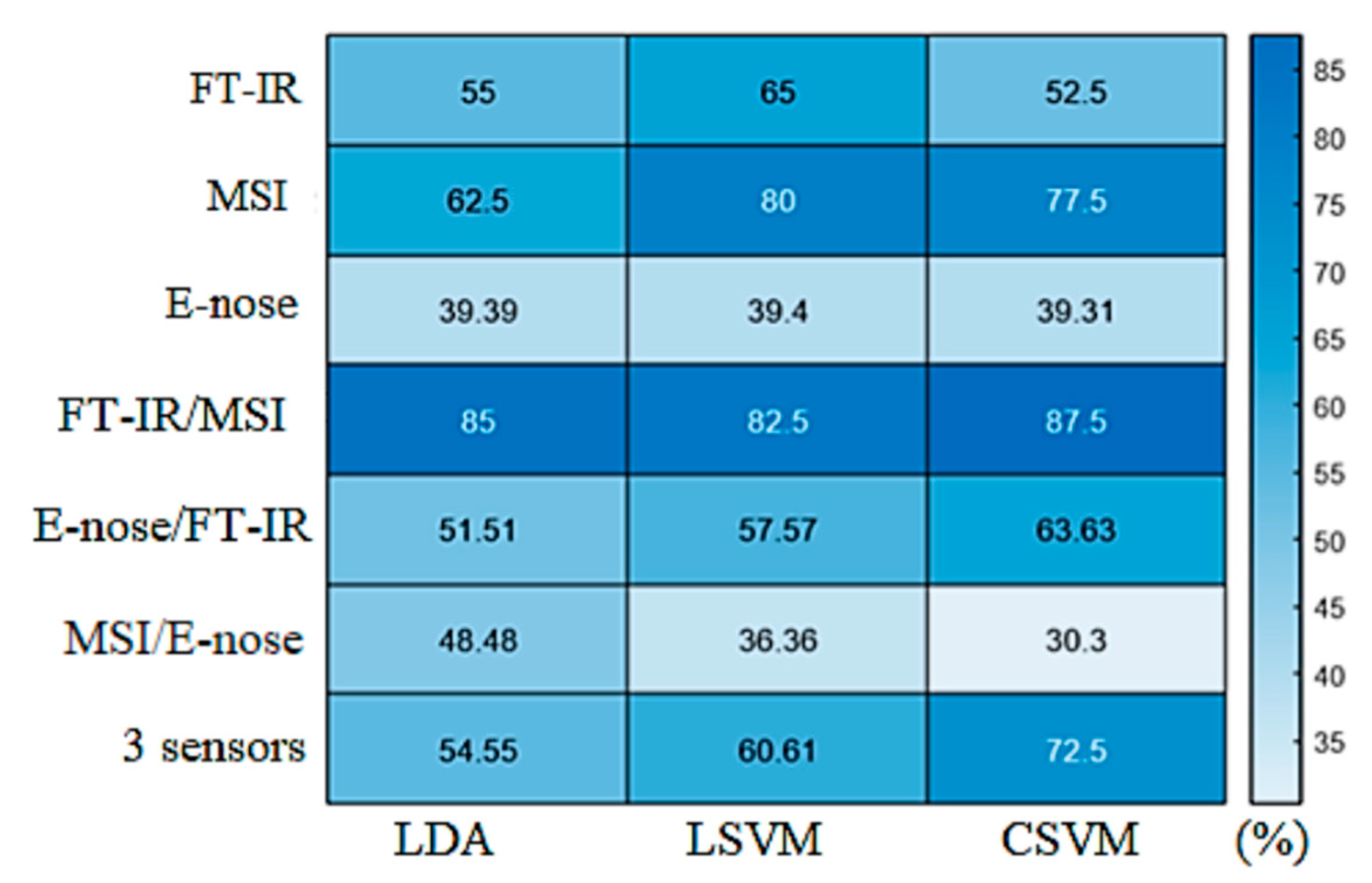
| Sensor | Process | Observations | Slope | Offset | Correlation Coefficient, r | Root Mean Squared Error, RMSE (Log CFU/g) |
|---|---|---|---|---|---|---|
| MSI | FCV 1 | 169 | 0.776 | 1.698 | 0.868 | 0.815 |
| Prediction | 40 | 0.511 | 3.419 | 0.803 | 0.998 | |
| FT-IR | FCV | 169 | 0.62 | 2.87 | 0.746 | 1.099 |
| Prediction | 40 | 0.374 | 4.902 | 0.497 | 1.627 | |
| E-nose | FCV | 169 | 0.576 | 3.232 | 0.757 | 1.12 |
| Prediction | 40 | 0.044 | 6.145 | 0.245 | 1.921 | |
| MSI/FT-IR | FCV | 169 | 0.687 | 2.363 | 0.818 | 0.941 |
| Prediction | 40 | 0.592 | 2.689 | 0.783 | 0.983 | |
| FT-IR/E-nose | FCV | 169 | 0.598 | 3.055 | 0.758 | 1.131 |
| Prediction | 40 | 0.171 | 6.245 | 0.222 | 1.757 | |
| MSI/E-nose | FCV | 169 | 0.596 | 3.061 | 0.75 | 1.149 |
| Prediction | 40 | 0.503 | 3.498 | 0.727 | 1.373 | |
| Three sensors | FCV | 169 | 0.596 | 3.056 | 0.751 | 1.148 |
| Prediction | 40 | 0.474 | 3.821 | 0.722 | 1.367 |
| Sensor | ||||||
|---|---|---|---|---|---|---|
| E-Nose | FT-IR | MSI | ||||
| Step | k-CV 1 | Prediction | k-CV | Prediction | k-CV | Prediction |
| RMSE (log CFU/g) | 1.311 | 1.921 | 1.846 | 3.583 | 0.832 | 0.973 |
| E-nose/FT-IR | FT-IR/MSI | MSI/E-nose | ||||
| Step | k-CV | Prediction | k-CV | Prediction | k-CV | Prediction |
| RMSE (log CFU/g) | 1.06 | 1.579 | 0.953 | 0.999 | 1.134 | 1.658 |
| 3-sensors | ||||||
| Step | k-CV | Prediction | ||||
| RMSE (log CFU/g) | 1.022 | 1.938 | ||||
| Sensor | Model | Step | Confusion Matrix | Performance Metrics | |||
|---|---|---|---|---|---|---|---|
| MSI | LSVM | k-CV | o/p | Class 1 1 | Class 2 2 | Sensitivity (%) | Precision (%) |
| Class 1 | 64 | 7 | 90.14 | 83.12 | |||
| Class 2 | 13 | 85 | 86.75 | ||||
| Prediction | o/p | Class 1 | Class 2 | Sensitivity (%) | Precision (%) | ||
| Class 1 | 17 | 5 | 77.27 | 85 | |||
| Class 2 | 3 | 15 | 83.33 | ||||
| Model | Step | Confusion Matrix | Performance metrics | ||||
| CSVM | k-CV | o/p | Class 1 | Class 2 | Sensitivity (%) | Precision (%) | |
| Class 1 | 53 | 18 | 74.65 | 68.83 | |||
| Class 2 | 24 | 74 | 75.51 | ||||
| Prediction | o/p | Class 1 | Class 2 | Sensitivity (%) | Precision (%) | ||
| Class 1 | 21 | 1 | 95.45 | 72.41 | |||
| Class 2 | 8 | 10 | 55.55 | ||||
| Sensor | Model | Step | Confusion Matrix | Performance Metrics | |||
|---|---|---|---|---|---|---|---|
| FT-IR/MSI | LDA | k-CV | o/p | Class 1 | Class 2 | Sensitivity (%) | Precision (%) |
| Class 1 | 56 | 14 | 80 | 80 | |||
