Oral Microbiota and Tumor—A New Perspective of Tumor Pathogenesis
Abstract
1. Introduction
2. Systemic Involvement of Oral Microecology
3. Dysregulated Oral Microbiota Poses a Challenge to the Immune System
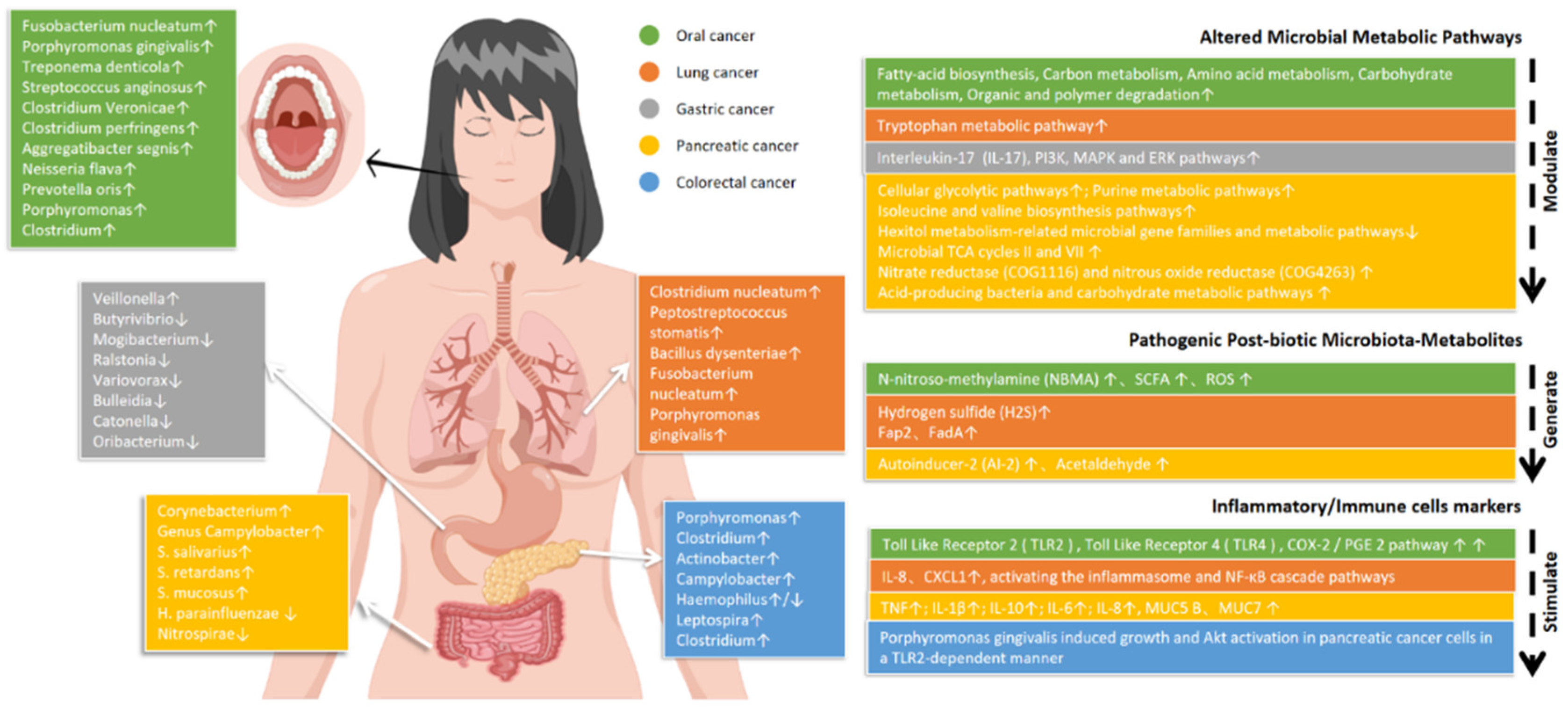
4. Is the Oral Microbiota a Cause or a Consequence of Tumorigenesis?
4.1. Oral Cancer and Oral Microbiota
4.2. Gastric Cancer and Oral Microbiota
4.3. Colorectal Cancer and Oral Microbiota
4.4. Pancreatic Cancer and Oral Microbiota
4.5. Lung Cancer and Oral Microbiota
4.6. Breast Cancer and Oral Microbiota
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Klünemann, M.; Andrejev, S.; Blasche, S.; Mateus, A.; Phapale, P.; Devendran, S.; Vappiani, J.; Simon, B.; Scott, T.A.; Kafkia, E.; et al. Bioaccumulation of therapeutic drugs by human gut bacteria. Nature 2021, 597, 533–538. [Google Scholar] [CrossRef]
- Wu, W.L.; Adame, M.D.; Liou, C.W.; Barlow, J.T.; Lai, T.T.; Sharon, G.; Schretter, C.E.; Needham, B.D.; Wang, M.I.; Tang, W.; et al. Microbiota regulate social behaviour via stress response neurons in the brain. Nature 2021, 595, 409–414. [Google Scholar] [CrossRef]
- Rizvi, Z.A.; Dalal, R.; Sadhu, S.; Kumar, Y.; Kumar, S.; Gupta, S.K.; Tripathy, M.R.; Rathore, D.K.; Awasthi, A. High-salt diet mediates interplay between NK cells and gut microbiota to induce potent tumor immunity. Sci. Adv. 2021, 7, eabg5016. [Google Scholar] [CrossRef]
- Tuominen, H.; Rautava, J. Oral Microbiota and Cancer Development. Pathobiology 2021, 88, 116–126. [Google Scholar] [CrossRef]
- López-Santacruz, H.D.; López-López, A.; Revilla-Guarinos, A.; Camelo-Castillo, A.; Esparza-Villalpando, V.; Mira, A.; Aranda-Romo, S. Streptococcus dentisani is a common inhabitant of the oral microbiota worldwide and is found at higher levels in caries-free individuals. Int. Microbiol. 2021, 24, 619–629. [Google Scholar] [CrossRef]
- Kong, J.; Zhang, G.; Xia, K.; Diao, C.; Yang, X.; Zuo, X.; Li, Y.; Liang, X. Tooth brushing using toothpaste containing theaflavins reduces the oral pathogenic bacteria in healthy adults. 3 Biotech 2021, 11, 150. [Google Scholar] [CrossRef]
- Pomeroy, C.; Shanholtzer, C.J.; Peterson, L.R. Selenomonas bacteraemia—Case report and review of the literature. J. Infect. 1987, 15, 237–242. [Google Scholar] [CrossRef]
- Kim, Y.C.; Choi, S.; Sohn, K.H.; Bang, J.Y.; Kim, Y.; Jung, J.W.; Kim, H.Y.; Park, J.; Kim, K.; Kang, M.G.; et al. Selenomonas: A marker of asthma severity with the potential therapeutic effect. Allergy 2022, 77, 317–320. [Google Scholar] [CrossRef]
- Casasanta, M.A.; Yoo, C.C.; Udayasuryan, B.; Sanders, B.E.; Umaña, A.; Zhang, Y.; Peng, H.; Duncan, A.J.; Wang, Y.; Li, L.; et al. Fusobacterium nucleatum host-cell binding and invasion induces IL-8 and CXCL1 secretion that drives colorectal cancer cell migration. Sci. Signal. 2020, 13, eaba9157. [Google Scholar] [CrossRef]
- Aas, J.A.; Paster, B.J.; Stokes, L.N.; Olsen, I.; Dewhirst, F.E. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 2005, 43, 5721–5732. [Google Scholar] [CrossRef]
- Bai, H.; Yang, J.; Meng, S.; Liu, C. Oral Microbiota-Driven Cell Migration in Carcinogenesis and Metastasis. Front. Cell. Infect. Microbiol. 2022, 12, 864479. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Hwang, B.O.; Lim, M.; Ok, S.H.; Lee, S.K.; Chun, K.S.; Park, K.K.; Hu, Y.; Chung, W.Y.; Song, N.Y. Oral-Gut Microbiome Axis in Gastrointestinal Disease and Cancer. Cancers 2021, 13, 2124. [Google Scholar] [CrossRef]
- Seidel, C.L.; Gerlach, R.G.; Wiedemann, P.; Weider, M.; Rodrian, G.; Hader, M.; Frey, B.; Gaipl, U.S.; Bozec, A.; Cieplik, F.; et al. Defining Metaniches in the Oral Cavity According to Their Microbial Composition and Cytokine Profile. Int. J. Mol. Sci. 2020, 21, 8218. [Google Scholar] [CrossRef]
- Chen, H.; Xie, H.; Shao, D.; Chen, L.; Chen, S.; Wang, L.; Han, X. Oral Microbiota, a Potential Determinant for the Treatment Efficacy of Gastric Helicobacter pylori Eradication in Humans. Pol. J. Microbiol. 2022, 71, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.H.; Chen, H.M.; Yang, S.F.; Liang, C.; Peng, C.Y.; Lin, F.M.; Tsai, L.L.; Wu, B.C.; Hsin, C.H.; Chuang, C.Y.; et al. Bacterial alterations in salivary microbiota and their association in oral cancer. Sci. Rep. 2017, 7, 16540. [Google Scholar] [CrossRef] [PubMed]
- Durack, J.; Christian, L.S.; Nariya, S.; Gonzalez, J.; Bhakta, N.R.; Ansel, K.M.; Beigelman, A.; Castro, M.; Dyer, A.M.; Israel, E.; et al. Distinct associations of sputum and oral microbiota with atopic, immunologic, and clinical features in mild asthma. J. Allergy Clin. Immunol. 2020, 146, 1016–1026. [Google Scholar] [CrossRef]
- Mohammed, H.; Varoni, E.M.; Cochis, A.; Cordaro, M.; Gallenzi, P.; Patini, R.; Staderini, E.; Lajolo, C.; Rimondini, L.; Rocchetti, V. Oral Dysbiosis in Pancreatic Cancer and Liver Cirrhosis: A Review of the Literature. Biomedicines 2018, 6, 115. [Google Scholar] [CrossRef]
- Jiménez, C.; Garrido, M.; Pussinen, P.; Bordagaray, M.J.; Fernández, A.; Vega, C.; Chaparro, A.; Hoare, A.; Hernández, M. Systemic burden and cardiovascular risk to Porphyromonas species in apical periodontitis. Clin. Oral Investig. 2021, 26, 993–1001. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, T.; Wu, K.; Li, Z.; Chen, X.; Jiang, S.; Du, L.; Lu, S.; Lin, C.; Wu, J.; et al. Metagenomic Analysis Reveals a Possible Association Between Respiratory Infection and Periodontitis. Genom. Proteom. Bioinform. 2021, 16, 252–261. [Google Scholar] [CrossRef]
- Bregaint, S.; Boyer, E.; Fong, S.B.; Meuric, V.; Bonnaure-Mallet, M.; Jolivet-Gougeon, A. Porphyromonas gingivalis outside the oral cavity. Odontology 2021, 110, 1–19. [Google Scholar] [CrossRef]
- Si, J.; Lee, C.; Ko, G. Oral Microbiota: Microbial Biomarkers of Metabolic Syndrome Independent of Host Genetic Factors. Front. Cell. Infect. Microbiol. 2017, 7, 516. [Google Scholar] [CrossRef]
- Ribeiro, A.S.; Silva, D.A.; Silva, F.P.; Santos, G.C.; Campos, L.M.; Oliveira, L.V.; Santos, D.A. Epidemiology and phospholipase activity of oral Candida SPP. Among patients with central nervous system diseases before and after dental cleaning procedure. Braz. J. Microbiol. 2010, 41, 19–23. [Google Scholar] [CrossRef][Green Version]
- Karpiński, T.M. Role of Oral Microbiota in Cancer Development. Microorganisms 2019, 7, 20. [Google Scholar] [CrossRef]
- He, X.; McLean, J.S.; Guo, L.; Lux, R.; Shi, W. The social structure of microbial community involved in colonization resistance. ISME J. 2014, 8, 564–574. [Google Scholar] [CrossRef]
- Willems, H.M.; Kos, K.; Jabra-Rizk, M.A.; Krom, B.P. Candida albicans in oral biofilms could prevent caries. Pathog. Dis. 2016, 74, ftw039. [Google Scholar] [CrossRef]
- Iwase, T.; Uehara, Y.; Shinji, H.; Tajima, A.; Seo, H.; Takada, K.; Agata, T.; Mizunoe, Y. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 2010, 465, 346–349. [Google Scholar] [CrossRef]
- Wen, Z.T.; Yates, D.; Ahn, S.J.; Burne, R.A. Biofilm formation and virulence expression by Streptococcus mutans are altered when grown in dual-species model. BMC Microbiol. 2010, 10, 111. [Google Scholar] [CrossRef]
- Howard, K.C.; Gonzalez, O.A.; Garneau-Tsodikova, S. Porphyromonas gingivalis: Where do we stand in our battle against this oral pathogen? RSC Med. Chem. 2021, 12, 666–704. [Google Scholar] [CrossRef]
- Abdulkareem, A.A.; Shelton, R.M.; Landini, G.; Cooper, P.R.; Milward, M.R. Periodontal pathogens promote epithelial-mesenchymal transition in oral squamous carcinoma cells in vitro. Cell Adhes. Migr. 2018, 12, 127–137. [Google Scholar] [CrossRef]
- Abdulkareem, A.A.; Shelton, R.M.; Landini, G.; Cooper, P.R.; Milward, M.R. Potential role of periodontal pathogens in compromising epithelial barrier function by inducing epithelial-mesenchymal transition. J. Periodontal Res. 2018, 53, 565–574. [Google Scholar] [CrossRef]
- Brock, M.; Bahammam, S.; Sima, C. The Relationships Among Periodontitis, Pneumonia and COVID-19. Front. Oral Health 2021, 2, 801815. [Google Scholar] [CrossRef] [PubMed]
- Diaz, P.I.; Hoare, A.; Hong, B.Y. Subgingival Microbiome Shifts and Community Dynamics in Periodontal Diseases. J. Calif. Dent. Assoc. 2016, 44, 421–435. [Google Scholar] [PubMed]
- Radstake, T. Extraintestinal translocation of microbes and tissue specificity in rheumatic musculoskeletal disease (RMD): Its more than a gut feeling. Ann. Rheum. Dis. 2018, 77, 1702–1704. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Cong, Y. Gut microbiota-derived metabolites in the regulation of host immune responses and immune-related inflammatory diseases. Cell. Mol. Immunol. 2021, 18, 866–877. [Google Scholar] [CrossRef] [PubMed]
- Kitamoto, S.; Nagao-Kitamoto, H.; Jiao, Y.; Gillilland, M.G., 3rd; Hayashi, A.; Imai, J.; Sugihara, K.; Miyoshi, M.; Brazil, J.C.; Kuffa, P.; et al. The Intermucosal Connection between the Mouth and Gut in Commensal Pathobiont-Driven Colitis. Cell 2020, 182, 447–462.e414. [Google Scholar] [CrossRef]
- Groeger, S.; Meyle, J. Oral Mucosal Epithelial Cells. Front. Immunol. 2019, 10, 208. [Google Scholar] [CrossRef]
- Tuganbaev, T.; Yoshida, K.; Honda, K. The effects of oral microbiota on health. Science 2022, 376, 934–936. [Google Scholar] [CrossRef]
- Serapio-Palacios, A.; Woodward, S.E.; Vogt, S.L.; Deng, W.; Creus-Cuadros, A.; Huus, K.E.; Cirstea, M.; Gerrie, M.; Barcik, W.; Yu, H.; et al. Type VI secretion systems of pathogenic and commensal bacteria mediate niche occupancy in the gut. Cell Rep. 2022, 39, 110731. [Google Scholar] [CrossRef]
- Fan, X.; Alekseyenko, A.V.; Wu, J.; Peters, B.A.; Jacobs, E.J.; Gapstur, S.M.; Purdue, M.P.; Abnet, C.C.; Stolzenberg-Solomon, R.; Miller, G.; et al. Human oral microbiome and prospective risk for pancreatic cancer: A population-based nested case-control study. Gut 2018, 67, 120–127. [Google Scholar] [CrossRef]
- Mascitti, M.; Togni, L.; Troiano, G.; Caponio, V.C.A.; Gissi, D.B.; Montebugnoli, L.; Procaccini, M.; Lo Muzio, L.; Santarelli, A. Beyond Head and Neck Cancer: The Relationship Between Oral Microbiota and Tumour Development in Distant Organs. Front. Cell. Infect. Microbiol. 2019, 9, 232. [Google Scholar] [CrossRef]
- Wang, X.; Jia, Y.; Wen, L.; Mu, W.; Wu, X.; Liu, T.; Liu, X.; Fang, J.; Luan, Y.; Chen, P.; et al. Porphyromonas gingivalis Promotes Colorectal Carcinoma by Activating the Hematopoietic NLRP3 Inflammasome. Cancer Res. 2021, 81, 2745–2759. [Google Scholar] [CrossRef]
- Yang, Y.; Jobin, C. Far reach of Fusobacterium nucleatum in cancer metastasis. Gut 2020, 70, 1427–1429. [Google Scholar] [CrossRef]
- Sun, J.; Tang, Q.; Yu, S.; Xie, M.; Xie, Y.; Chen, G.; Chen, L. Role of the oral microbiota in cancer evolution and progression. Cancer Med. 2020, 9, 6306–6321. [Google Scholar] [CrossRef]
- Koliarakis, I.; Messaritakis, I.; Nikolouzakis, T.K.; Hamilos, G.; Souglakos, J.; Tsiaoussis, J. Oral Bacteria and Intestinal Dysbiosis in Colorectal Cancer. Int. J. Mol. Sci. 2019, 20, 4146. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, J.; Zhang, C.; Yu, N.; Lu, Z.; Zhang, S.; Li, Y.; Li, Q.; Liu, J.; Liu, D.; et al. Porphyromonas gingivalis exacerbates ulcerative colitis via Porphyromonas gingivalis peptidylarginine deiminase. Int. J. Oral Sci. 2021, 13, 31. [Google Scholar] [CrossRef]
- Hayes, C.; Donohoe, C.L.; Davern, M.; Donlon, N.E. The oncogenic and clinical implications of lactate induced immunosuppression in the tumour microenvironment. Cancer Lett. 2021, 500, 75–86. [Google Scholar] [CrossRef]
- Frankel, T.L.; Pasca di Magliano, M. Immune sensing of microbial metabolites: Action at the tumor. Immunity 2022, 55, 192–194. [Google Scholar] [CrossRef]