| Class 2 | 14 | 85 | 85.86 | ||||
| Prediction | o/p | Class 1 | Class 2 | Sensitivity (%) | Precision (%) | ||
| Class 1 | 19 | 3 | 86.36 | 86.36 | |||
| Class 2 | 3 | 15 | 83.33 | ||||
| Model | Step | Confusion Matrix | Performance metrics | ||||
| LSVM | k-CV | o/p | Class 1 | Class 2 | Sensitivity (%) | Precision (%) | |
| Class 1 | 61 | 9 | 87.14 | 78.20 | |||
| Class 2 | 17 | 82 | 82.83 | ||||
| Prediction | o/p | Class 1 | Class 2 | Sensitivity (%) | Precision (%) | ||
| Class 1 | 17 | 5 | 77.27 | 89.47 | |||
| Class 2 | 2 | 16 | 88.89 | ||||
| Model | Step | Confusion Matrix | Performance metrics | ||||
| CSVM | k-CV | o/p | Class 1 | Class 2 | Sensitivity (%) | Precision (%) | |
| Class 1 | 54 | 16 | 77.14 | 75 | |||
| Class 2 | 18 | 81 | 81.82 | ||||
| Prediction | o/p | Class 1 | Class 2 | Sensitivity (%) | Precision (%) | ||
| Class 1 | 20 | 2 | 90 | 86.95 | |||
| Class 2 | 3 | 15 | 83.33 | ||||
| Three sensors | Model | Step | Confusion Matrix | Performance metrics | |||
| CSVM | o/p | Class 1 | Class 2 | Sensitivity (%) | Precision (%) | ||
| k-CV | Class 1 | 59 | 6 | 90.77 | 86.76 | ||
| Class 2 | 9 | 95 | 91.34 | ||||
| Prediction | o/p | Class 1 | Class 2 | Sensitivity (%) | Precision (%) | ||
| Class 1 | 15 | 2 | 88.23 | 68.18 | |||
| Class 2 | 7 | 14 | 66.67 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Spyrelli, E.D.; Nychas, G.-J.E.; Panagou, E.Z. Assessment of the Microbial Spoilage and Quality of Marinated Chicken Souvlaki through Spectroscopic and Biomimetic Sensors and Data Fusion. Microorganisms 2022, 10, 2251. https://doi.org/10.3390/microorganisms10112251
Spyrelli ED, Nychas G-JE, Panagou EZ. Assessment of the Microbial Spoilage and Quality of Marinated Chicken Souvlaki through Spectroscopic and Biomimetic Sensors and Data Fusion. Microorganisms. 2022; 10(11):2251. https://doi.org/10.3390/microorganisms10112251
Chicago/Turabian StyleSpyrelli, Evgenia D., George-John E. Nychas, and Efstathios Z. Panagou. 2022. "Assessment of the Microbial Spoilage and Quality of Marinated Chicken Souvlaki through Spectroscopic and Biomimetic Sensors and Data Fusion" Microorganisms 10, no. 11: 2251. https://doi.org/10.3390/microorganisms10112251
APA StyleSpyrelli, E. D., Nychas, G.-J. E., & Panagou, E. Z. (2022). Assessment of the Microbial Spoilage and Quality of Marinated Chicken Souvlaki through Spectroscopic and Biomimetic Sensors and Data Fusion. Microorganisms, 10(11), 2251. https://doi.org/10.3390/microorganisms10112251