- Vinasco, K.; Mitchell, H.M.; Kaakoush, N.O.; Castaño-Rodríguez, N. Microbial carcinogenesis: Lactic acid bacteria in gastric cancer. Biochim. Biophys. Acta Rev. Cancer 2019, 1872, 188309. [Google Scholar] [CrossRef]
- Khurshid, Z.; Zafar, M.S.; Khan, R.S.; Najeeb, S.; Slowey, P.D.; Rehman, I.U. Role of Salivary Biomarkers in Oral Cancer Detection. Adv. Clin. Chem. 2018, 86, 23–70. [Google Scholar] [CrossRef]
- Chattopadhyay, I.; Verma, M.; Panda, M. Role of Oral Microbiome Signatures in Diagnosis and Prognosis of Oral Cancer. Technol. Cancer Res. Treat. 2019, 18, 1533033819867354. [Google Scholar] [CrossRef]
- Amer, A.; Galvin, S.; Healy, C.M.; Moran, G.P. The Microbiome of Potentially Malignant Oral Leukoplakia Exhibits Enrichment for Fusobacterium, Leptotrichia, Campylobacter, and Rothia Species. Front. Microbiol. 2017, 8, 2391. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Gong, D.; Shi, C.; Shao, F.; Shi, J.; Fei, J. Dysbiosis of oral buccal mucosa microbiota in patients with oral lichen planus. Oral Dis. 2017, 23, 674–682. [Google Scholar] [CrossRef] [PubMed]
- Gopinath, D.; Menon, R.K.; Wie, C.C.; Banerjee, M.; Panda, S.; Mandal, D.; Behera, P.K.; Roychoudhury, S.; Kheur, S.; Botelho, M.G.; et al. Differences in the bacteriome of swab, saliva, and tissue biopsies in oral cancer. Sci. Rep. 2021, 11, 1181. [Google Scholar] [CrossRef]
- Guo, N.; Zhou, L.X.; Meng, N.; Shi, Y.P. Associations of oral and intestinal florae and serum inflammatory factors with pathogenesis of oral cancer. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 11090–11095. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Hao, Y.; Peng, X.; Li, B.; Han, Q.; Ren, B.; Li, M.; Li, L.; Li, Y.; Cheng, G.; et al. The Clinical Potential of Oral Microbiota as a Screening Tool for Oral Squamous Cell Carcinomas. Front. Cell. Infect. Microbiol. 2021, 11, 728933. [Google Scholar] [CrossRef] [PubMed]
- Al-Hebshi, N.N.; Nasher, A.T.; Idris, A.M.; Chen, T. Robust species taxonomy assignment algorithm for 16S rRNA NGS reads: Application to oral carcinoma samples. J. Oral Microbiol. 2015, 7, 28934. [Google Scholar] [CrossRef]
- Rai, A.K.; Panda, M.; Das, A.K.; Rahman, T.; Das, R.; Das, K.; Sarma, A.; Kataki, A.C.; Chattopadhyay, I. Dysbiosis of salivary microbiome and cytokines influence oral squamous cell carcinoma through inflammation. Arch. Microbiol. 2021, 203, 137–152. [Google Scholar] [CrossRef]
- Wang, Y.; Liu, S.; Li, B.; Jiang, Y.; Zhou, X.; Chen, J.; Li, M.; Ren, B.; Peng, X.; Zhou, X.; et al. Staphylococcus aureus induces COX-2-dependent proliferation and malignant transformation in oral keratinocytes. J. Oral Microbiol. 2019, 11, 1643205. [Google Scholar] [CrossRef]
- Arzmi, M.H.; Dashper, S.; McCullough, M. Polymicrobial interactions of Candida albicans and its role in oral carcinogenesis. J. Oral Pathol. Med. 2019, 48, 546–551. [Google Scholar] [CrossRef]
- Zhou, Y.; Cheng, L.; Lei, Y.L.; Ren, B.; Zhou, X. The Interactions Between Candida albicans and Mucosal Immunity. Front. Microbiol. 2021, 12, 652725. [Google Scholar] [CrossRef]
- Falsetta, M.L.; Klein, M.I.; Colonne, P.M.; Scott-Anne, K.; Gregoire, S.; Pai, C.H.; Gonzalez-Begne, M.; Watson, G.; Krysan, D.J.; Bowen, W.H.; et al. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect. Immun. 2014, 82, 1968–1981. [Google Scholar] [CrossRef]
- Janus, M.M.; Crielaard, W.; Volgenant, C.M.; van der Veen, M.H.; Brandt, B.W.; Krom, B.P. Candida albicans alters the bacterial microbiome of early in vitro oral biofilms. J. Oral Microbiol. 2017, 9, 1270613. [Google Scholar] [CrossRef]
- Wu, J.S.; Zheng, M.; Zhang, M.; Pang, X.; Li, L.; Wang, S.S.; Yang, X.; Wu, J.B.; Tang, Y.J.; Tang, Y.L.; et al. Porphyromonas gingivalis Promotes 4-Nitroquinoline-1-Oxide-Induced Oral Carcinogenesis with an Alteration of Fatty Acid Metabolism. Front. Microbiol. 2018, 9, 2081. [Google Scholar] [CrossRef]
- Wang, Y.C.; Lu, S.; Zhou, X.J.; Yang, L.; Liu, P.; Zhang, L.; Hu, Y.; Dong, X.Z. miR-1273h-5p suppresses CXCL12 expression and inhibits gastric cancer cell invasion and metastasis. Open Med. 2022, 17, 930–946. [Google Scholar] [CrossRef]
- Zhang, X.; Li, C.; Cao, W.; Zhang, Z. Alterations of Gastric Microbiota in Gastric Cancer and Precancerous Stages. Front. Cell. Infect. Microbiol. 2021, 11, 559148. [Google Scholar] [CrossRef]
- Huang, K.; Gao, X.; Wu, L.; Yan, B.; Wang, Z.; Zhang, X.; Peng, L.; Yu, J.; Sun, G.; Yang, Y. Salivary Microbiota for Gastric Cancer Prediction: An Exploratory Study. Front. Cell. Infect. Microbiol. 2021, 11, 640309. [Google Scholar] [CrossRef]
- Coker, O.O.; Dai, Z.; Nie, Y.; Zhao, G.; Cao, L.; Nakatsu, G.; Wu, W.K.; Wong, S.H.; Chen, Z.; Sung, J.J.Y.; et al. Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut 2018, 67, 1024–1032. [Google Scholar] [CrossRef]
- Chen, X.H.; Wang, A.; Chu, A.N.; Gong, Y.H.; Yuan, Y. Mucosa-Associated Microbiota in Gastric Cancer Tissues Compared with Non-cancer Tissues. Front. Microbiol. 2019, 10, 1261. [Google Scholar] [CrossRef]
- Sun, J.H.; Li, X.L.; Yin, J.; Li, Y.H.; Hou, B.X.; Zhang, Z. A screening method for gastric cancer by oral microbiome detection. Oncol. Rep. 2018, 39, 2217–2224. [Google Scholar] [CrossRef]
- Wu, J.; Xu, S.; Xiang, C.; Cao, Q.; Li, Q.; Huang, J.; Shi, L.; Zhang, J.; Zhan, Z. Tongue Coating Microbiota Community and Risk Effect on Gastric Cancer. J. Cancer 2018, 9, 4039–4048. [Google Scholar] [CrossRef]
- Cui, J.; Cui, H.; Yang, M.; Du, S.; Li, J.; Li, Y.; Liu, L.; Zhang, X.; Li, S. Tongue coating microbiome as a potential biomarker for gastritis including precancerous cascade. Protein Cell 2019, 10, 496–509. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Long, J.; Wang, C.; Blot, W.J.; Pei, Z.; Shu, X.; Wu, F.; Rothman, N.; Wu, J.; Lan, Q.; et al. Prospective study of oral microbiome and gastric cancer risk among Asian, African American and European American populations. Int. J. Cancer 2022, 150, 916–927. [Google Scholar] [CrossRef] [PubMed]
- Castaño-Rodríguez, N.; Goh, K.L.; Fock, K.M.; Mitchell, H.M.; Kaakoush, N.O. Dysbiosis of the microbiome in gastric carcinogenesis. Sci. Rep. 2017, 7, 15957. [Google Scholar] [CrossRef] [PubMed]
- Yee, J.K.C. Are the view of Helicobacter pylori colonized in the oral cavity an illusion? Exp. Mol. Med. 2017, 49, e397. [Google Scholar] [CrossRef] [PubMed]
- Silva, D.G.; Stevens, R.H.; Macedo, J.M.; Hirata, R.; Pinto, A.C.; Alves, L.M.; Veerman, E.C.; Tinoco, E.M. Higher levels of salivary MUC5B and MUC7 in individuals with gastric diseases who harbor Helicobacter pylori. Arch. Oral Biol. 2009, 54, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Doherty, J.R.; Cleveland, J.L. Targeting lactate metabolism for cancer therapeutics. J. Clin. Investig. 2013, 123, 3685–3692. [Google Scholar] [CrossRef]
- Lee, H.; Woo, S.M.; Jang, H.; Kang, M.; Kim, S.Y. Cancer depends on fatty acids for ATP production: A possible link between cancer and obesity. Semin. Cancer Biol. 2022, 86, 347–357. [Google Scholar] [CrossRef]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef]
- Mirzaei, R.; Dehkhodaie, E.; Bouzari, B.; Rahimi, M.; Gholestani, A.; Hosseini-Fard, S.R.; Keyvani, H.; Teimoori, A.; Karampoor, S. Dual role of microbiota-derived short-chain fatty acids on host and pathogen. Biomed. Pharm. 2022, 145, 112352. [Google Scholar] [CrossRef]
- Yao, Y.; Cai, X.; Fei, W.; Ye, Y.; Zhao, M.; Zheng, C. The role of short-chain fatty acids in immunity, inflammation and metabolism. Crit. Rev. Food Sci. Nutr. 2022, 62, 1–12. [Google Scholar] [CrossRef]
- Kim, M.H.; Kang, S.G.; Park, J.H.; Yanagisawa, M.; Kim, C.H. Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 2013, 145, 396–406.e391-310. [Google Scholar] [CrossRef]
- Matsushita, M.; Fujita, K.; Hayashi, T.; Kayama, H.; Motooka, D.; Hase, H.; Jingushi, K.; Yamamichi, G.; Yumiba, S.; Tomiyama, E.; et al. Gut Microbiota-Derived Short-Chain Fatty Acids Promote Prostate Cancer Growth via IGF1 Signaling. Cancer Res. 2021, 81, 4014–4026. [Google Scholar] [CrossRef]
- Moritani, K.; Takeshita, T.; Shibata, Y.; Ninomiya, T.; Kiyohara, Y.; Yamashita, Y. Acetaldehyde production by major oral microbes. Oral Dis. 2015, 21, 748–754. [Google Scholar] [CrossRef]
- Engstrand, L.; Graham, D.Y. Microbiome and Gastric Cancer. Dig. Dis. Sci. 2020, 65, 865–873. [Google Scholar] [CrossRef]
- Wang, Z.; Gao, X.; Zeng, R.; Wu, Q.; Sun, H.; Wu, W.; Zhang, X.; Sun, G.; Yan, B.; Wu, L.; et al. Changes of the Gastric Mucosal Microbiome Associated With Histological Stages of Gastric Carcinogenesis. Front. Microbiol. 2020, 11, 997. [Google Scholar] [CrossRef]
- Han, Y.W.; Wang, X. Mobile microbiome: Oral bacteria in extra-oral infections and inflammation. J. Dent. Res. 2013, 92, 485–491. [Google Scholar] [CrossRef]
- The global, regional, and national burden of colorectal cancer and its attributable risk factors in 195 countries and territories, 1990-2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Gastroenterol. Hepatol. 2019, 4, 913–933. [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Zhang, X.; Liu, T.; Huang, J.; He, J. PICALM exerts a role in promoting CRC progression through ERK/MAPK signaling pathway. Cancer Cell Int. 2022, 22, 178. [Google Scholar] [CrossRef]
- Kaźmierczak-Siedlecka, K.; Dvořák, A.; Folwarski, M.; Daca, A.; Przewłócka, K.; Makarewicz, W. Fungal Gut Microbiota Dysbiosis and Its Role in Colorectal, Oral, and Pancreatic Carcinogenesis. Cancers 2020, 12, 1326. [Google Scholar] [CrossRef]
- Di Spirito, F.; Toti, P.; Pilone, V.; Carinci, F.; Lauritano, D.; Sbordone, L. The Association between Periodontitis and Human Colorectal Cancer: Genetic and Pathogenic Linkage. Life 2020, 10, 211. [Google Scholar] [CrossRef] [PubMed]
- Momen-Heravi, F.; Babic, A.; Tworoger, S.S.; Zhang, L.; Wu, K.; Smith-Warner, S.A.; Ogino, S.; Chan, A.T.; Meyerhardt, J.; Giovannucci, E.; et al. Periodontal disease, tooth loss and colorectal cancer risk: Results from the Nurses’ Health Study. Int. J. Cancer 2017, 140, 646–652. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhang, Y.; Qian, Y.; Xie, Y.H.; Jiang, S.S.; Kang, Z.R.; Chen, Y.X.; Chen, Z.F.; Fang, J.Y. Alterations in the oral and gut microbiome of colorectal cancer patients and association with host clinical factors. Int. J. Cancer 2021, 149, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Ray, K. Gut microbiota: Oral microbiome could provide clues to CRC. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 690. [Google Scholar] [CrossRef] [PubMed]
- Loftus, M.; Hassouneh, S.A.; Yooseph, S. Bacterial community structure alterations within the colorectal cancer gut microbiome. BMC Microbiol. 2021, 21, 98. [Google Scholar] [CrossRef] [PubMed]
- Komiya, Y.; Shimomura, Y.; Higurashi, T.; Sugi, Y.; Arimoto, J.; Umezawa, S.; Uchiyama, S.; Matsumoto, M.; Nakajima, A. Patients with colorectal cancer have identical strains of Fusobacterium nucleatum in their colorectal cancer and oral cavity. Gut 2019, 68, 1335–1337. [Google Scholar] [CrossRef]
- Yang, Y.; Cai, Q.; Shu, X.O.; Steinwandel, M.D.; Blot, W.J.; Zheng, W.; Long, J. Prospective study of oral microbiome and colorectal cancer risk in low-income and African American populations. Int. J. Cancer 2019, 144, 2381–2389. [Google Scholar] [CrossRef]
- Flemer, B.; Warren, R.D.; Barrett, M.P.; Cisek, K.; Das, A.; Jeffery, I.B.; Hurley, E.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. The oral microbiota in colorectal cancer is distinctive and predictive. Gut 2018, 67, 1454–1463. [Google Scholar] [CrossRef]
- Flemer, B.; Lynch, D.B.; Brown, J.M.; Jeffery, I.B.; Ryan, F.J.; Claesson, M.J.; O’Riordain, M.; Shanahan, F.; O’Toole, P.W. Tumour-associated and non-tumour-associated microbiota in colorectal cancer. Gut 2017, 66, 633–643. [Google Scholar] [CrossRef]
- Hale, V.L.; Chen, J.; Johnson, S.; Harrington, S.C.; Yab, T.C.; Smyrk, T.C.; Nelson, H.; Boardman, L.A.; Druliner, B.R.; Levin, T.R.; et al. Shifts in the Fecal Microbiota Associated with Adenomatous Polyps. Cancer Epidemiol. Biomark. Prev. 2017, 26, 85–94. [Google Scholar] [CrossRef]
- Yu, M.R.; Kim, H.J.; Park, H.R. Fusobacterium nucleatum Accelerates the Progression of Colitis-Associated Colorectal Cancer by Promoting EMT. Cancers 2020, 12, 2728. [Google Scholar] [CrossRef]
- Gur, C.; Ibrahim, Y.; Isaacson, B.; Yamin, R.; Abed, J.; Gamliel, M.; Enk, J.; Bar-On, Y.; Stanietsky-Kaynan, N.; Coppenhagen-Glazer, S.; et al. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity 2015, 42, 344–355. [Google Scholar] [CrossRef]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef]
- Gur, C.; Maalouf, N.; Shhadeh, A.; Berhani, O.; Singer, B.B.; Bachrach, G.; Mandelboim, O. Fusobacterium nucleatum supresses anti-tumor immunity by activating CEACAM1. Oncoimmunology 2019, 8, e1581531. [Google Scholar] [CrossRef]
- Kudo, Y.; Tada, H.; Fujiwara, N.; Tada, Y.; Tsunematsu, T.; Miyake, Y.; Ishimaru, N. Oral environment and cancer. Genes Environ. 2016, 38, 13. [Google Scholar] [CrossRef]
- Hong, J.; Guo, F.; Lu, S.Y.; Shen, C.; Ma, D.; Zhang, X.; Xie, Y.; Yan, T.; Yu, T.; Sun, T.; et al. F. nucleatum targets lncRNA ENO1-IT1 to promote glycolysis and oncogenesis in colorectal cancer. Gut 2021, 70, 2123–2137. [Google Scholar] [CrossRef]
- Cueva, C.; Silva, M.; Pinillos, I.; Bartolomé, B.; Moreno-Arribas, M.V. Interplay between Dietary Polyphenols and Oral and Gut Microbiota in the Development of Colorectal Cancer. Nutrients 2020, 12, 625. [Google Scholar] [CrossRef]
- Li, B.; Ge, Y.; Cheng, L.; Zeng, B.; Yu, J.; Peng, X.; Zhao, J.; Li, W.; Ren, B.; Li, M.; et al. Oral bacteria colonize and compete with gut microbiota in gnotobiotic mice. Int. J. Oral Sci. 2019, 11, 10. [Google Scholar] [CrossRef]
- Narengaowa; Kong, W.; Lan, F.; Awan, U.F.; Qing, H.; Ni, J. The Oral-Gut-Brain AXIS: The Influence of Microbes in Alzheimer’s Disease. Front. Cell. Neurosci. 2021, 15, 633735. [Google Scholar] [CrossRef]
- Bullman, S.; Pedamallu, C.S.; Sicinska, E.; Clancy, T.E.; Zhang, X.; Cai, D.; Neuberg, D.; Huang, K.; Guevara, F.; Nelson, T.; et al. Analysis of Fusobacterium persistence and antibiotic response in colorectal cancer. Science 2017, 358, 1443–1448. [Google Scholar] [CrossRef]
- Mo, S.; Ru, H.; Huang, M.; Cheng, L.; Mo, X.; Yan, L. Oral-Intestinal Microbiota in Colorectal Cancer: Inflammation and Immunosuppression. J. Inflamm. Res. 2022, 15, 747–759. [Google Scholar] [CrossRef] [PubMed]
- Kuraji, R.; Sekino, S.; Kapila, Y.; Numabe, Y. Periodontal disease-related nonalcoholic fatty liver disease and nonalcoholic steatohepatitis: An emerging concept of oral-liver axis. Periodontol 2000 2021, 87, 204–240. [Google Scholar] [CrossRef] [PubMed]
- Milella, L. The Negative Effects of Volatile Sulphur Compounds. J. Vet. Dent. 2015, 32, 99–102. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhang, A.H.; Wu, F.F.; Wang, X.J. Alterations in the Gut Microbiota and Their Metabolites in Colorectal Cancer: Recent Progress and Future Prospects. Front. Oncol. 2022, 12, 841552. [Google Scholar] [CrossRef] [PubMed]
- Villéger, R.; Lopès, A.; Veziant, J.; Gagnière, J.; Barnich, N.; Billard, E.; Boucher, D.; Bonnet, M. Microbial markers in colorectal cancer detection and/or prognosis. World J. Gastroenterol. 2018, 24, 2327–2347. [Google Scholar] [CrossRef] [PubMed]
- Wong, M.C.S.; Jiang, J.Y.; Liang, M.; Fang, Y.; Yeung, M.S.; Sung, J.J.Y. Global temporal patterns of pancreatic cancer and association with socioeconomic development. Sci. Rep. 2017, 7, 3165. [Google Scholar] [CrossRef]
- Zhou, Q.; Tao, X.; Xia, S.; Guo, F.; Pan, C.; Xiang, H.; Shang, D. T Lymphocytes: A Promising Immunotherapeutic Target for Pancreatitis and Pancreatic Cancer? Front. Oncol. 2020, 10, 382. [Google Scholar] [CrossRef]
- Gaiser, R.A.; Halimi, A.; Alkharaan, H.; Lu, L.; Davanian, H.; Healy, K.; Hugerth, L.W.; Ateeb, Z.; Valente, R.; Fernández Moro, C.; et al. Enrichment of oral microbiota in early cystic precursors to invasive pancreatic cancer. Gut 2019, 68, 2186–2194. [Google Scholar] [CrossRef]
- Tiffon, C. Defining Parallels between the Salivary Glands and Pancreas to Better Understand Pancreatic Carcinogenesis. Biomedicines 2020, 8, 178. [Google Scholar] [CrossRef]
- Wei, A.L.; Li, M.; Li, G.Q.; Wang, X.; Hu, W.M.; Li, Z.L.; Yuan, J.; Liu, H.Y.; Zhou, L.L.; Li, K.; et al. Oral microbiome and pancreatic cancer. World J. Gastroenterol. 2020, 26, 7679–7692. [Google Scholar] [CrossRef]
- Lu, H.; Ren, Z.; Li, A.; Li, J.; Xu, S.; Zhang, H.; Jiang, J.; Yang, J.; Luo, Q.; Zhou, K.; et al. Tongue coating microbiome data distinguish patients with pancreatic head cancer from healthy controls. J. Oral Microbiol. 2019, 11, 1563409. [Google Scholar] [CrossRef]
- Olson, S.H.; Satagopan, J.; Xu, Y.; Ling, L.; Leong, S.; Orlow, I.; Saldia, A.; Li, P.; Nunes, P.; Madonia, V.; et al. The oral microbiota in patients with pancreatic cancer, patients with IPMNs, and controls: A pilot study. Cancer Causes Control 2017, 28, 959–969. [Google Scholar] [CrossRef]
- Michaud, D.S.; Izard, J.; Wilhelm-Benartzi, C.S.; You, D.H.; Grote, V.A.; Tjønneland, A.; Dahm, C.C.; Overvad, K.; Jenab, M.; Fedirko, V.; et al. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut 2013, 62, 1764–1770. [Google Scholar] [CrossRef]
- Farrell, J.J.; Zhang, L.; Zhou, H.; Chia, D.; Elashoff, D.; Akin, D.; Paster, B.J.; Joshipura, K.; Wong, D.T. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 2012, 61, 582–588. [Google Scholar] [CrossRef]
- Alkharaan, H.; Lu, L.; Gabarrini, G.; Halimi, A.; Ateeb, Z.; Sobkowiak, M.J.; Davanian, H.; Fernández Moro, C.; Jansson, L.; Del Chiaro, M.; et al. Circulating and Salivary Antibodies to Fusobacterium nucleatum Are Associated with Cystic Pancreatic Neoplasm Malignancy. Front. Immunol. 2020, 11, 2003. [Google Scholar] [CrossRef]
- Dickson, I. Microbiome promotes pancreatic cancer. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 328. [Google Scholar] [CrossRef]
- Li, S.; Fuhler, G.M.; Bn, N.; Jose, T.; Bruno, M.J.; Peppelenbosch, M.P.; Konstantinov, S.R. Pancreatic cyst fluid harbors a unique microbiome. Microbiome 2017, 5, 147. [Google Scholar] [CrossRef]
- Gnanasekaran, J.; Binder Gallimidi, A.; Saba, E.; Pandi, K.; Eli Berchoer, L.; Hermano, E.; Angabo, S.; Makkawi, H.A.; Khashan, A.; Daoud, A.; et al. Intracellular Porphyromonas gingivalis Promotes the Tumorigenic Behavior of Pancreatic Carcinoma Cells. Cancers 2020, 12, 2331. [Google Scholar] [CrossRef]
- Kiss, B.; Mikó, E.; Sebő, É.; Toth, J.; Ujlaki, G.; Szabó, J.; Uray, K.; Bai, P.; Árkosy, P. Oncobiosis and Microbial Metabolite Signaling in Pancreatic Adenocarcinoma. Cancers 2020, 12, 1068. [Google Scholar] [CrossRef]
- Chen, J.; Domingue, J.C.; Sears, C.L. Microbiota dysbiosis in select human cancers: Evidence of association and causality. Semin. Immunol. 2017, 32, 25–34. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, T.; Liu, Y.; Chen, S.; Yu, Z. Application of Nanoparticles in the Treatment of Lung Cancer with Emphasis on Receptors. Front. Pharmacol. 2021, 12, 781425. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Phillips, S.; Gail, M.H.; Goedert, J.J.; Humphrys, M.S.; Ravel, J.; Ren, Y.; Caporaso, N.E. The effect of cigarette smoking on the oral and nasal microbiota. Microbiome 2017, 5, 3. [Google Scholar] [CrossRef]
- Lim, M.Y.; Hong, S.; Hwang, K.H.; Lim, E.J.; Han, J.Y.; Nam, Y.D. Diagnostic and prognostic potential of the oral and gut microbiome for lung adenocarcinoma. Clin. Transl. Med. 2021, 11, e508. [Google Scholar] [CrossRef] [PubMed]
- Hosgood, H.D.; Cai, Q.; Hua, X.; Long, J.; Shi, J.; Wan, Y.; Yang, Y.; Abnet, C.; Bassig, B.A.; Hu, W.; et al. Variation in oral microbiome is associated with future risk of lung cancer among never-smokers. Thorax 2021, 76, 256–263. [Google Scholar] [CrossRef] [PubMed]
- Vogtmann, E.; Hua, X.; Yu, G.; Purandare, V.; Hullings, A.G.; Shao, D.; Wan, Y.; Li, S.; Dagnall, C.L.; Jones, K.; et al. The oral microbiome and lung cancer risk: An analysis of 3 prospective cohort studies. J. Natl. Cancer Inst. 2022, 104, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Tsay, J.J.; Wu, B.G.; Sulaiman, I.; Gershner, K.; Schluger, R.; Li, Y.; Yie, T.A.; Meyn, P.; Olsen, E.; Perez, L.; et al. Lower Airway Dysbiosis Affects Lung Cancer Progression. Cancer Discov. 2021, 11, 293–307. [Google Scholar] [CrossRef]
- Liu, Y.; Yuan, X.; Chen, K.; Zhou, F.; Yang, H.; Yang, H.; Qi, Y.; Kong, J.; Sun, W.; Gao, S. Clinical significance and prognostic value of Porphyromonas gingivalis infection in lung cancer. Transl. Oncol. 2021, 14, 100972. [Google Scholar] [CrossRef]
- Yan, C.; Luo, Z.; Lin, Z.; He, H.; Luo, Y.; Chen, J. Shear Wave Elastography-Assisted Ultrasound Breast Image Analysis and Identification of Abnormal Data. J. Healthc. Eng. 2022, 2022, 5499354. [Google Scholar] [CrossRef]
- Shao, J.; Wu, L.; Leng, W.D.; Fang, C.; Zhu, Y.J.; Jin, Y.H.; Zeng, X.T. Periodontal Disease and Breast Cancer: A Meta-Analysis of 1,73,162 Participants. Front. Oncol. 2018, 8, 601. [Google Scholar] [CrossRef]
- Wu, Z.; Byrd, D.A.; Wan, Y.; Ansong, D.; Clegg-Lamptey, J.N.; Wiafe-Addai, B.; Edusei, L.; Adjei, E.; Titiloye, N.; Dedey, F.; et al. The oral microbiome and breast cancer and nonmalignant breast disease, and its relationship with the fecal microbiome in the Ghana Breast Health Study. Int. J. Cancer 2022, 151, 1248–1260. [Google Scholar] [CrossRef]
- Parhi, L.; Alon-Maimon, T.; Sol, A.; Nejman, D.; Shhadeh, A.; Fainsod-Levi, T.; Yajuk, O.; Isaacson, B.; Abed, J.; Maalouf, N.; et al. Breast cancer colonization by Fusobacterium nucleatum accelerates tumor growth and metastatic progression. Nat. Commun. 2020, 11, 3259. [Google Scholar] [CrossRef]
- Snider, E.J.; Freedberg, D.E.; Abrams, J.A. Potential Role of the Microbiome in Barrett’s Esophagus and Esophageal Adenocarcinoma. Dig. Dis. Sci. 2016, 61, 2217–2225. [Google Scholar] [CrossRef]
- Park, O.J.; Kim, A.R.; So, Y.J.; Im, J.; Ji, H.J.; Ahn, K.B.; Seo, H.S.; Yun, C.H.; Han, S.H. Induction of Apoptotic Cell Death by Oral Streptococci in Human Periodontal Ligament Cells. Front. Microbiol. 2021, 12, 738047. [Google Scholar] [CrossRef] [PubMed]
- Inaba, H.; Sugita, H.; Kuboniwa, M.; Iwai, S.; Hamada, M.; Noda, T.; Morisaki, I.; Lamont, R.J.; Amano, A. Porphyromonas gingivalis promotes invasion of oral squamous cell carcinoma through induction of proMMP9 and its activation. Cell. Microbiol. 2014, 16, 131–145. [Google Scholar] [CrossRef] [PubMed]
- Muto, M.; Hitomi, Y.; Ohtsu, A.; Shimada, H.; Kashiwase, Y.; Sasaki, H.; Yoshida, S.; Esumi, H. Acetaldehyde production by non-pathogenic Neisseria in human oral microflora: Implications for carcinogenesis in upper aerodigestive tract. Int. J. Cancer 2000, 88, 342–350. [Google Scholar] [CrossRef]
- Park, D.Y.; Park, J.Y.; Lee, D.; Hwang, I.; Kim, H.S. Leaky Gum: The Revisited Origin of Systemic Diseases. Cells 2022, 11, 1079. [Google Scholar] [CrossRef]
- Schiffman, J.D.; Fisher, P.G.; Gibbs, P. Early detection of cancer: Past, present, and future. Am. Soc. Clin. Oncol. Educ. Book 2015, 35, 57–65. [Google Scholar] [CrossRef]
- Nonaka, T.; Wong, D.T.W. Saliva-Exosomics in Cancer: Molecular Characterization of Cancer-Derived Exosomes in Saliva. Enzymes 2017, 42, 125–151. [Google Scholar] [CrossRef]
- Cheng, J.; Nonaka, T.; Wong, D.T.W. Salivary Exosomes as Nanocarriers for Cancer Biomarker Delivery. Materials 2019, 12, 654. [Google Scholar] [CrossRef]
- Bracci, P.M. Oral Health and the Oral Microbiome in Pancreatic Cancer: An Overview of Epidemiological Studies. Cancer J. 2017, 23, 310–314. [Google Scholar] [CrossRef]
- Pandya, G.; Kirtonia, A.; Singh, A.; Goel, A.; Mohan, C.D.; Rangappa, K.S.; Pandey, A.K.; Kapoor, S.; Tandon, S.; Sethi, G.; et al. A comprehensive review of the multifaceted role of the microbiota in human pancreatic carcinoma. Semin. Cancer Biol. 2021, 86, 682–692. [Google Scholar] [CrossRef] [PubMed]
| Subject | Organisms (Oral Bacteria) | Sample Type | Reference |
|---|---|---|---|
| 47 OSCC patients and 48 healthy individuals as controls. | The proportions of Actinobacteria, Fusobacterium, Moraxella, Bacillus, and Veillonella species were higher in the disease group than they were in the control group. | saliva, subgingival plaque, the tumor surface, tumor tissue samples | [55] |
| 60 OSCC patients and 120 gender and age-matched controls. | The proportions of Prevotella oris, Neisseria flava, Neisseria flavescens/subflava, F. nucleatum, Aggregatibacter segnis, Streptococcus mitis, and Fusobacterium periodontium were higher in the disease group than they were in the control group. | fresh OSCC biopsies samples | [56] |
| 48 OSCC patients and 46 controls. | The proportions of Prevotella, Campylobacter, Capnocytophaga, Solobacteria, Peptostreptococcus, and Catonella were higher in the disease group than they were in the control group. | whole mouth fluid (WMF) and swab samples | [53] |
| Oral cancer patients (n = 50) and healthy subjects (n = 50). | The proportions of Staphylococcus and Rothia were higher in the disease group than they were in the control group. | swab samples | [54] |
| 25 patients with OSCC and 24 healthy controls were recruited from Dr. B. Borooah Cancer Institute (BBCI), Guwahati, Assam, India. | The proportions of P. melaninogenica, Streptococcus anginosus, Veillonella parvula, Prevotella pallens, Porphyromonas endodontalis, Prevotella nanceiensis, Dialister sp., Campylobacter ureolyticus, Fusobacterium sp., P. nigrescens, Neisseria bacilliformis, and Peptostreptococcus anaerobius were higher in the disease group than they were in the control group. | samples of the whole saliva | [57] |
| Patients presenting with OLK (n = 36, average age: 60.6). | The proportions of Fusobacterium, Leptotrichia, Campylobacter, and Rothia were higher in the disease group than they were in the control group. | swabs | [51] |
| 43 oral lichen planus patients and 21 mucosal healthy volunteers. | The proportions of Fusobacterium, Leptotrichia, and Lautropia were higher in the disease group than they were in the control group. | buccal scraping samples | [52] |
| Subject | Organisms (Oral Bacteria) | Sample Type | Reference |
|---|---|---|---|
| 293 patients included superficial gastritis (SG; n = 101), atrophic gastritis (AG; n = 93), and gastric cancer (GC; n = 99). | The proportions of presumed proinflammatory taxa, including Corynebacterium and Streptococcus were higher in the disease group than they were in the control group. | saliva sample | [66] |
| 81 cases including SG, AG, intestinal metaplasia (IM) and GC from Xi’an, China. | Oral bacteria such as Peptostreptococcus stomatis, Streptococcus anginosus, Parvimonas micra, Slackia exigua and Dialister pneumosintes were enriched in cancerous tissues. | gastric mucosal samples | [67] |
| 62 GC patients who underwent subtotal gastrectomy at The First Hospital of China Medical University. | Oral bacteria such as Fusobacterium, Streptococcus, Peptostreptococcus, and Prevotella were enriched in cancerous tissues. | gastric tissue samples | [68] |
| 37 individuals with GC and 13 controls. | The proportions of Veillonella, Prevotella, Aggregatibacter, and Megasphaera increased were higher in the disease group than they were in the control group, while the proportions of Leptotrichia, Rothia, Capnocytophaga, Campylobacter, Tannerella and Granulicatella were lower. | saliva and plaque samples | [69] |
| 57 newly diagnosed gastric adenocarcinomas and 80 healthy controls. | The proportion of Firmicutes was higher in the disease group than it was in the control group, while the proportion of Bacteroidetes was lower. | tongue coating sample | [70] |
| 78 gastritis patients and 50 healthy individuals. | The proportion of Campylobacter concisus was higher in the disease group than it was in the control group. | tongue-coating samples | [71] |
| 165 GC cases and 323 matched controls from Asian, African American, and European American populations. | The proportions of Neisseria mucosa and Prevotella pleuritidis were higher in the disease group than they were in the control group, while the proportions of Mycoplasma orale and Eubacterium yurii were lower. | pre-diagnostic buccal samples | [72] |
| 12 GC cases and 20 matched controls (functional dyspepsia) in Singapore and Malaysia. | Oral bacteria such as Lactococcus, Veilonella, and Fusobacteriaceae (Fusobacterium and Leptotrichia) were enriched in cancerous tissues. | antral gastric biopsies | [73] |
| 47 patients including SG, AG, gastric intraepithelial neoplasia (GIN), and GC. | Oral bacteria such as Slackia, Selenomonas, Bergeyella and Capnocytophaga were enriched in cancerous tissues. | gastric mucosal specimens | [65] |
| Subject | Organisms (Oral Bacteria) | Sample Type | Reference |
|---|---|---|---|
| 1165 cases with CRC and 739 cases for the periodontal bone loss. | Oral bacteria such as Fusobacteria were enriched in stool samples of CRC patients. | tissue sample | [92] |
| Matching samples of unstimulated saliva, cancer tissues or biopsies and stools were collected from 30 CRC and 30 HC patients. | The proportion of Salivary Firmicutes-to-Bacteroides ratio was higher in the disease group than it was in the control group. | unstimulated saliva, cancer tissues, or biopsies and stool samples | [93] |
| Individuals with either CRC (n = 99), colorectal polyps (n = 32) or healthy individuals as controls (n = 103). | Oral bacteria such as Fusobacterium, Peptostreptococcus, Porphyromonas, and Micromonas were enriched in the stool of patients with CRC or adenomas. | oral swabs, colonic mucosa, and stool samples | [94] |
| 252 healthy and advanced CRC subjects. | Oral bacteria such as F. nucleatum, Peptostreptococcus stomatis, Gemella morbillorum, and Parvimonas micra were enriched in fecal samples of CRC patients. | fecal sample | [95] |
| Saliva samples from 14 CRC patients were collected. | The proportion of F. nucleatum was higher in the disease group than it was in the control group. | saliva samples | [96] |
| Individuals including 231 incident CRC cases and 462 controls. | The proportion of Bifidobacteriaceae was higher in the disease group than it was in the control group. | mouth rinse samples | [97] |
| Populations including CRC (99 subjects), colorectal polyps (32), or controls (103). | The proportions of Haemophilus, Micromonas, Prevotella, Heterobacterium, Anaerobic, Neisseria, and Streptococcus were lower in the disease group than they were in the control group. | oral swabs, colonic mucosal and stool samples | [98] |
| Mucosal samples from 59 patients undergoing surgery for CRC, 21 individuals with polyps and 56 healthy controls. | Bacteroidetes Cluster 1 and Firmicutes Cluster 1 were reduced in fecal samples of CRC patients, whereas Bacteroidetes Cluster 2, Firmicutes Cluster 2, Pathogen Cluster and Prevotella Cluster were enriched in them. | fecal and mucosal samples | [99] |
| Fecal microbiota in patients with adenomas (n = 233) and those without adenomas (n = 547) were analyzed. | Pro-inflammatory bacteria of the genera Biliophilus, Desulfovibrio, Mogibacterium and various Bacteroidetes were enriched in fecal samples of CRC patients. | fecal sample | [100] |
| Scheme | Organisms (Oral Bacteria) | Sample Type | Reference |
|---|---|---|---|
| 30 PHC patients and 25 healthy controls. | The proportions of Firmicutes, Fusobacteria, and Actinobacteria were higher in the disease group than they were in the control group. | tongue coat samples | [121] |
| 361 incident adenocarcinoma of pancreas and 371 matched controls. | The proportions of P. gingivalis and A. actinomycetemcomitans were higher in the disease group than they were in the control group, while the proportion of Leptotrichia was lower. | oral wash samples | [39] |
| Forty newly diagnosed PDAC patients, 39 IPMN patients, and 58 controls. | The proportions of Firmicutes and related taxa (Bacilli, Lactobacillales, Streptococcaceae, Streptococcus, and Streptococcus thermophilus) were higher in the disease group than they were in the control group. | saliva samples | [122] |
| 405 pancreatic cancer cases and 416 matched controls. | The proportion of antibodies against P. gingivalis ATTC 53978 was higher in the disease group than it was in the control group. | blood samples | [123] |
| 10 resectable patients with pancreatic cancer and 10 matched healthy controls. | The proportions of N. elongata and S. mitis were lower in the disease group than they were in the control group, while the proportion of G. adiacens was higher. | saliva samples | [124] |
| patients with pancreatic cancer (n = 41) and healthy individuals (n = 69). | The proportions of Lactobacillales, Bacilli, Streptococcus, Firmicutes, Actinomyces, Rothia, Leptotrichia, Lactobacillus, E. coli, and Enterobacteriales were higher in the disease group than they were in the control group. | saliva samples | [120] |
| patients with suspected PCN (n = 105). | The proportion of F. nucleatum genome was higher in the disease group than it was in the control group. | cyst fluid and peripheral blood liquid biopsies | [118] |
| IPMN pancreatic cystic tumor cases and controls. | The reactivity of salivary IgA to F. nucleatum and the Fap2 mimotope increased. | paired plasma and saliva samples | [125] |
| Subject | Organisms (Oral Bacteria) | Sample Type | Reference |
|---|---|---|---|
| Lung adenocarcinoma patients who did not smoke (cancer, n = 91) and healthy controls (control, n = 91). | The proportion of Eillonella was higher in the disease group than it was in the control group, while the proportions of Mogibacterium, Butyrivibrio, Variovorax, Ralstonia, Catonella, Bulleidia, and Oribacterium were lower. | saliva | [133] |
| Cases were subjects who were diagnosed with incident lung cancer (n = 114) and with controls (n = 114). | The proportions of Bacilli class and Lactobacillales order were higher in the disease group than they were in the control group, while the proportions of Spirochaetia and Bacteroidetes were lower. | mouth rinse samples | [134] |
| 148 subjects with lung nodules from the NYU Lung Cancer Biomarker Center. | Oral bacteria such as Veillonella, Streptococcus, Prevotella, and Haemophilus were enriched in LC. | lower airway brushes | [136] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; He, M.; Lei, Y.; Liu, Y.; Li, X.; Xiang, X.; Wu, Q.; Wang, Q. Oral Microbiota and Tumor—A New Perspective of Tumor Pathogenesis. Microorganisms 2022, 10, 2206. https://doi.org/10.3390/microorganisms10112206
Li S, He M, Lei Y, Liu Y, Li X, Xiang X, Wu Q, Wang Q. Oral Microbiota and Tumor—A New Perspective of Tumor Pathogenesis. Microorganisms. 2022; 10(11):2206. https://doi.org/10.3390/microorganisms10112206
Chicago/Turabian StyleLi, Simin, Mingxin He, Yumeng Lei, Yang Liu, Xinquan Li, Xiaochen Xiang, Qingming Wu, and Qiang Wang. 2022. "Oral Microbiota and Tumor—A New Perspective of Tumor Pathogenesis" Microorganisms 10, no. 11: 2206. https://doi.org/10.3390/microorganisms10112206
APA StyleLi, S., He, M., Lei, Y., Liu, Y., Li, X., Xiang, X., Wu, Q., & Wang, Q. (2022). Oral Microbiota and Tumor—A New Perspective of Tumor Pathogenesis. Microorganisms, 10(11), 2206. https://doi.org/10.3390/microorganisms10112206






